Abstract
1-Aminocyclopropane-1-carboxylate synthase (ACS) catalyzes the rate-limiting step in the ethylene biosynthetic pathway in plants. The Arabidopsis genome encodes nine ACS polypeptides that form eight functional (ACS2, ACS4-9, and ACS11) homodimers and one nonfunctional (ACS1) homodimer. Transgenic Arabidopsis lines were constructed expressing the β-glucuronidase (GUS) and green fluorescence protein (GFP) reporter genes from the promoter of each of the gene family members to determine their patterns of expression during plant development. All genes, except ACS9, are expressed in 5-d-old etiolated or light-grown seedlings yielding distinct patterns of GUS staining. ACS9 expression is detected later in development. Unique and overlapping expression patterns were detected for all the family members in various organs of adult plants. ACS11 is uniquely expressed in the trichomes of sepals and ACS1 in the replum. Overlapping expression was observed in hypocotyl, roots, various parts of the flower (sepals, pedicle, style, etc.) and in the stigmatic and abscission zones of the silique. Exogenous indole-3-acetic acid (IAA) enhances the constitutive expression of ACS2, 4, 5, 6, 7, 8, and 11 in the root. Wounding of hypocotyl tissue inhibits the constitutive expression of ACS1 and ACS5 and induces the expression of ACS2, 4, 6, 7, 8, and 11. Inducers of ethylene production such as cold, heat, anaerobiosis, and Li+ ions enhance or suppress the expression of various members of the gene family in the root of light-grown seedlings. Examination of GUS expression in transverse sections of cotyledons reveals that all ACS genes, except ACS9, are expressed in the epidermis cell layer, guard cells, and vascular tissue. Similar analysis with root tip tissue treated with IAA reveals unique and overlapping expression patterns in the various cell types of the lateral root cap, cell division, and cell expansion zones. IAA inducibility is gene-specific and cell type-dependent across the root tip zone. This limited comparative exploration of ACS gene family expression reveals constitutive spatial and temporal expression patterns of all gene family members throughout the growth period examined. The unique and overlapping gene activity pattern detected reveals a combinatorial code of spatio-temporal coexpression among the various gene family members during plant development. This raises the prospect that functional ACS heterodimers may be formed in planta.
The gas ethylene (C2H4) has been known since the beginning of the past century to be used by plants as a signaling molecule for regulating a variety of developmental processes and stress responses (Abeles et al., 1992). These include seed germination, leaf and flower senescence, fruit ripening, cell elongation, nodulation, and wound and pathogen responses. Ethylene production is induced by a variety of external factors, including wounding, viral infection, elicitors, auxin treatment, chilling injury, drought, Cd2+ and Li+ ions, O3, SO2, and other pollutants (Yang and Hoffman, 1984; Abeles et al., 1992; Liang et al., 1996; Bleecker and Kende, 2000; Thomma et al., 2001). Enhancement in ethylene production serves as a signaling mechanism with profound physiological consequences.
Ethylene is biosynthesized from Met, which is converted to S-adenosylmethionine (AdoMet) by the enzyme S-adenosylmethionine synthase. AdoMet is converted by the enzyme 1-aminocyclopropane-1-carboxylate synthase (ACS) to methylthioadenosine and 1-aminocyclopropane-1-carboxylic acid (ACC), the precursor of ethylene (Zarembinski and Theologis, 1994; Bleecker and Kende, 2000; Wang et al., 2002). ACC is oxidized to CO2, HCN, and ethylene by ACC oxidase. ACS is a cytosolic enzyme with a short half life that requires pyridoxal phosphate (PLP) as a cofactor (Yang and Hoffman, 1984; Yip et al., 1990; Bleecker and Kende, 2000). The activity of ACS is regulated at the transcriptional (Liang et al., 1992; Zarembinski and Theologis, 1994; Bleecker and Kende, 2000; Wang et al., 2002) and posttranscriptional levels (Woeste et al., 1999; Chae et al., 2003; Wang et al., 2004).
ACS is encoded by a multigene family in every plant species examined (Liang et al., 1992; Zarembinski and Theologis, 1994; Bleecker and Kende, 2000). In Arabidopsis, the ACS gene family encodes nine polypeptides, of which eight form functional homodimers and one, a nonfunctional homodimer (Arabidopsis Genome Initiative, 2000; Yamagami et al., 2003). The primary sequence encoded by these genes shows sequence conservation ranging from 50% to 96% amino acid sequence identity with the highest variability at the carboxylic end of the protein (Yamagami et al., 2003). The variable carboxyl terminus is unimportant for enzyme activity but serves as a regulatory domain responsible for posttranslational regulation of the enzyme (Tatsuki and Mori, 2001; Chae et al., 2003; Wang et al., 2004).
ACS shares sequence similarity with other PLP-dependent enzymes and is most closely related to the subgroup 1 aminotransferases, which includes Asp aminotransferase. ACS contains all 11 invariant residues in this subgroup, including four conserved residues (Gly-197, Asp-222, Lys-258, and Arg-386) present in all aminotransferases (Mehta et al., 1989; Rottmann et al., 1991). Complementation studies with ACS mutants have shown that the enzyme functions as a homodimer whose active site is formed from the interaction of residues from the monomeric subunits similar to Asp aminotransferase (Li et al., 1997; Tarun and Theologis, 1998). In particular, the Y92 residue that helps in anchoring the PLP cofactor to the ACS apoenzyme interacts with active-site residue K278, which forms a covalent Schiff base with the PLP cofactor from the adjacent subunit. The three-dimensional structure of ACS has confirmed this model (Capitani et al., 1999) and together with available biochemical data explain the catalytic roles of the conserved and nonconserved active site residues (White et al., 1994; Tarun and Theologis, 1998; Tarun et al., 1998).
The biological significance of multigene families in general and of the ACS gene family in particular is unknown. It has been postulated that the presence of ACS isozymes may reflect tissue specific expression that satisfies the biochemical environment of the cells or tissues in which each isozyme is expressed (Rottmann et al., 1991). For example, if a group of cells or tissues have low concentrations of the ACS substrate, AdoMet, then these cells express a high affinity (low Km) ACS isozyme. Accordingly, the distinct biological function of each isozyme is defined by its biochemical properties, which in turn define its tissue specific expression. Such a concept enhances the physiological fine-tuning of the cell and demands that the enzymatic properties of each isozyme be distinct. Biochemical characterization of the ACSs supports this proposition (Yamagami et al., 2003). Since the isozymes are homodimers, the question arises whether their subunits can form functional heterodimers and thus further enhance isozyme diversity. Recently, we have used a complementation assay in Escherichia coli JAde6 (Tarun et al., 1998) for testing the formation of functional heterodimers by intermolecular complementation of K278A and Y92A mutants of various isozymes (Tsuchisaka and Theologis, 2004). The analysis reveals that the subunits of all isozymes can form heterodimers; however, enzymatically active heterodimers are formed only among isozymes that belong to one or the other of the two phylogenetic branches. This finding suggests that the shared active sites formed between the heterodimeric subunits of the same branch are structurally similar to those of the corresponding homodimers and dissimilar to those from different branches. However, ACS7 is an exception to this rule, as it forms functional heterodimers with some members of both branches when it provides the unmutated “K278” residue. ACS1, the nonfunctional isozyme, can also form functional heterodimers with members of its phylogenetic branch when its partners provide the unmutated K278 residue. The ACSs can potentially form 45 homo- and heterodimers of which 25 are functional (8 homodimers and 17 heterodimers). And bimolecular fluorescence complementation (BiFC; Hu et al., 2002) and biochemical coaffinity purification assays show the inactivity of certain heterodimers is not due to the absence of heterodimerization but rather to structural restrain(s) that prevents the shared active sites to be functional. From these results, we proposed that functional heterodimerization enhances the isozyme diversity of the ACS gene family and provides physiological versatility by being able to operate in a broad gradient of AdoMet concentration in various cells/tissues during plant growth and development. It has not yet been determined whether ACS heterodimers are formed in planta.
Here, we present a limited comparative histochemical characterization of Arabidopsis transgenic lines expressing β-glucuronidase (GUS) from each promoter of the ACS gene family members to determine their patterns of expression during growth and development. Such an analysis will facilitate the search for potential formation of ACS heterodimers in planta using BiFC that is currently in progress in our laboratory. The results presented indicate that ACS heterodimers may be formed in planta because various ACS gene family members are expressed in the same cells/tissues during plant growth and development.
RESULTS
The Construct and Transgenic Lines
To monitor promoter activity of each ACS gene family member during plant growth and development, we introduced into the Arabidopsis genome by Agrobacterium mediated transformation (Clough and Bent, 1998) a double gene construct shown in Supplemental Figure 1, available at www.plantphysiol.org. The expression pattern of each gene can be monitored using either the E. coli gene uidA encoding GUS (Jefferson et al., 1987) or the green fluorescence protein (GFP; Siemering et al., 1996; Haseloff et al., 1997) as visual markers. Supplemental Figure 2 compares the promoter activity of seven of the nine gene family members in the root of 5-d-old light-grown seedlings using either GUS or GFP. Similar expression patterns are observed with both reporters. The expression of ACS1 and ACS9 is nil in that particular tissue, whereas ACS2, 6, 7, 8, and 11 are expressed in the vascular tissue with different intensities as detected by both reporters. Control plants transgenic for the promotorless GUS gene, do not show any detectable GUS expression (data not shown). Results for the ACS4 and ACS5 genes are not shown in Supplemental Figure 2 because transgenic lines have not been constructed with both reporters. We have used previously made single gene constructs with GUS only as a reporter (R. Dent and A. Theologis, unpublished data). The analysis presented in this study was carried out using GUS as the monitoring reporter because the lab had more experience with this visual marker. In addition the visual intensity of the GFP reporter in some tissues was lower compared with that of the GUS reporter. The background was higher due to tissue autofluorescence. Heterozygous transgenic lines selected among three to five independent transformants for each gene promoter were used during the course of this study. Similar patterns of expression were observed among the various independent transformants (see Supplemental Figs. 3–6). T2 lines with the strongest expression were chosen for this analysis. The locations of the transgenes in the Arabidopsis genome are shown in Supplemental Figure 7.
Expression in Etiolated and Light-Grown Seedlings
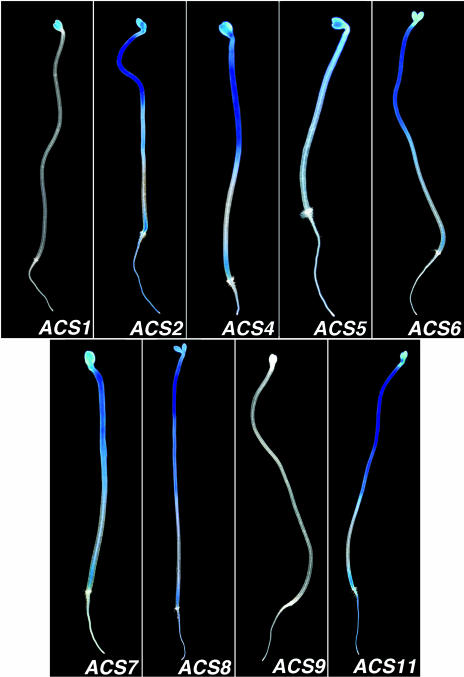
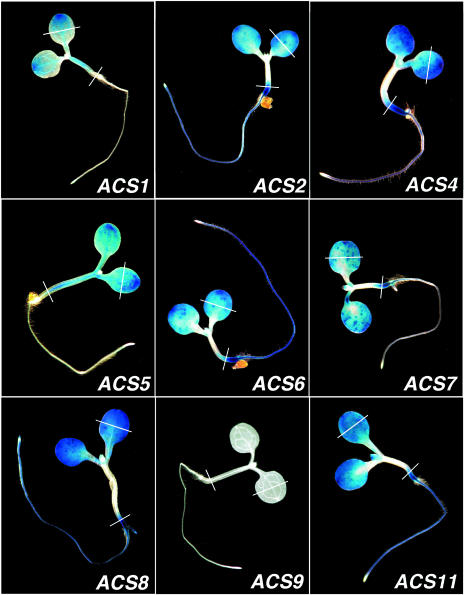
First, we studied the expression of ACS genes in 5-d-old etiolated and light-grown seedlings after 12 h of histochemical staining. The results are shown in Figures 1 and 2, respectively. In etiolated seedlings, the expression of ACS2, 4, 6, 7, 8, and 11 is confined to the elongation zone of the hypocotyl, at the embryonic root region, the cotyledons, and the root vascular tissue (Fig. 1). The expression pattern of ACS5 is similar but of lower intensity (Fig. 1; compare ACS5 with ACS2, 4, 6, 7, 8, and 11). The ACS1 gene is weakly expressed in the cotyledons and the expression of ACS9 is nil (Fig. 1). In light-grown seedlings, the expression patterns of ACS2, 4, 6, 7, 8, and 11 are similar but not identical (Fig. 2). These genes are expressed in the cotyledons, the embryonic root and in the root. ACS8 is the only gene expressed in the root tip. In addition, we see expression of ACS2, 4, 6, 7, 8, and 11 in the primary leaves, and of ACS2, 5, 8, and 11 in the shoot apex (Fig. 3). The nonfunctional ACS1 is not expressed in the roots, but together with ACS5, is expressed throughout the hypocotyl (Fig. 2). The expression of ACS9 is also nil in 5-d-old light-grown seedlings (Fig. 2). Subsequently, we investigated the possibility that the absence of ACS9 expression in 5-d-old dark- and light-grown seedlings was due to the developmental regulation of ACS9 gene expression. This idea was reinforced by our previous finding that the ACS9 transcript could be detected in root and silique tissue from 21-d-old light-grown seedlings (Yamagami et al., 2003). Supplemental Figure 8 compares the expression pattern of ACS9 in 5-, 10-, and 15-d-old light-grown seedlings. ACS9 expression is detected in the leaf blades of 10-d-old seedlings (Supplemental Fig. 8). After 15 d, ACS9 expression is detected in all the petioles and leaf blades of younger leaves. Expression of ACS9 is nil in the root (Supplemental Fig. 8). Similar expression patterns were observed in various independent ACS9 transgenic lines (Supplemental Fig. 6).
Figure 1.
Expression of the ACS gene family members in 5-d-old etiolated seedlings.
Figure 2.
Table 1.
| ACS1 | ACS2 | ACS4 | ACS5 | ACS6 | ACS7 | ACS8 | ACS9 | ACS11 | |
|---|---|---|---|---|---|---|---|---|---|
| Cotyledon | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| Primary leaves | ○ | ○ | ○ | ○ | ○ | ○ | |||
| Shoot apex | ○ | ○ | ○ | ○ | |||||
| Hypocotyl | ○ | ○ | |||||||
| Embryonic root | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||
| Root | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||
| Root tip | ○ |
Figure 3.
Expression of the ACS gene family members in the shoot apex regions of Figure 2.
ACS Expression in Mature Plants and Their Organs
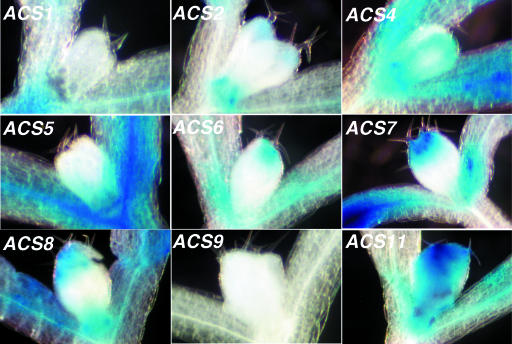
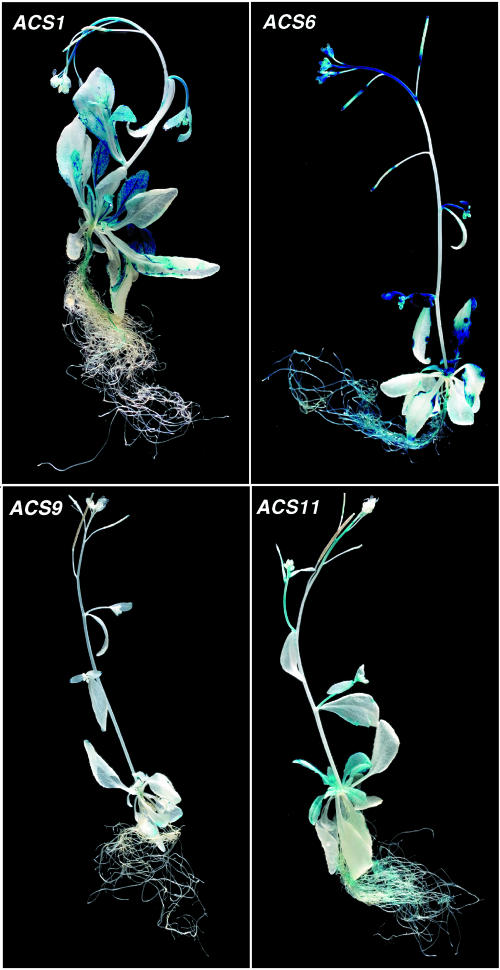
To investigate the expression of the ACS gene family members in mature plants, we grew them hydroponically for 1 month and subsequently stained them for 12 h. The results are shown in Figure 4. The expression patterns among the various family members are dissimilar and fall into four classes: ACS1-, ACS6-, ACS9-, and ACS11-like. The ACS6-like class contains all the rest of the gene family members whose expression patterns are shown in Supplemental Figure 9. ACS1 is expressed in the leaf vascular tissue, central leaf veins, and flower stem (Fig. 4). ACS9 is barely expressed at this stage of development. ACS11 is expressed in the inflorescence stem, younger leaves, cauline leaves, and in the roots (Fig. 4). All the members of the ACS6-like class are expressed in the roots, inflorescence stem, siliques, and younger leaves. Next, we examined the expression of the family members in various organs of the mature plants.
Figure 4.
Expression of ACS1, ACS6, ACS9, and ACS11 gene family members in 1-month-old plants.
Roots
Supplemental Figure 10 shows that all ACS genes except ACS1 and ACS9 are expressed in the maturation zone of the roots and primarily in the vascular tissue. Among the remaining group of genes (ACS2, 4, 5, 6, 7, 8, and 11), ACS5 and ACS11 are the weakest expressors. Interestingly, ACS8 is the only one that is expressed at the root cup of the root tip region.
Rosette Leaves
A close examination of the rosette leaves shows that ACS1 is expressed in the vascular tissue of younger leaves, whereas ACS2, 4, 5, 6, 7, and 8 are primarily expressed in the younger rosette leaves. ACS11 expression is restricted to the trichomes and ACS9 appears to be almost inactive (Supplemental Fig. 11).
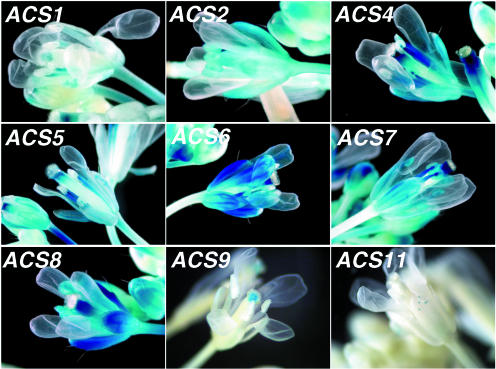
Flower
A quite diverse expression pattern is observed among the various gene family members. The simplest expression pattern is seen with ACS11, which is expressed in the trichomes of the sepals (Fig. 5). Quite a few members such as ACS1, 2, 4, 5, and 8 are expressed in the pedicel. The carpel tissue expresses ACS1 and ACS9 at the stigmatic region and ACS2, 4, 5, 6, 7, and 8 at the style. ACS1, 2, 4, 5, 6, 7, and 8 expression can be detected in the filament of the stamen, whereas ACS2, 7, and 8 are also expressed in the anthers. All members except ACS9 and 11 are expressed in the sepals of the flower. Expression of some ACS genes is detected in petals in more advanced stages of flower development (data not shown).
Figure 5.
Table 2.
| ACS1 | ACS2 | ACS4 | ACS5 | ACS6 | ACS7 | ACS8 | ACS9 | ACS11 | |
|---|---|---|---|---|---|---|---|---|---|
| Carpel | |||||||||
| Stigma | ○ | ○ | |||||||
| Style | ○ | ○ | ○ | ○ | ○ | ○ | |||
| Stamens | |||||||||
| Anther | ○ | ○ | ○ | ||||||
| Filament | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||
| Sepals | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||
| Petals | |||||||||
| Pedicel | ○ | ○ | ○ | ○ | ○ | ||||
| Trichome | ○ |
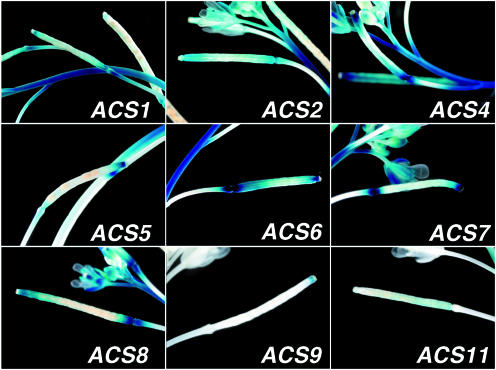
Silique
Figure 6 shows the highly localized expression of ACS9 at the style of the silique. A similar, but broader expression pattern is expressed with ACS5. Many ACS members, such as ACS1, 2, 4, 6, 7, and 8, are expressed in the abscission zone and in the valve of the silique. Unique patterns are observed with ACS1, which is expressed in the replum, and ACS7 expressed in the stigmatic tissue. ACS11 is not expressed in the silique (Fig. 6).
Figure 6.
Table 3.
| ACS1 | ACS2 | ACS4 | ACS5 | ACS6 | ACS7 | ACS8 | ACS9 | ACS11 | |
|---|---|---|---|---|---|---|---|---|---|
| Style | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |
| Stigmatic tissue | ○ | ||||||||
| Abscission zone | ○ | ○ | ○ | ○ | ○ | ○ | |||
| Replum | ○ |
Effect of Auxin, Wounding, Stress Conditions, and Li+ Ions
Ethylene production is known to be induced by auxin, a variety of stress conditions, such as heat and cold, wounding, and chemicals (Yang and Hoffman, 1984; Abeles et al., 1992; Liang et al., 1996; Bleecker and Kende, 2000). We examined the effect of some of these inducers on the expression of the ACS gene family members in the root tip zone of 5-d-old light-grown seedlings.
Auxin
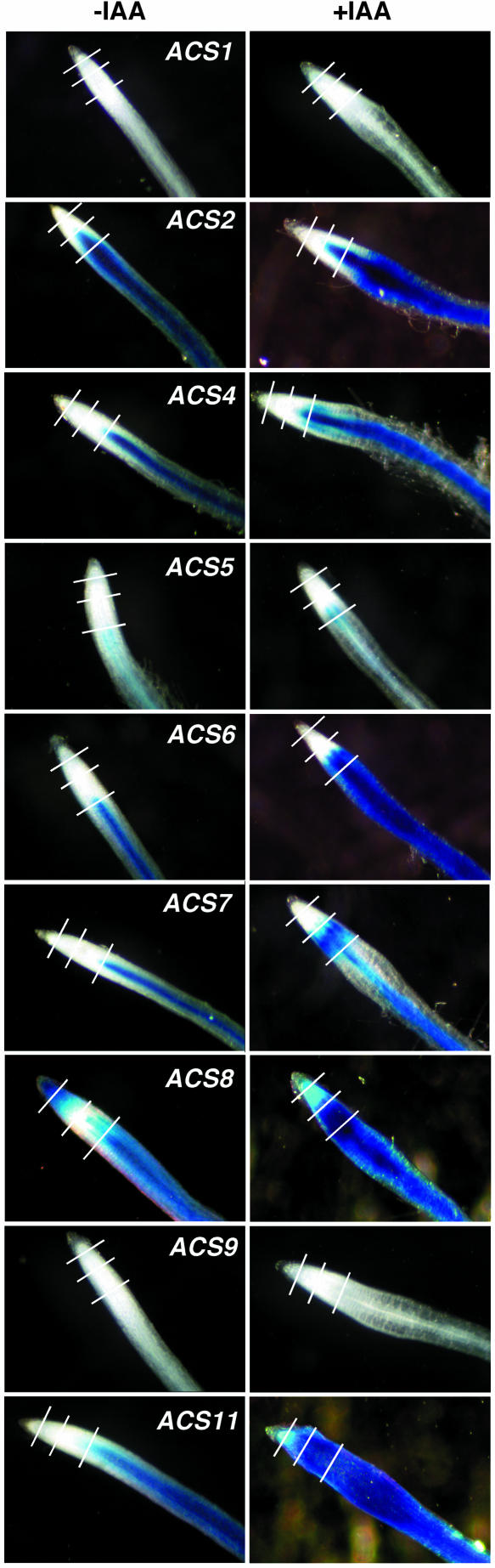
A well-known inducer of ethylene production is auxin (Yang and Hoffman, 1984; Abeles et al., 1992; Liang et al., 1996; Bleecker and Kende, 2000), which enhances the transcriptional activity of a set of ACS gene family members (ACS2, 4, 5, 6, 8, and 11; Yamagami et al., 2003). Figure 7 shows the effect of 20 μm IAA on the expression of the various ACS gene family members. All ACS genes, except ACS1 and ACS9, are expressed constitutively in the root tip of the 5-d-old seedlings, with ACS5 being the least expressed (Fig. 7). ACS8 is the only one among the various members that is expressed in the root tip (Fig. 7). IAA treatment not only enhances the expression of the genes that were constitutively expressed (ACS2, 4, 5, 6, 7, 8, and 11) but also alters their pattern of expression (Fig. 7; compare the expression pattern with and without IAA). The expression of ACS1 and ACS9 is not affected by IAA. IAA has a localized effect on the expression of ACS7 (Fig. 7). The hormone alters the pattern of its constitutive expression that is primarily in the vascular zone of the root tip. This gene has not been previously identified as auxin regulated using intact seedlings (Yamagami et al., 2003).
Figure 7.
Effect of IAA on the expression of the ACS gene family members in the root tip of 5-d-old light-grown seedlings. IAA concentration, 20 μm; duration of treatment, 24 h. The white lines indicate the location of the transverse sectioning for obtaining the results shown in Figure 11.
Wounding
The effect of cutting on the expression of the ACS gene family members was studied in the hypocotyl tissue of 5-d-old light-grown seedlings. The results are shown in Figure 8 and in Supplemental Figure 8. The expression patterns of the ACS gene family members fall into three groups regarding their expression in the hypocotyl tissue. ACS1/ACS5 are expressed throughout the hypocotyl tissue (Fig. 8), whereas ACS2, 4, 6, 7, 8, and 11 are expressed only at the basal part of the hypocotyl tissue (Fig. 8; Supplemental Fig. 12); ACS9 is not expressed in the hypocotyl (Fig. 8). Cutting the hypocotyl tissue where it is indicated with a white line in Figure 8 and Supplemental Figure 8 and subsequent staining the tissues after 4 h from cutting shows the following. Cutting inhibits the expression of the genes that are constitutively expressed in the intact tissue like ACS1 and ACS5 (Fig. 8) and greatly enhances the expression of genes whose expression is nil in the hypocotyl (Fig. 8; Supplemental Fig. 12), such as ACS2, 4, 6, 7, and 8. Cutting has no effect on ACS9 gene expression (Fig. 8).
Figure 8.
Effect of cutting on the expression of ACS1, ACS5, ACS6, and ACS9 in 5-d-old light-grown seedlings. Each seedling was cut with a blade in the middle of the hypocotyl (see white line in A) and incubated for 4 h on Murashige and Skoog media prior to staining. A, Intact seedlings. B, Upper part of seedlings after 4 h from cutting. C, Magnified view of B. D, Lower part of seedlings after 4 h from cutting. E, Magnified view of D.
Cold and Heat Treatment
Supplemental Figure 13 compares the expression pattern of the various ACS gene family members in the root tip region of cold and heat treated 5-d-old light grown seedlings. Cold treatment inhibits the expression of ACS5 and ACS11 and also alters the pattern of ACS8 expression (Supplemental Fig. 13; compare control with cold treatment). On the other hand, heat enhances the expression of ACS4 and alters the pattern of ACS8 and ACS11 gene expression. The expression of the remaining ACS genes is not altered.
Anaerobiosis
Treatment of 5-d-old light-grown seedlings with N2 gas for 24 h inhibits the expression of all constitutively expressed ACS gene family member with different degrees of intensity (Supplemental Fig. 13). For example, the expression of ACS5, 6, 7, and 11 is completely inhibited, whereas the expression of ACS2, 4, and 8 is reduced considerably (Supplemental Fig. 13). The expression of the nonconstitutively expressed genes, ACS1 and ACS9, is nil after N2 treatment.
Li+ Ions
Lithium treatment alters the expression pattern of ACS2 and ACS8, whereas it induces the expression of ACS5 and inhibits the expression of ACS11 (Supplemental Fig. 13). The expression of ACS4 and ACS7 is reduced considerably. The results in Supplemental Figure 13 indicate that the expression of ACS9 is not enhanced by any of the treatments mentioned above. Subsequently, we tested whether these various treatments had any effect on the expression of ACS9 in older seedlings. We found that IAA and lithium treatments induce ACS gene expression in the roots of 15-d-old seedlings (Supplemental Fig. 14).
Cell Specific Expression
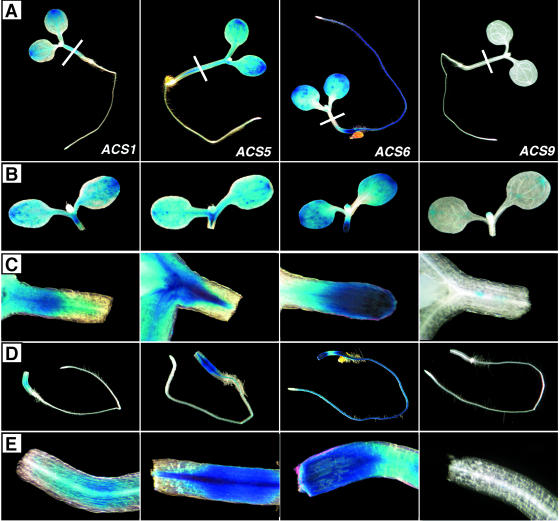
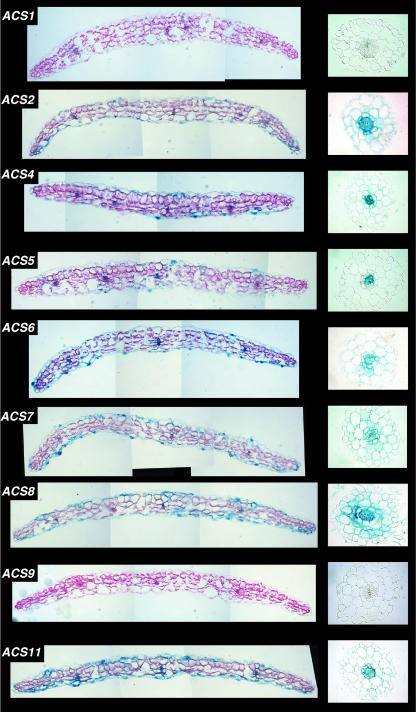
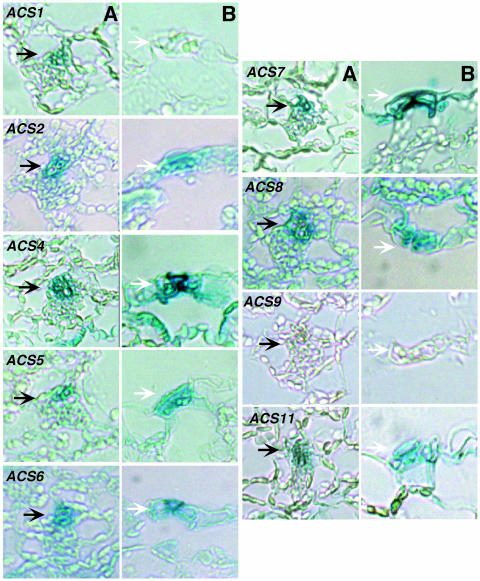
The analysis with intact seedlings revealed that many ACS gene family members have overlapping expression patterns. The question arises whether all the cells in the tissues/organs that show overlapping expression patterns also have overlapping expression. We addressed this question by examining transverse sections of cotyledons and hypocotyls of 5-d-old light-grown seedlings. We also examined transverse sections of the root tip zone treated with or without auxin. The tissue choice for this analysis was based on the suitability of the tissue for cross-sectioning. The results are presented in Figures 9, 10, and 11, respectively. Figure 2 shows that all the ACS-GUS transgenic lines except ACS9-GUS express GUS reporter in the cotyledons and in various regions of the hypocotyl in 5-d-old light-grown seedlings. To determine more precisely which cells expressed GUS, stained cotyledon and hypocotyl tissues were sectioned and analyzed (the position of sectioning is indicated by a white line on each seedling in Fig. 2). Figure 9 shows that GUS expression of all genes, except ACS1 (ACS1 is expressed in the vascular tissue) and ACS9, is restricted in the epidermal cell layer, guard cells, and the vascular bundles (Fig. 9, left column). The expression of the ACS genes is nil in the mesophyll cells. Figure 9 also shows that GUS activity is restricted primarily in the vascular tissue of the 5-d-old hypocotyl for all ACS genes except ACS1 and ACS9 for which expression is nil (Fig. 9, right column). The expression of ACS2, ACS7, and ACS8 extends beyond the vascular zone to some layer of the parenchymatic tissue (Fig. 9, right column). Closer investigation of the GUS expression in the guard cells at higher magnification shows that all ACS gene family members (except ACS1 and ACS9) are expressed in the guard cells (Fig. 10).
Figure 9.
Expression of ACS gene family members observed in transverse sections of the cotyledons (left column) and hypocotyls (right column) of 5-d-old light-grown seedlings. Tissue sectioning was carried out after 2 h of GUS staining. Section thickness, cotyledon, 20 μm; hypocotyl, 8 μm.
Figure 10.
Expression of the ACS gene family members in the vascular tissue (A) and guard cells (B) observed in transverse sections of cotyledons of 5-d-old light-grown transgenic seedlings. Tissue sectioning was performed after 2 h of GUS staining. Section thickness, 8 μm.
Figure 11.
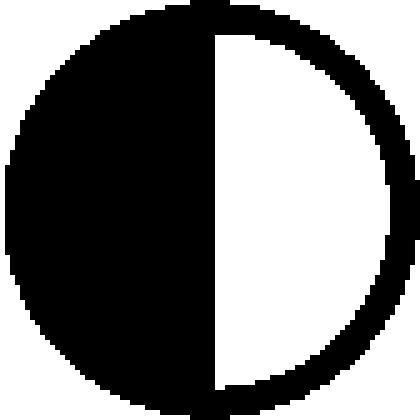 denotes −IAA and • denotes +IAA. Arrows (↑) denote enhanced expression in response to auxin.
denotes −IAA and • denotes +IAA. Arrows (↑) denote enhanced expression in response to auxin.Table 4.
| ACS1 | ACS2 | ACS4 | ACS5 | ACS6 | ACS7 | ACS8 | ACS9 | ACS11 | |
|---|---|---|---|---|---|---|---|---|---|
| Root cap | |||||||||
| Lateral |
 •↑ •↑ |
||||||||
| Columella | |||||||||
| Cell Division | |||||||||
| Lateral | • | • | |||||||
| Epidermis | • | • | • | ||||||
| Cortex | • | ||||||||
| Endodermis | • | ||||||||
| Pericycle | • | ||||||||
| Procambium | • | ||||||||
| Protophloem | • | ||||||||
| Protoxylem |
 • • |
• | • | ||||||
| Cell expansion | |||||||||
| Epidermis | • | ||||||||
| Cortex | • | ||||||||
| Endodermis |
 • • |
• | • | • | • | • | |||
| Pericycle |
 • • |
• | • | • | • | ||||
| Procambium |
 • • |
 • • |
• | • | • | ||||
| Protophloem |
 • • |
 • • |
• | • | • | ||||
| Protoxylem |
 • • |
 • • |
 • • |
 •↑ •↑ |
 • • |
 •↑ •↑ |
 •↑ •↑ |
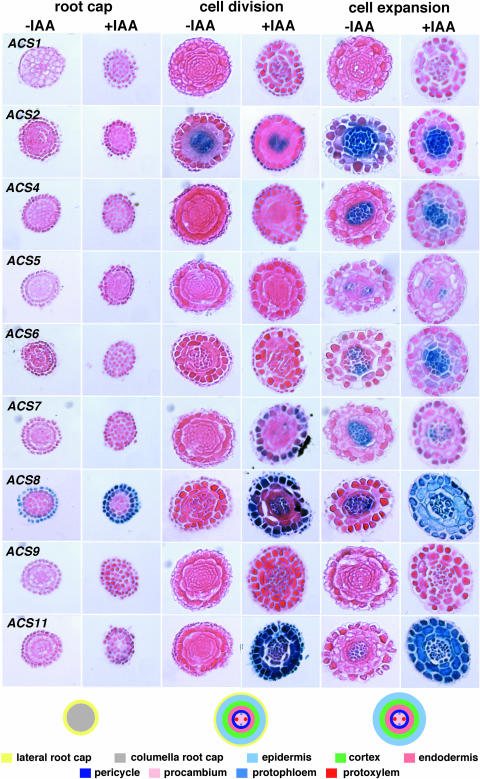
Next, we examined the cell specific expression of the ACS gene family members by sectioning the root tip of stained control and IAA treated 5-d-old light-grown seedlings (the location of the tissue cross-sectioning shown in Fig. 7). Three different zones were examined: lateral root cap, cell division, and cell expansion (Dolan et al., 1993, 1994; Dolan and Davies, 2004). The results are shown in Figure 11. Unique and overlapping patterns of ACS gene expression were observed in these three zones. Exogenous IAA induces the expression of various ACS genes (ACS6, 7, 8, and 11) in specific cell layers in these three root tip zones. Specifically, among the various ACS genes ACS8 is uniquely expressed in the lateral root cap zone. Very weak expression is also seen in the second cell layer of the lateral root cap zone (Fig. 11). Exogenous IAA enhances the preexisting ACS8-GUS expression in lateral root cap zone and greatly induces its activity in the second layer of the root cap zone. Moving upwards to the cell division zone, we only see expression of ACS2, which is restricted in the protoxylem cell files (Fig. 11). IAA treatment does not affect ACS2 expression; however, it induces the expression of ACS7 in the epidermis of the cell division zone, ACS8 in the lateral root cap, epidermis, and protoxylem, and ACS11 is induced in all cell types of the cell division zone.
Moving further upstream in the cell expansion zone, we see ACS2 expression in the endodermis, pericycle, procambium, protophloem, and protoxylem; IAA neither affects the pattern of expression nor enhances its intensity. ACS4 is primarily expressed in the procambium, protoxylem, and protophloem cell files, and in the presence of auxin the GUS expression is detected in endodermis (a few cells), pericycle, procambium, protoxylem, and protophloem. We observed weak expression of ACS5 in protoxylem that is not affected by auxin. ACS6 is expressed in protoxylem and in the presence of auxin the GUS expression is detected in endodermis (a few cells), pericycle, procambium, protoxylem, and protophloem. ACS7 is expressed primarily in protoxylem, and upon IAA treatment its expression is reduced. ACS8 is expressed at low level in protoxylem, and IAA treatment results in strong expression in all cell layers of this zone. The expression of ACS9 is nil in this zone in the presence and absence of IAA. Finally, ACS11 behaves similarly to ACS8, but its expression intensity is higher (Fig. 11).
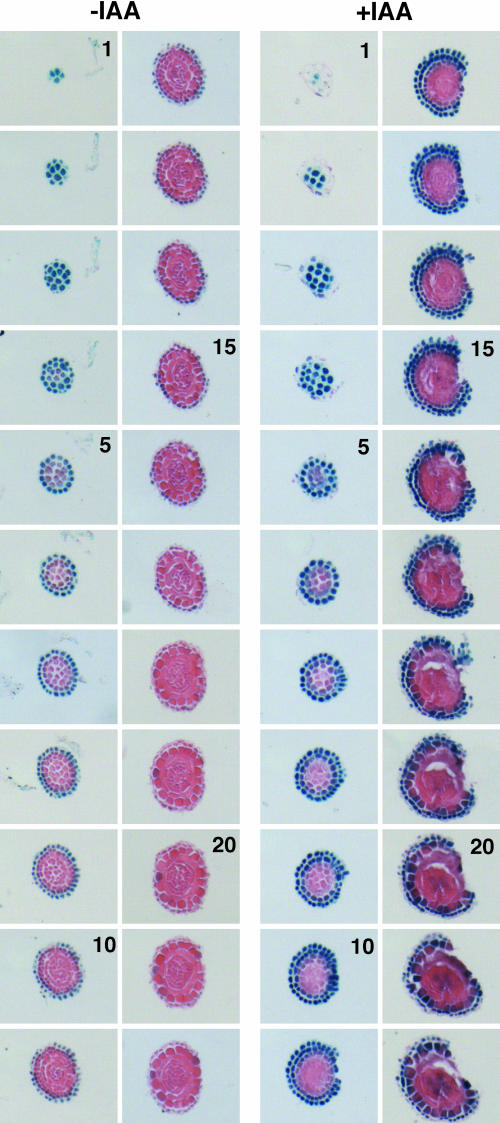
The unique expression of ACS8 in the root cap zone in the presence and absence of auxin was further investigated in great detail by serial sectioning in a direction moving away from the tip. The results are shown in Figure 12. The expression of ACS8 is highly restricted in the root cap cells and the first cell layer of the lateral root cap. The columella cells do not express ACS8. Auxin enhances the constitutive expression of ACS8 but also induces its activity in the second cell layer of the lateral root cap. These results clearly demonstrate that the effects of ACS8 shown in Figure 11 in the absence or presence of auxin are not due to a difference in the sectioning regions of the treated and untreated tissue. The absence of ACS8-GUS expression in the second layer of the lateral root cap is attributed to the suboptimal auxin concentration for transcriptional activation in the untreated tissue (Figs. 11 and 12).
Figure 12.
Expression of ACS8 in the lateral root cap zone of the primary root. GUS activity staining in several transverse sections (8 μm) of the primary root tip is shown for a 5-d-old light-grown transgenic for ACS8-GUS/GFP. The numbers show the order of sectioning away from the root tip.
DISCUSSION
Ethylene and Plant Growth and Development
As part of a continuing effort to elucidate the molecular details of ethylene biosynthesis at the level of the ACS, we examined the spatio-temporal expression patterns of the Arabidopsis ACS gene family members during Arabidopsis plant growth and development. We introduced into the Arabidopsis genome promoter GUS or GFP fusions that also contain DNA sequences located 3′ of the protein coding region to ensure cell and tissue specific expression (Dietrich et al., 1992). This visual global comparative and qualitative exploration of GUS expression patterns implies transcriptional activation of the endogenous ACS gene family members in specific tissues and cells throughout development. Ten years ago, a similar analysis was performed with ACS2, one of the first Arabidopsis gene family members to be cloned (Rodrigues-Pousada et al., 1993). The results are very similar to those presented here. Despite the fact that the exploration is highly limited and static, reminiscent of planetary explorations, the findings clearly show unique and overlapping patterns of expression among the various ACS gene family members in specific tissues and specialized cells. The majority of expression patterns correlate well with physiological target sites of ethylene action accumulated from extensive physiological and genetic evidence during the past century (Harvey and Rose, 1915; Goeschl et al., 1966, 1967; Smith and Robertson, 1971; Bucher and Pilet, 1982; Eisinger, 1983; Osborne et al., 1985; Guzman and Ecker, 1990; M.B. Jackson 1991; Osborne, 1991; Raskin, 1991; Schiefelbein and Benfey, 1991; Abeles et al., 1992; Kieber et al., 1993; Smalle et al., 1997; Achard et al., 2003; Dolan and Davies, 2004). Differential expression patterns among the various members were detected in response to wounding and various stress conditions. The findings reinforce once more the view of the central role of ethylene in a variety of physiological and developmental processes throughout the plant life cycle. A striking feature of ACS gene family expression is its overlapping pattern detected in the guard cells of the stomata. Surprisingly, very little is known about the role of ethylene in stomata physiology (Serna and Fenoll, 1997).
The exploration also revealed that among the nine gene family members, ACS9 has the most limited expression repertoire. The precise biological function of each ACS gene family member will be defined in the future by single or higher order loss-of-function mutations. The results of this analysis have the potential to serve as a guide during the exploration of the biological consequences of loss-of-function mutations for each ACS isozyme. If ethylene is an essential element for plant development, the expectation will be that the construction of a null Arabidopsis plant for all ACS gene family members will be lethal. However, the possibility exists that nonlethality may suggest that an as yet unidentified gaseous ligand is sensed by the ethylene receptors. It is of great interest that a penta-ACS mutant is healthy, taller, and lives longer (Tsuchisaka and Theologis, unpublished data). The verdict on the essential role of ethylene on plant growth and development will await the construction of an ennead-mutant. The current exploration also indicates the urgent need for development of high throughput technologies for monitoring in planta gene expression patterns of gene family members simultaneously throughout development. New technology is needed for creating a “movie” rather than simply “snapshots” (Birnbaum et al., 2003; Scheres et al., 2004).
Auxin and ACS
Auxin regulated ethylene production is one of the best known hormone interactions in plant biology (Yang and Hoffman, 1984; Abeles et al., 1992). Many auxin effects have been attributed to be mediated by ethylene (Abeles et al., 1992; Klee and Romano, 1994). The well-known tomato dgt mutant supports this view. It is defective in sensing auxin for enhancing ethylene production necessary for proper root gravitropic response (Bradford and Yang, 1980). Auxin induced ethylene production is due to transcriptional enhancement of the ACS genes (Bleecker and Kende, 2000). Reverse transcription-PCR analysis with RNA from auxin treated etiolated Arabidopsis seedlings identified six auxin regulated ACS gene family members (ACS2, 4, 5, 6, 8, and 11; Yamagami et al., 2003). The results presented here confirmed this observation and also added two additional members in the list of auxin regulated genes, ACS7 (Figs. 7 and 11) and ACS9 (Supplemental Fig. 14). ACS1 is the only one not yet found to be regulated by IAA. It is of great interest that all ACS genes, except ACS1, are induced by short treatment by cyclohexamide (Yamagami et al., 2003). This suggests that all auxin-regulated ACS gene family members are under the control of a fast turning over repressor(s) molecule. This type of transcriptional regulation is reminiscent of that seen for the Aux/IAA proteins (Abel et al., 1995a; Koshiba et al., 1995; Abel and Theologis, 1996). The prospect arises that the putative repressor(s) of the auxin-regulated ACS gene family members are the Aux/IAA proteins. Auxin induced ACS4 gene expression is inhibited by the axr1-12 and axr2 mutations (Abel et al., 1995b), components of the Aux/IAA-auxin response factor signaling apparatus (Frugis and Chua, 2002; Dharmasiri and Estelle, 2004).
The results of cell specific expression of the various ACS genes in the root tip region in response to auxin revealed the complexity of the ACS gene expression at the level of the individual cells by the hormone. The auxin inducibility is not only gene specific, but also cell type specific (Fig. 11). The possibility exists that the inability of certain root cell types to induce various ACS genes in response to auxin may be due to the absence of expression of specific Aux/IAA or ARF genes responsible for transcriptional enhancement of specific ACS gene family members. If indeed all ACS gene family members (except ACS1) are under the control of the Aux/IAA-ARF signaling apparatus, it must also sense other signaling pathways that are responsible for transcriptional activation of ACS genes (stress, N2, Li+, disease resistance, etc.). The sensing mechanism may be the activation of the proteolytic pathway for the degradation of the Aux/IAA repressors. Whereas transcriptional regulation of ACS gene family members is central for enhancement of ethylene production, recent findings indicate the central role of ACS protein turnover as a key regulator of ethylene production in plants (Chae et al., 2003; Wang et al., 2004; Woeste et al., 1999). The steady-state levels of ethylene production during plant development in individual cells and tissues must be regulated by a “sensor,” which controls the transcriptional and posttranscriptional regulation of ACS gene family members by various signaling pathways.
Unique and Overlapping Patterns of Expression: Implications
A major discovery of the Arabidopsis genome sequencing project was the finding that many gene products encode isoforms of the same polypeptide (Arabidopsis Genome Initiative, 2000). While the various ACS isozymes catalyze the same biochemical reaction, it is not known whether their biological function(s) are distinct or overlapping. Genetic evidence (Vogel et al., 1998) and evolutionary considerations (Nowak et al., 1997) support the view that each member of the ACS gene family may have a distinct biological function. It has been suggested that tissue specific expression of a particular ACS isozyme satisfies the biochemical environment of the cells and tissues in which each isozyme is expressed (Rottmann et al., 1991). According to this view, the distinct biological function of each isozyme is defined by its biochemical properties. Such a concept enhances the physiological fine-tuning of the cell and demands that the enzymatic properties of each isozyme be distinct (Graur and Li, 2000). The results presented herein as well as the recent biochemical characterization of the eight functional isozymes (Yamagami et al., 2003) support this proposition.
ACS is a homodimer with shared active sites (Li et al., 1997; Tarun and Theologis, 1998). Intermolecular complementation experiments in E. coli provided evidence of functional heterodimeric interactions among the various ACS gene family members (Tsuchisaka and Theologis, 2004). These findings raise the question of where similar interactions take place in planta. A prerequisite for ACS heterodimer formation in planta is the existence of overlapping expression among various ACS gene family members. The results presented in this communication provide strong support for potential ACS heterodimer(s) formation in specific cells and tissues.
Overlapping gene expression patterns provide potentially additional metabolic flexibility in individual cell tissue if the heterodimers formed have different biochemical properties than the corresponding homodimers. We do not yet know whether the enzymatic properties of the various heterodimers are distinct. This is a task for the future. The capacity of the various isozymes to form active heterodimers further enhances the biochemical diversity of the ACS gene family. It provides an extensive repertoire of ACS isozymes capable of operating under a very broad spectrum of AdoMet concentration during the plant cycle (Graur and Li, 2000). Gene redundancy provides biochemical and metabolic flexibility. The driving force for conducting the analysis presented in this communication was to determine whether overlapping gene expression patterns can be detected among the ACS gene family members. It is a pilot experiment that will serve as a beacon during our exploration for ACS heterodimer formation using BiFC which currently is in progress in our laboratory.
Utility of Resources Generated
The resources generated during the course of this study together with the available ACS open reading frame (ORF) clones (Yamagami et al., 2003), and the T-DNA insertion lines (Borevitz and Ecker, 2004) constitute a complete set of molecular genomic tools for future functional and proteomics research in ethylene biosynthesis and of the ACS gene family in particular. These tools have been deposited in the ABRC (http://www.biosci.ohio-state.edu/plantbio/Facilities/abrc/abrchome.htm) and are available without any restrictions to the entire plant biology community worldwide. The transgenic lines have the potential to provide useful information on the regulation of each ACS gene family member by a variety of signaling pathways (Alonso and Ecker, 2001; Thomma et al., 2001; Wang et al., 2002). Genetic crosses of the ACS-GUS lines with known mutants that affect the pathway have the potential to define the signaling pathways sensed transcriptionally by each ACS gene family member. For example, it is not yet known which ACS gene(s) are transcriptionally regulated by ethylene positively or negatively (Yang and Hoffman, 1984). The observation that the ethylene response mutants overproduce ethylene (Guzman and Ecker, 1990) suggests the presence of a molecular link between ethylene perception and biosynthesis. The link may affect ACS gene transcription, ACS protein stability, or both. Genetic crosses of the ACS-GUS/GFP or ACS-GUS/GFP-ACS ORF fusion lines with ethylene response mutants have the potential to identify genetically the ACS gene family members that communicate with the ethylene signaling apparatus. The lines may also be used as visual markers for various developmental studies as well as for reverse genetic screens to isolate mutations for trans acting factors responsible for the regulation of each family member to various inducers (Susek et al., 1993; Oono et al., 1998). Similar resources have to be generated for all the genes of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). A complete set a promoters, 3′ untranslated regions, and ORF clones will allow the new generation of plant biologists to carry out functional and proteomics experiments on a global scale. The days of one by one belong to the past.
MATERIALS AND METHODS
Enzymes and Chemicals
Restriction and DNA modifying enzymes were obtained from New England Biolabs (Beverly, MA) and Roche Diagnostics (Indianapolis). All other chemicals used for biochemical analysis were of analytical grade and purchased from Sigma-Aldrich (St. Louis). Oligonucleotides were purchased from Operon Technologies (Alameda, CA) or synthesized in house with a polyplex oligonucleotide synthesizer (GeneMachines, San Carlos, CA).
Plant Material and Treatment
Arabidopsis ecotype Columbia was used throughout this study. One-month-old plants were grown as described in the manual of the Arabidospsis Biological Resources Center (ABRC) and used for transformation. All seeds were surface sterilized for 8 min in 5% NaOCl and 0.15% Tween 20, rinsed excessively in distilled water, and cold treated at 4°C for 2 d by plating them on plates containing Murashige and Skoog medium containing 0.8% Select agar (Life Technologies, Rockville, MD), 0.5 mm MES, pH 5.7, 1% Suc, 1× B5 vitamins with 100 μg/mL gentamicin, and 100 μg/mL carbenicillin. The cold treated seeds were subsequently germinated under various conditions to obtain transformed seedlings or mature plants for experimentation.
Mature T2 transgenic plants were grown hydroponically for determining GUS expression in roots as follows. Gentamicin resistant seedlings were obtained by germinating seeds at 25°C under 16-h-light/8-h-dark cycle for 10 d. The seedlings were placed in the hydroponic culture solution (Baba and Takahashi, 1979; 1 L) by dilution of the following solutions. Solution A, 0.18 m (NH4)2SO4, 0.045 m K2SO4, 0.27 m MgSO4, 0.09 m KNO3, 0.09 m KH2PO4; solution B, 0.18 m Ca(NO3)2 and 0.03 m C6H5FeO7; and solution C, 46.26 mm H3BO4, 9.15 mm MnCl2, 0.77 mm ZnSO4, 0.32 mm CuSO4, and 0.52 mm Na2MoO4. The dilution ratio is: A, 1:500; B, 1:500; C, 1:1,000. The pH was adjusted to 5.7 with NaOH. The culture solution was aerated with a fish-tank pump and refilled daily. It was replaced with fresh solution weekly. The plants were grown in a greenhouse at 25°C under a 16-h-light/8-h-dark light cycle.
Five-day-old light-grown T2 transgenic seedlings were obtained by incubation of seeds at 25°C under a 16-h-light/8-h-dark light cycle. IAA treated seedlings were obtained by placing them on top of MS plates containing 20 μm IAA, and incubating for 24 h under the conditions described above. Similarly, cold and heat treated seedlings were obtained by placing them on Murashige and Skoog plates and incubating at 4°C and 37°C, respectively, for 24 h under light conditions described above. LiCl treated seedlings were obtained by placing them on MS plates containing 50 mm LiCl and incubating at 25°C for 24 h. Anaerobiotically treated seedlings were obtained by treating 5-d-old seedlings with N2 for 24 h at 25°C. Wounding experiments were carried out by cutting hypocotyls in the middle with a razor blade and incubating the cut tissue for 4 h at 25°C.
Molecular Biology Details
The construction of the transgenes used in this study is described in the supplemental data.
Histochemical GUS Assay
Histochemical assays of GUS activity in transgenic lines were performed as described (Jefferson et al., 1987). Treated seedlings or adult plants were incubated at 37°C in 100 mm sodium phosphate pH 7.5, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 10 mm EDTA, and 0.1% Triton X-100 containing 1 mm 5-bromo-4-chloro-β-d-gluclonide. Incubation times varied from 2 to 12 h following vacuum infiltration. Subsequently, the samples were then transferred to 70% ethanol to remove the chlorophyll.
Tissue traverse sections were prepared as follows: Stained samples were infiltrated with a series of ethanol, Histoclear (National Diagnostics, Atlanta, GA), Paraplast X-tra (Kendall, Mansfield, MA) solutions as previously described (Jackson, 1991). Ribbons of 8- to 20-μm sections were cut on a rotary microtome (MICROM HM340; Carl Zeiss, San Leandro, CA), floated on 42°C water in the water bass for about 10 min, and transferred to slides and dried overnight on a 42°C block. Sections were dewaxed with Histoclear and mounted permanently with Hydromount Aqueous Non-Fluorescing Mounting media (National Diagnostics, Atlanta).
Imaging of Green Fluorescence in Plants
Five-day-old seedlings were placed in a drop of water on a glass slide, and directly examined with an Axioplan Zeiss Fluorescence Microscopy system (Zeiss, Jena, Germany) with SPOT 2.2 software (Diagnostic Instruments, Sterling Heights, MI). The excitation filter was D480/30, the emission filter was D535/40, and the beamsplitter was 505dclp (Chroma Technology, Rockingham, VT).
Mapping the Insertion Sites of the GUS/GFP Transgenes
We used thermal asymmetric interlaced PCR for mapping the integration site of the ACS promoter-GUS/GFP-3′ fusions in the transgenic lines following the procedure described by Liu et al. (1995; Liu and Whitter, 1995) and McElver et al. (2001). The primers are shown in the supplemental data.
Genomic Resources
Table I shows the plasmids and transgenic lines that have been deposited in the ABRC (http://www.biosci.ohio-state.edu/plantbio/Facilities/abrc/abrchome.htm) in order for the rest of the plant community to be able to use the resources generated in this study without any restrictions.
Table I.
Plasmids and transgenic lines deposited in ABRC
| Plasmids
| |||
|---|---|---|---|
| Promoters/5′ UTRs
|
3′ UTRs
|
||
| Name of Plasmid | Accession No. | Name of Plasmid | Accession No. |
| pTheo-ACS1-5′ | AY680407 | pTheo-ACS1-3′ | AY680416 |
| pTheo-ACS2-5′ | AY680408 | pTheo-ACS2-3′ | AY680417 |
| pTheo-ACS4-5′ | AY680409 | pTheo-ACS4-3′ | AY680418 |
| pTheo-ACS5-5′ | AY680410 | pTheo-ACS5-3′ | AY680419 |
| pTheo-ACS6-5′ | AY680411 | pTheo-ACS6-3′ | AY680420 |
| pTheo-ACS7-5′ | AY680412 | pTheo-ACS7-3′ | AY680421 |
| pTheo-ACS8-5′ | AY680413 | pTheo-ACS8-3′ | AY680422 |
| pTheo-ACS9-5′ | AY680414 | pTheo-ACS9-3′ | AY680423 |
| pTheo-ACS11-5′ | AY680415 | pTheo-ACS11-3′ | AY680424 |
| Plasmids
|
Transgenic Linesa
|
||
| Name of Plasmids
|
Name of At Line
|
||
| pTheo-At-ACS1-GUS/GFP | Theo-At-ACS1-GUS/GFP | ||
| pTheo-At-ACS2-GUS/GFP | Theo-At-ACS2-GUS/GFP | ||
| pTheo-At-ACS4-GUS | Theo-At-ACS4-GUS | ||
| pTheo-At-ACS5-GUS | Theo-At-ACS5-GUS | ||
| pTheo-At-ACS6-GUS/GFP | Theo-At-ACS6-GUS/GFP | ||
| pTheo-At-ACS7-GUS/GFP | Theo-At-ACS7-GUS/GFP | ||
| pTheo-At-ACS8-GUS/GFP | Theo-At-ACS8-GUS/GFP | ||
| pTheo-At-ACS9-GUS/GFP | Theo-At-ACS9-GUS/GFP | ||
| pTheo-At-ACS11-GUS/GFP | Theo-At-ACS11-GUS/GFP | ||
The GenBank accession numbers of the DNA sequences in each plasmid are shown.
The lines are homozygous for the insertion, except ACS7.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY680407 to AY680424.
Supplementary Material
Acknowledgments
We thank Dr. Rachel Dent for constructing the ACS4- and ACS5-GUS transgenic lines; Drs. Jim Haseloff and Sarah Hodge for the generous gift of pBIN-mgfp5-ER plasmid; Dr. Jennifer Fletcher for her advice on Arabidopsis plant morphology; Drs. Cristel Carles and Leor Eshed-Williams for their help with tissue sectioning; Dr. Yutaka Oono for help and advice with thermal asymmetric interlaced-PCR; and Mr. David Hantz, Mrs. Julia Calfas, and Mr. Vinnie Pirozzi for greenhouse support.
This work was supported by the National Science Foundation (grant nos. MCB–9982895, IBN–0211421) and by the U.S. Department of Agriculture-Agricultural Research Service (CRIS no. 5335–21430–005–00D to A.T.).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.049999.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Theologis A (1995. a) The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Abel S, Nguyen MD, Chow W, Theologis A (1995. b) ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin. J Biol Chem 270: 19093–19099 [DOI] [PubMed] [Google Scholar]
- Abeles FB, Morgan PW, Saltveit ME Jr (1992) Ethylene in Plant Biology. Academic Press, New York
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Ecker JR (2001) The ethylene pathway: a paradigm for plant hormone signaling and interaction. Science's STKE 2001: 1–10 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baba K, Takahashi Y (1979) Suikou-hou oyobi sakou-hou. In Y Tokari, T Matsuo, Y Hatamura, N Yamada, T Harada, N Suzuki, eds, Sakumotsu-shiken-hou. Nougyo-gijutu-kyokai Press, Tokyo
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Ecker JR (2004) Plant Genomics: the third wave. Annu Rev Genomics Hum Genet 5: 443–477 [DOI] [PubMed] [Google Scholar]
- Bradford KJ, Yang SF (1980) Stress-induced ethylene production in the ethylene-requiring tomato mutant Diageotropica. Plant Physiol 65: 327–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Pilet P-E (1982) Ethylene effects on growing and gravi reacting maize zea-mays cultivar lg-11 root segments. Physiol Plant 55: 1–4 [Google Scholar]
- Capitani G, Hohenester E, Feng L, Storici P, Kirsch JF, Jansonius JN (1999) Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J Mol Biol 294: 745–756 [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Radke SE, Harada JJ (1992) Downstream DNA sequences are required to activate a gene expressed in the root cortex of embryos and seedlings. Plant Cell 4: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Davies J (2004) Cell expansion in roots. Curr Opin Plant Biol 7: 33–39 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K (1994) Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120: 2465–2474 [Google Scholar]
- Eisinger W (1983) Regulation of pea internode expansion in ethylene. Annu Rev Plant Physiol 34: 225–240 [Google Scholar]
- Frugis G, Chua N-H (2002) Ubiquitin-mediated proteolysis in plant hormone signal transduction. Trends Cell Biol 12: 308–311 [DOI] [PubMed] [Google Scholar]
- Goeschl JD, Rappaport L, Pratt HK (1966) Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol 41: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeschl JD, Pratt HK, Bonner BA (1967) An effect of light on the production of ethylene and the growth of the plumular portion of etiolated pea seedlings. Plant Physiol 42: 1077–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Li W-H (2000) Gene duplication, exon shuffling, and concerted evolution. In Fundamentals of Molecular Evolution, Ed 2. Sinauer Associates, Sunderland, MA, pp 249–322
- Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey EM, Rose RC (1915) The effects of illuminating gas on root systems. Bot Gaz 60: 27–44 [Google Scholar]
- Haseloff SR, Meyer SE, Callis J (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Jackson DP (1991) In situ hybridization in plants. In DJ Bowles, SJ Gurr, M McPherson, eds, Molecular Plant Pathology: A Practical Approach. Oxford, Oxford Univeristy Press, pp 163–174
- Jackson MB (1991) Ethylene in root growth and development. In AK Mattoo, JC Suttle, eds, The Plant Hormone Ethylene. CRC Press, Boca Raton, FL, pp 169–181
- Jefferson RA, Kavanagh TA, Bevan MV (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Klee HJ, Romano CP (1994) The roles of phytochromes in development as studied in transgenic plants. CRC Crit Rev Plant Sci 13: 311–324 [Google Scholar]
- Koshiba T, Ballas N, Wong L-M, Theologis A (1995) Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J Mol Biol 253: 396–413 [DOI] [PubMed] [Google Scholar]
- Li Y, Feng L, Kirsch JF (1997) Kinetic and spectroscopic investigatioins of wild-type and mutant forms of apple 1-aminocyclopropane-1-carboxylate synthase. Biochemistry 36: 15477–15488 [DOI] [PubMed] [Google Scholar]
- Liang X-W, Abel S, Keller JA, Shen NF, Theologis A (1992) The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Proc Natl Acad Sci USA 89: 11046–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Shen NF, Theologis A (1996) Li+-regulated 1-aminocyclopropane-1-carboxylate synthase gene expression in Arabidopsis thaliana. Plant J 10: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu YG, Whitter RF (1995) Thermal asymmetric interlaced PCR: automutable amplification and sequencing of insert end fragments from Y1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- McElver JA, Tzafrir IA, Aux GA, Rogers RA, Ashby CA, Smith KA, Thomas CA, Schetter AA, Zhou QA, Cushman MAA, et al (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta PK, Hale TI, Christen P (1989) Evolutionary relationships among aminotransferases. Tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. Eur J Biochem 186: 249–253 [DOI] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388: 167–171 [DOI] [PubMed] [Google Scholar]
- Oono Y, Chen QG, Overvoorde PJ, Köhler C, Theologis A (1998) age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne D (1991) Ethylene in leaf ontogeny and abscission. In AK Mattoo, JC Suttle, eds, The Plant Hormone Ethylene. CRC Press, Boca Raton, FL, pp 193–214
- Osborne DJ, McManus MT, Webb J (1985) Target cells for ethylene action. In JA Roberts and GA Tucker, eds, Ethylene and Plant Development. Butterworths, London, pp 197–212
- Raskin I (1991) Ethylene in vegetative growth. In AK Mattoo, JC Suttle, eds, The Plant Hormone Ethylene. CRC Press, Boca Raton, FL, pp 183–192
- Rodrigues-Pousada RA, De RR, Dedonder A, Van CW, Engler G, Van MM, Van DSD (1993) The Arabidopsis 1-aminocyclopropane-1-carboxylate synthase gene 1 is expressed during early development. Plant Cell 5: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann WE, Peter GF, Oeller PW, Keller JA, Shen NF, Nagy BP, Taylor LP, Campbell AD, Theologis A (1991) 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J Mol Biol 222: 937–961 [DOI] [PubMed] [Google Scholar]
- Scheres B, van den Toorn H, Heidstra R (2004) Root genomics: towards digital in situ hybridization. Genome Biol 5: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Benfey PN (1991) The development of plant roots new approaches to underground problems. Plant Cell 3: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Fenoll C (1997) Tracing the ontogeny of stomatal clusters in Arabidopsis with molecular markers. Plant J 12: 747–755 [DOI] [PubMed] [Google Scholar]
- Siemering KR, Golbik R, Sever R, Haseloff J (1996) Mutations that suppress the thermosensitivity of green fluorescent protein. Curr Biol 6: 1653–1663 [DOI] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van MM, Van DSD (1997) Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proc Natl Acad Sci USA 94: 2756–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KA, Robertson PD (1971) Effect of ethylene on root extension of cereals. Nature 234: 148–149 [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FA, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Tarun AS, Theologis A (1998) Complementation analysis of mutants of 1-aminocyclopropane-1-carboxylate synthase reveals the enzyme is a dimer with shared active sites. J Biol Chem 273: 12509–12514 [DOI] [PubMed] [Google Scholar]
- Tarun AS, Lee JS, Theologis A (1998) Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: a key enzyme in ethylene biosynthesis. Proc Natl Acad Sci USA 94: 9796–9801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki M, Mori H (2001) Phosphorylation of tomato 1-aminocyclopropane-1-carboxylic acid synthase, LE-ACS2, at the C-terminal region. J Biol Chem 276: 28051–28057 [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Penninckx IAMA, Broekaert WF, Cammue BPA (2001) The complexity of disease signaling in Arabidopsis. Curr Opin Immunol 13: 63–68 [DOI] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A (2004) Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc Natl Acad Sci USA 101: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Woeste K, Theologis A, Kieber JJ (1998) Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc Natl Acad Sci USA 95: 4766–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Yoshida H, Lurin C, Ecker JR (2004) Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950 [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14 (Supple): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Vasquez J, Yang SF, Kirsch JF (1994) Expression of apple 1-aminocyclopropane-1-carboxylate synthase in Escherichia coli: kinetic characterization of wild-type and active-site mutant forms. Proc Natl Acad Sci USA 91: 12428–12432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeste KE, Ye C, Kieber JJ (1999) Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol 119: 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami T, Tsuchisaka A, Yamada K, Haddon WF, Harden LA, Theologis A (2003) Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem 278: 49102–49112 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yip W-K, Dong J-G, Kenny JW, Thompson GA, Yang SF (1990) Characterization and sequencing of the active site of 1-aminocyclopropane-1-carboxylate synthase. Proc Natl Acad Sci USA 87: 7930–7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A (1994) Ethylene biosynthesis and action: a case of conservation. Plant Mol Biol 26: 1579–1597 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.