Abstract
Background
In this study, we measured the latent HIV-1 reservoir harboring replication-competent HIV-1 in resting CD4+ T cells in participants on highly active antiretroviral therapy (HAART), quantitating the frequency of latent infection through the use of a Primer ID-based Ultra Deep Sequencing Assay (UDSA), in comparison to the readout of the quantitative viral outgrowth assay (QVOA).
Methods
Viral RNA derived from culture wells of QVOA that scored as HIV-1 p24 capsid (CA) antigen-positive were tagged with a specific barcode during cDNA synthesis, and the sequences within the V1–V3 region of the HIV-1 env gene were analyzed for diversity using the Primer ID-based paired-end MiSeq platform. We analyzed samples from a total of 19 participants, 2 initially treated with HAART in acute infection and 17 treated during chronic infection. Phylogenetic trees were generated with all viral lineages detected from culture wells derived from each participant to determine the number of distinct viral lineages (DVLs) growing out in each well, thus capturing another level of information beyond the well being positive for viral antigen. The infectious units per million cells (IUPM) values estimated using a maximum likelihood approach, based on the number of DVLs detected (VOA-UDSA), were compared with those obtained from QVOA measured using limiting dilution.
Results
IUPM estimates determined by VOA-UDSA ranged from 0.14 to 3.66 and strongly correlated with the IUPM estimates determined by QVOA (r=0.94; p<0.0001).
Conclusions
VOA-UDSA may be an alternative readout for that currently used for QVOA.
Keywords: HIV-1, latent reservoir, quantitative viral outgrowth assay, infectious units per million cells, ultra deep sequencing
INTRODUCTION
Eradication of HIV-1 from infected individuals on suppressive therapy has been hampered due to HIV-1 persistence in viral reservoirs. Among potential cellular reservoirs contributing to HIV-1 persistence,1–3 latent infection of resting memory CD4+ T cells is well demonstrated.4–8 Assays have been developed to quantify resting CD4+ T cells that harbor replication-competent HIV-1 in people on HAART. The current most widely recognized culture assay, the quantitative viral outgrowth assay (QVOA), measures infectious units per million (IUPM) resting CD4+ T cell. Although QVOA is the current gold standard for measuring the latent HIV-1 reservoir, the assay has limited throughput capacity due to the need for large numbers of cells and the need to wait for the outgrowth of virus from single cells to assess the titer by limiting dilution.9 Using QVOA it has been possible to show that a small fraction of the resting CD4+ T cells harbor replication-competent HIV-1 (approximately 0.1 to 10 per million cells).4,10,11 However, Ho et al. in a recent study, demonstrated that this number is likely an underestimate as some cells that encode an HIV-1 provirus and can be induced to express HIV-1 RNA do not produce a replication-competent virus that can be recovered following a single activation event during the QVOA.12 A modified version of QVOA has recently been developed. In this modified assay, a CD4+/CCR5+ cell line (MOLT-4/CCR5) replaces primary cells to support virus outgrowth, and an HIV-specific RT-PCR assay replaces p24 ELISA for virus detection in the supernatant, which allows earlier detection of virus outgrowth.13 These modifications shorten the time required to complete the QVOA, however, the modified assay still requires large amounts of blood from a patient to allow limiting dilution of resting CD4+ T cells to measure an IUPM value.
PCR-based assays detecting HIV-1 DNA are the most sensitive assays for detecting viral sequences in a cell population, but these assays greatly overestimate the number of infectious proviruses as they detect all forms of HIV-1 DNA, including replication-incompetent proviruses and unintegrated viral genomes that dominate the proviral pool.14–17 Compared to the frequency of resting CD4+ T cells harboring inducible, replication-competent HIV-1, an approximately ~300-fold excess amount of mostly defective HIV-1 proviral DNA has been detected by PCR-based assays.18,19 Assays detecting cell-associated HIV-1 RNA after the induction of viruses in resting CD4+ T cells have increased sensitivity, however, these assays also overestimate the number of inducible, infectious proviruses because not all transcribed viral RNAs give rise to replication competent virus.20,21 Recently, a new RNA assay termed TILDA (tat/rev Induced Limiting Dilution Assay) was proposed to measure the frequency of latently infected resting CD4+ T cells by quantifying tat/rev multiply-spliced (ms) HIV-1 RNA upon induction.22 The median frequency of latently infected CD4+ T cells estimated by TILDA was 48 times more than the frequency measured by QVOA.22 The assay requires significantly fewer cells from a patient and takes less time than does QVOA, however, it is likely that the latent HIV-1 reservoir estimated by TILDA is also an overestimate as not all cells expressing tat/rev ms RNA are likely to produce replication-competent viruses.
Recent efforts directed at ultimately curing an HIV-1 infection have led to the identification of latency reversing agents such as histone deacetylase (HDAC) inhibitors that reverse HIV-1 latency by inducing HIV-1 transcription.23–26 These drugs are currently being tested in clinical trials, now in combination with immunotherapies to speed the clearance of infected cells. To assess the effectiveness of these drugs, it is necessary to have established assays that can precisely record changes in the frequency of latent infection. It has been difficult to demonstrate that other assays directed at assessing the size of the latent reservoir are accurate surrogates for QVOA measurements.19
The viral population found in resting CD4+ T cells of viremic patients can be very diverse.9 In an attempt to develop an alternative readout for the QVOA, we quantified the frequency of latent HIV-1 infection by employing a Primer ID-based Ultra Deep Sequencing Assay of the outgrowth virus (VOA-UDSA). VOA-UDSA uses the differences in viral sequence to identify the number of distinct viruses induced to replicate in a culture supernatant, thereby yielding an estimate of the frequency of latently HIV-infected resting CD4+ T cells in the culture, given the assumption that most latently infected cells are infected by a single viral genome. Thus, VOA-UDSA could eliminate the limiting dilution culture protocol that is typically required for estimating a titer of latently infected cells using QVOA. In our current study, the IUPM values determined by VOA-UDSA from p24+ wells were strongly correlated with the IUPM values determined by QVOA (r=0.94; p<0.0001), suggesting that this approach can provide an alternative readout for the QVOA.
METHODS
Participants, QVOA, and Viral Samples
All participants were on suppressive antiretroviral therapy (ART) with plasma HIV-1 RNA levels of <50 copies/ml for a minimum of 6 months prior to enrollment. Studies were approved by the University of North Carolina at Chapel Hill Biomedical Review Board. Informed consent was obtained from all participants prior to study enrollment. Lymphocytes were obtained by continuous-flow leukapheresis, and resting CD4+ T cells were isolated by negative selection as previously described.27 QVOA to recover replication competent HIV-1 from resting CD4+ T cells was performed as reported elsewhere.28–30 This included inducing expression of the latent virus and co-culture with allogeneic CD4+ T cells. Supernatants from the co-cultures were harvested at day 15 and at day 19 to test for p24 using an ELISA, with excess supernatant medium frozen. The titer of resting CD4+ T cells that could be induced to produce infectious virus was reported as infectious units per million cells (IUPM) and was estimated via maximum likelihood as previously described; this approach requires that multiple wells at various dilutions of cells be tested until wells negative for viral outgrowth are obtained, i.e. limiting dilution.28–30 The 95% confidence intervals for the IUPM estimates were calculated by using the program IUPMStats v1.0 Infection Frequency calculator.31
MiSeq Library Preparation and Sequencing
The 300 base paired-end multiplex Illumina MiSeq library preparation and sequencing was performed as previously described.32 Briefly, viral RNA was extracted from the culture supernatants derived from VOA using a QIAamp viral RNA kit (Qiagen) and quantified by quantitative real-time RT-PCR using a TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). The sequences of the primers and probe to detect the HIV-1 gag region were previously described.33 The cDNA reaction was carried out with approximately 10,000 copies of HIV-1 RNA and a Primer-ID cDNA primer composed of an HIV-1 gene-specific sequence for priming at the 3’ end, a four nucleotide spacer, an eight or ten nucleotide randomized sequence (Primer ID), and a PCR priming site at the 5’ end (see Supplemental Digital Content 1). The HIV-1 gene-specific sequence for priming corresponds to the V3 region of the HIV-1 env gene (nt position from 7238 to 7209, HBX2 numbering). The reverse primer used in the 2nd round PCR contained a specific Barcode to assign the culture supernatant, i.e. the well derived from the VOA. A maximum of 24 indexed reverse primers were used to multiplex samples in a single deep sequencing run. The final PCR products corresponding to the HIV-1 env V1–V3 region were subjected to deep sequencing analysis.
For viral samples derived from participants C1, C2, C7, C8, and C12, the cDNA reaction was carried out using one of the four Primer ID cDNA primers containing a specific Barcode (see Supplemental Digital Content 1). After the cDNA reaction step, four cDNA reactions each with a different Barcode were pooled before cDNA purification to result in 96 libraries in a single deep sequencing run.
Distinct Viral Lineages (DVLs)
Primer IDs with multiple sequence reads detected in each well were used to create consensus sequences as previously described.32 Consensus sequences with an abundance higher than the arbitrary cut-off value, 2.5% (Fig. 1A and see Supplemental Digital Content 2), were processed to detect viral lineages in each well and to determine DVLs by using an in-house pipeline (available at https://github.com/SwanstromLab/DVL). Viral lineages and DVLs were also manually determined using MUSCLE (v3.8.1)34,35 and neighbor-joining phylogenetic tree analysis (MEGA 5.10), respectively. The total number of DVLs divided by the total number of viral lineages was used to report % DVL detected for each participants.
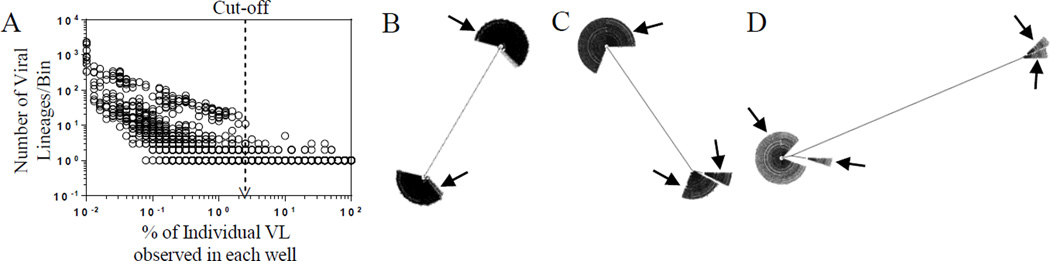
Figure 1.
A, Arbitrary abundance cut-off for the minimum abundance of individual consensus sequences. The arbitrary cut-off value, 2.5%, for the minimum abundance of individual consensus sequences is shown in dotted line with an arrowhead. B, C, and D, Viral Lineages detected in a single well. Phylogenetic trees showing examples of 2, 3, and 4 viral lineages detected in a single well are shown in B, C, and D, respectively. Trees were generated using MUSCLE (v3.8.1). Arrows indicate distinct viral lineages detected in each well.
IUPM Calculation For VOA-UDSA
The IUPM was estimated using maximum likelihood assuming the number of infected cells per well followed a Poisson distribution for each DVL. Assuming independence among wells and different lineages, the likelihood of the observed data is proportional to:
| (Equation 1) |
where M is the total number of wells used in the QVOA, xM is the total number of wells scored as p24 positive in the QVOA, m is the number of wells analyzed by VOA-UDSA, n is the total number of DVLs detected, yi is the number of wells expressing ith DVL to correct the potential of two identical viral lineages present in the same well, and λi is the mean of the ith DVL. When all QVOA positive wells were analyzed using UDSA (m = xM), the maximum likelihood estimator (MLE) of λi equals . Otherwise (m < xM), the MLE was obtained numerically using the Equation 1. IUPM estimates were obtained by summing the MLEs of the DVL specific means and then multiplying by the appropriate scaling factor according to the number of cells per well used in the QVOA. Wald-type confidence intervals were computed using the observed information to estimate the standard error of the IUPM estimate.
Simulation of Recombination Errors
Two wells derived from participant C16 were selected to simulate method-introduced recombination errors. Each well contained only one viral lineage and there were 9 nucleotide differences between the two viral lineages within the V1–V3 region of the HIV-1 env sequences. The viral RNAs extracted from each well were mixed at a molar ratio of 1:1 for the cDNA reaction that was then subjected to UDS to result in approximately 4000 template consensus sequences, and this was repeated as four sets of duplicate cDNA reactions. Consensus sequences were generated without using the arbitrary cut-off value to maximize the detection of minor viral lineages. Highlighter plots were generated using all viral lineages to identify recombinants.
Statistical Analysis
To measure the correlation between the IUPMs obtained from the two assays, VOA-UDSA and QVOA, correlation analysis was performed to calculate the Spearman rank correlation coefficient (r) and the best-fitting straight line was formed using linear regression analysis. The Wilcoxon matched-pairs test was used to compare the mean ranks of the IUPM values obtained from the two assays, VOA-UDSA and QVOA. All statistical analyses were performed using GraphPad Prism software version 6 (San Diego, CA) and all exact p values are two-tailed.
RESULTS
VOA-UDSA Can Detect Multiple Viral Lineages In A Single VOA Well
The goal of this work was to develop an assay that eliminates the limiting dilution step of the QVOA. VOA-UDSA uses viral diversity and Primer ID-based UDS to quantify the number of resting CD4+ T cells harboring inducible, replication-competent HIV-1 in participants on HAART. VOA-UDSA uses differences in viral sequences to count the number of different viruses induced from cultured resting CD4+ T cells based on the assumption that individual cells carry distinct proviruses.
VOA-UDSA involves induction of proviruses in latently HIV-infected resting CD4+ T cells and deep sequencing analysis of viral samples of the culture supernatants to determine distinct viral lineages (DVLs) (see Supplemental Digital Content 3). We analyzed 146 culture supernatants derived from QVOA, representing cells from 2 participants in acute infection and 17 participants who were chronically infected. Individual viral RNA templates were tagged with a Primer ID, allowing the construction of a template consensus sequence for each RNA copied in the cDNA step. In addition, a separate barcode was added to the PCR products of each well to designate the well of origin. An amplicon representing the region from V1–V3 of the viral env gene was sequenced.
Figure 1A shows the distribution of the number of viral lineage as a function of abundance from all of the wells analyzed in this study. The number of observed lineages increases significantly as the abundance decreases below 2.5%. Since most of the wells analyzed in pools of 2.5 million resting CD4+ T cells should have only a few viral lineages, the use of UDS results in high levels of redundancy in the sequencing of these few lineages. As a result of this, sequencing errors are extensively represented in the data set. However, the sequencing errors are much less abundant than the true viral lineages, which we attribute to the significant increases seen in lineages of abundance below 2.5% (Fig. 1A). For this reason we count as viral lineages only those homogeneous viral populations that comprise at least 2.5% of the population. Each consensus template sequence that passed the abundance cut-off and that differed by one or more nucleotides within the HIV-1 env V1–V3 region was defined as a viral lineage. Using this cut-off we would ignore a slow growing virus that failed to reach at least 2.5% of the viral population in a well of the QVOA that had multiple viruses induced to replicate.
Figures 1B, C, and D show examples of 2, 3, and 4 viral lineages, respectively, identified in single wells of QVOA from chronically infected participants. We were able to detect up to 8 viral lineages in a well of the QVOA in this setting. In this study we analyzed wells containing either 2.5 or 0.5 million resting CD4+ T cells in the QVOA culture wells.
Largely Identical Viral Lineages Are Detected In QVOA In Participants Treated Since Acute Infection, Whereas The Majority Of Viral Lineages In QVOA In Participants Treated During Chronic Infection Are Distinct
Our assay is based on the assumption that the viral sequences derived from different proviruses are genetically distinct. However, it is possible that viral sequences in a participant display some degree of homogeneity. This would be the case if, for example, a cell with an inducible and replication-competent provirus were clonally expanded within the pool of latently infected cells. If two cells with identical inducible proviruses (at least as assessed using the V1–V3 sequence) are present in the same well, this would lead to an underestimate of IUPM values as the two cells would be recorded as producing a single viral lineage. To assess our assumption that each viral lineage would have a distinct sequence, all viral lineages detected in each patient were used to generate a neighbor-joining phylogenetic tree to determine distinct viral lineages (DVLs). Phylogenetic trees from a total of 19 participants, 2 in acute infection and 17 chronically infected, were examined for the degree of heterogeneity of viral sequence lineages. When the samples were derived from participants in acute infection, all viral lineages observed in different wells were homogeneous (Fig. 2A). This indicates that the readout of DVLs is not a viable approach in assessing the size of the latent reservoir in participants prior to the time when their reservoir has developed sequence diversity.
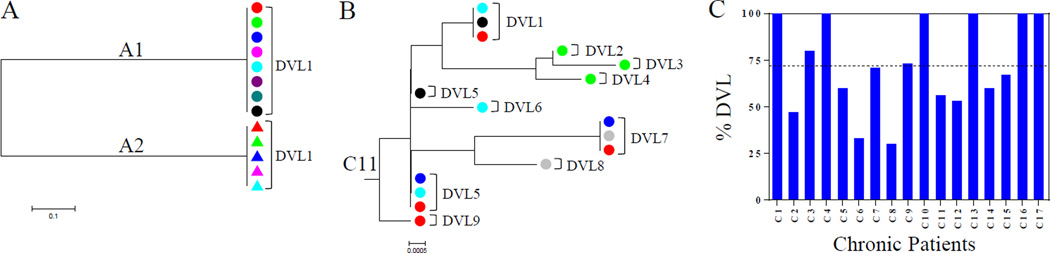
Figure 2.
Representative neighbor-joining phylogenetic trees of viral lineages to determine DVLs. A, The phylogenetic tree shows viral lineages detected in wells (n=8 for A1, n=5 for A2) derived from two participants in acute infection, A1 and A2. Individual wells are color coded. The tree was generated using MEGA 5.10. B, The phylogenetic tree shows viral lineages detected in wells (n=6) derived from a chronically infected participant, C11. Individual DVLs identified are indicated. Different wells are color coded and the tree was generated using MEGA 5.10. C, % DVL observed in each chronically infected participant. The horizontal dotted line indicates average % DVL.
In contrast, the majority of viral lineages observed from chronically infected participants were distinct. A phylogenetic tree showing viral lineage sequences from participant C11 reveals 9 DVLs among a total of 16 viral lineages detected in all wells, showing that 56% of the viral lineages detected among the wells analyzed represented distinct sequences (%DVL) (Fig. 2B and C). We observed that %DVL varied among chronically infected participants, ranging from 30% to 100%, with an average of 72% (Fig. 2C and Table 1). The lowest %DVL of 30% was seen in participant C8 who had the lowest CD4 and nadir CD4 counts among all patients (see Supplemental Digital Content 4). These results show that IUPM titers using VOA-UDSA requires a correction to account for the potential of two identical viral lineages being present in the same well.
Table 1.
Data obtained from VOA-UDSA and QVOA
| Patient ID |
No. of Cells (in million) |
QVOA Total Wells |
QVOA p24+ Wells |
VOA-UDSA Wells |
No. of VL |
No. of VL/well |
No. of DVL |
% DVL | IUPM-QVOA (95% CI) |
IUPM-VOA-UDSA (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| C1 | 2.5 | 36 | 4 | 4 | 12 | 3.00 | 12 | 100 | 0.05 (0.017–0.120) | 0.14 (0.077–0.238) |
| C2 | 2.5 | 36 | 5 | 5 | 15 | 3.00 | 7 | 47 | 0.07 (0.031–0.154) | 0.17 (0.104–0.287) |
| C3 | 2.5 | 18 | 5 | 5 | 5 | 1.00 | 4 | 80 | 0.12 (0.049–0.287) | 0.12 (0.048–0.278) |
| C4 | 2.5 | 18 | 5 | 3 | 3 | 1.00 | 3 | 100 | 0.14 (0.064–0.321) | 0.12 (0.051–0.293) |
| C5 | 2.5 | 14 | 4 | 3 | 5 | 1.67 | 3 | 60 | 0.15 (0.062–0.361) | 0.20 (0.087–0.437) |
| C6 | 2.5 | 18 | 7 | 6 | 12 | 2.00 | 4 | 33 | 0.18 (0.085–0.377) | 0.34(0.199–0.596) |
| C7 | 2.5 | 36 | 15 | 15 | 56 | 3.73 | 39 | 71 | 0.22 (0.134–0.360) | 0.64 (0.492–0.830) |
| C8 | 2.5 | 36 | 22 | 22 | 86 | 3.91 | 26 | 30 | 0.41 (0.273–0.617) | 1.06 (0.856–1.309) |
| C9 | 2.5 | 12 | 9 | 6 | 11 | 1.83 | 8 | 73 | 0.44 (0.221–0.873) | 0.60 (0.352–1.025) |
| C10 | 2.5 | 18 | 12 | 6 | 19 | 3.17 | 19 | 100 | 0.54 (0.323–0.912) | 0.73 (0.490–1.099) |
| C11 | 2.5 | 12 | 9 | 6 | 16 | 2.67 | 9 | 56 | 0.62 (0.337–1.137) | 0.89 (0.556–1.413) |
| C12 | 2.5 | 36 | 32 | 32 | 123 | 3.84 | 65 | 53 | 0.94 (0.632–1.393) | 1.42 (1.187–1.690) |
| C13 | 0.5 | 6 | 4 | 4 | 7 | 1.75 | 7 | 100 | 1.24 (0.743–2.084) | 2.55 (1.216–5.360) |
| C14 | 0.5 | 6 | 3 | 3 | 5 | 1.67 | 3 | 60 | 1.82 (0.902–3.658) | 2.12 (0.864–5.178) |
| C15 | 0.5 | 6 | 5 | 5 | 9 | 1.80 | 6 | 67 | 2.49 (1.103–5.625) | 3.66 (1.882–7.101) |
| C16 | 0.5 | 6 | 4 | 4 | 8 | 2.00 | 8 | 100 | 2.52 (1.158–5.462) | 2.92 (1.457–5.839) |
| C17 | 0.5 | 6 | 4 | 4 | 6 | 1.50 | 6 | 100 | 3.49 (1.689–7.196) | 2.19 (0.982–4.875) |
VOA-UDSA Titer Strongly Correlates With QVOA Titer
We estimated IUPM using VOA-UDSA for 17 chronically infected participants whose IUPM values had previously been determined by QVOA. The information of QVOA wells used for VOA-UDSA is listed in Table 1. For this analysis it was essential to correct for the potential of having two identical viral lineages in the same well by documenting the frequency of identical lineages between wells; this allowed us to use the Poisson Distribution to estimate then number of wells that had two identical lineages, which cannot be recorded by analyzing sequence diversity. In Figure 2B, DVL1 and DVL7 were detected in 3 wells, DVL5 was detected in 4 wells among the total of 6 wells analyzed by VOA-UDSA, and the remaining 6 DVLs were detected in a single well each. The sum of all individual DVL titers observed and inferred in each participant was adjusted for the initial number of resting CD4+ T cells seeded in the wells examined for viral outgrowth to express the IUPM estimates.
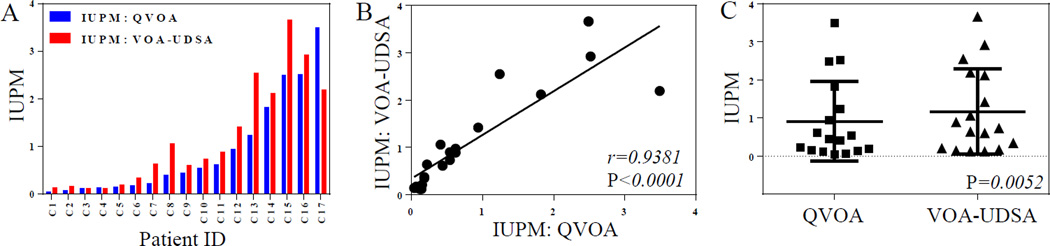
When the IUPM estimates obtained by VOA-UDSA were compared to those determined by QVOA, we observed a strong correlation between the two values (r=0.94, p<0.0001) (Fig. 3A and B). The estimated IUPM observed from VOA-UDSA on average tended to be slightly higher than that measured by QVOA, 1.17 IUPM versus 0.91 IUPM (p=0.0052), respectively (Fig. 3C).
Figure 3.
Correlation between VOA-UDSA and QVOA. A, The IUPM estimates of 17 chronically infected participants obtained independently by VOA-UDSA and QVOA are shown. B, The IUPM estimates obtained from VOA-UDSA strongly correlate with the IUPM estimates obtained from QVOA. The statistics were obtained from the Spearman rank correlation test and linear regression analysis was used to form the best-fitting straight line. C, The IUPM estimates obtained from the two assays, VOA-UDSA and QVOA were compared by the Wilcoxon matched-pairs test.
Frequency Of VOA-UDSA Introduced Recombination Events Is Low
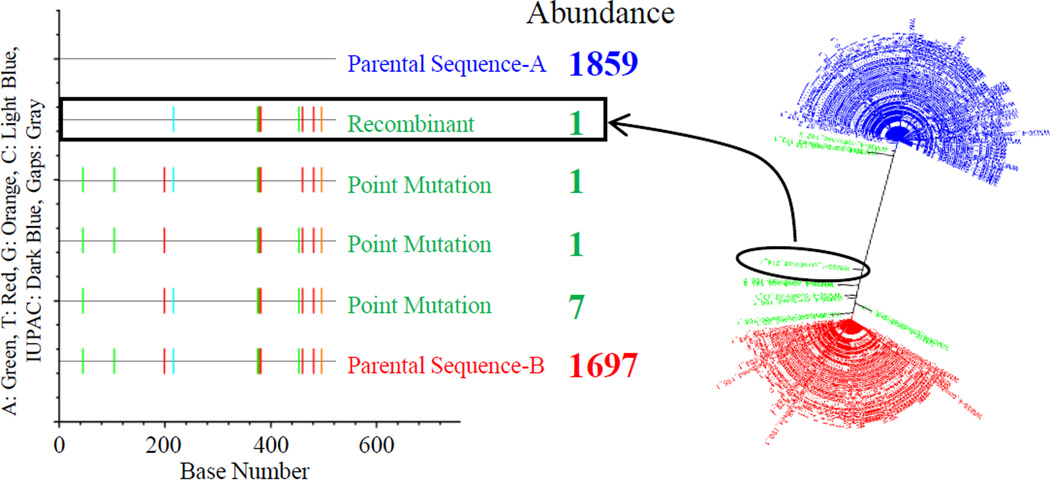
UDSA involves a cDNA reaction and PCR amplifications that can introduce errors as the result of nucleotide mis-incorporation and recombination, altering the true diversity of a viral population.36–40 To investigate whether recombination introduced during the reverse transcription or PCR steps can contribute to the apparent diversity of a viral population, thereby potentially overestimating the size of the latent reservoir, we simulated recombination by mixing two different RNA samples, each containing a single viral lineage where each lineage was distinct from the other. Four sets of duplicate cDNA reactions were carried out with the two RNAs mixed at a molar ratio of 1:1 to result in about 4,000 template consensus sequences from each cDNA reaction. Figure 4 shows the analysis of deep sequencing from one of the four cDNA reactions. Among all of the 3,566 template consensus sequences recovered, we detected only 1 template consensus sequence that was a recombinant between the two input RNAs, representing only 0.03% of all template consensus sequences (Fig. 4). On average, only 0.08% of all template consensus sequences were recombinants. This result indicates that the frequency of VOA-UDSA introduced recombination is very low and its contribution to the apparent diversity of a viral population is insignificant.
Figure 4.
Simulation of VOA-UDSA introduced recombination events. A representative highlighter plot and phylogenetic tree reveal the low frequency of method-introduced recombination events. Method-introduced recombination events was simulated by mixing two different RNA samples for cDNA reaction. Abundance, the number of consensus sequences, of parental sequence A and B is shown in blue and red, respectively. The number of consensus sequences of offspring generated by either recombination or point mutation is shown in green. The recombinant sequence shown in the highlighter plot and the phylogenetic tree is indicated with a box and a circle, respectively.
Recombination can also occur during the culture for viral outgrowth. To examine the frequency of recombination during the culture, we examined the total frequency of recombination by analyzing 68 wells containing at least 3 viral lineages by highlighter plot analysis. The result revealed that approximately 5% of all viral lineages analyzed were recombinants (data not shown). Considering the low frequency of VOA-UDSA introduced recombination, the majority of recombinants we detected were derived from the culture. These results suggest that the influence of recombination events during the culture or the assay on the size of HIV-1 latent reservoir is not substantial.
DISCUSSION
Assessing the effectiveness of strategies for HIV-1 eradication requires a robust assay applicable to clinical settings to measure changes in the size of the viral reservoir over the course of therapy. In this work we have explored an alternative readout to the current QVOA. The current forms of the QVOA rely on the limiting dilution of cells to detect individual cells that give rise to infectious virus with outgrowth detection by p24 or using real time PCR to detect viral RNA. Here we measured the number of resting CD4+ T cells harboring replication-competent HIV-1 by using a Primer ID-based Ultra Deep Sequencing Assay, VOA-UDSA, to count the number of distinct viruses that grow out in a culture. VOA-UDSA still involves viral outgrowth in directly analyzing the number of different sequences of the induced viruses in the culture supernatant to score the frequency of latently HIV-infected resting CD4+ T cells. However, it eliminates the need for the limiting dilution step and offers the possibility that a smaller number of outgrowth wells with multiple viruses could be analyzed to titer the number of virus-inducible cells. In addition, the use of viral RNA and PCR may allow assessment of viral outgrowth after a shorter period of time than needed to record p24 in the culture supernatant, at least with standard ELISA assays.
When we quantified IUPM estimates in 17 chronically infected participants by analyzing a total of 133 culture wells derived from QVOA scored as p24 positive, the IUPM estimates determined by VOA-UDSA were strongly correlated with the IUPM estimates determined by QVOA (r=0.94; p<0.0001). As such, VOA-UDSA is the first assay that strongly correlates with QVOA. With the multiplexing ability of next-generation sequencing (NGS), VOA-UDSA has the potential to be adapted to high throughput.
There are several features of using UDS in the outgrowth cultures that must be considered. First, viral population studies using the current UDS technology require preceding PCR amplification steps which can introduce PCR errors, altering diversity of a viral population.36,37 For example, PCR resampling can create biases artificially decreasing diversity while PCR misincorporation/sequencing errors can add artifactual diversity to the existing viral population.41 To overcome these limitations in deep sequencing, we have incorporated Primer ID so that we can account for each individual template that was sequenced. It has been demonstrated that the Primer ID approach reduces PCR and sequencing errors significantly and achieves more accurate viral population sampling.32 Second, because there is so little diversity in these cultures, any residual errors introduced from PCR and sequencing must be accounted for so that it is not included as distinct lineages. In examining all of our data we chose a 2.5% abundance cutoff for inclusion as a viral lineage within a well. This may limit detection of slow growing lineages in a mixture with faster growing lineages although a strong correlation seen between two IUPM estimates obtained from VOA-UDSA and QVOA suggests that the cutoff seems unlikely to cause a significant underestimate of the size of the reservoir. Third, while the factors that determine the complexity of the latent reservoir are poorly understood, identical viruses can be detected as growing out from separate cells, likely due to clonal expansion of HIV-1-infected CD4+ T cells,42 as also assessed by sequencing the V1–V3 region of the env gene in this study. To account for this phenomenon in estimating IUPM, we have corrected for unobservable identical lineages in the same well. This correction is not applicable for participants with an absence of diversity in the latent reservoir, such as those who are acutely infected or initiate therapy during acute infection. However, in North Carolina for example, only about 1% of people who were newly diagnosed with HIV-1 were diagnosed during acute HIV infection.43 Thus, at least for now, the number of people where VOA-UDSA is inappropriate may not be significant even if all people start therapy when diagnosed with HIV-1. In our unpublished study of the generation of diversity after infection, we have found significant diversity in the V1–V3 region of plasma virus by one year of infection (SZ, unpublished observation); while this observation did not include virus in the latent reservoir, it does suggest that restricted diversity of the viral population will be relevant only for the relatively small fraction of people who start therapy within the first year or two of infection.
In this analysis we have evaluated the quality of the data that can be obtained using VOA-UDSA to examine the viral lineages in the QVOA outgrowth wells and thus IUPM. There are additional improvements that can be explored to determine the utility of this approach. First, it is necessary to define a VOA-UDSA protocol in terms of the number of cells, the number of wells, and the length of time for outgrowth so that there can be standardized reporting to allow comparison to other methods. Second, since VOA-UDSA could be done with as few as several hundred copies of viral RNA for a cDNA reaction, it may be possible to detect induced viruses shortly after T cell activation, perhaps even without viral expansion. This would eliminate the necessity of using either PBMCs from healthy donors or a T-cell line expressing CD4, CCR5, and CXCR4 for outgrowth. This approach, however, may lead to the overestimation of IUPM titers since VOA-UDSA would detect all genomes able to produce virus particles including those from replication defective proviruses. Third, because identical sequences are a challenge for this assay, IUPM estimates using other regions such as gag or a longer env region, V1–V5, need to be explored to compare with IUPM estimates obtained from the V1–V3 region. This could enhance the accuracy of the IUPM estimates by reducing the frequency of detecting identical sequences between wells due to the relatively short amplicon, V1–V3.
While the accuracy of the QVOA is determined by limiting dilution, the VOA-UDSA generates a specific count of outgrowth events given the number of cells tested linking the accuracy to the total number of independent outgrowth events recorded. Future work will determine if this approach can improve the accuracy of measurements of the latent reservoir and/or reduce the number of cells needed to attain a desired level of accuracy.
Supplementary Material
Acknowledgments
We thank Brigitte Allard, Rosalie Bateson, and Katherine Sholtis for their technical assistance. We also thank the study participants for their contribution to this study.
Source of Funding: This work was supported by NIH grants R21 AI113124, the UNC Center for AIDS Research (CFAR P30 AI50410), and the Collaboratory of AIDS Researchers for Eradication (U19 AI096113).
RS is an inventor on Intellectual Property related to this research (Primer ID). This IP has been licensed to Cellular Research.
Footnotes
Partial data of this study were presented at the Towards an HIV Cure Symposium, July 18 to 19, 2015, Vancouver, Canada.
The other authors have no other conflicts of interest to disclose.
REFERENCES
- 1.Carter CC, Onafuwa-Nuga A, McNamara LA, et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med. 2010;16(4):446–451. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orenstein JM, Fox C, Wahl SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276(5320):1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 3.Wightman F, Solomon A, Khoury G, et al. Both CD31(+) and CD31(−) naive CD4(+) T cells are persistent HIV type 1-infected reservoirs in individuals receiving antiretroviral therapy. J Infect Dis. 2010;202(11):1738–1748. doi: 10.1086/656721. [DOI] [PubMed] [Google Scholar]
- 4.Chun TW, Carruth L, Finzi D, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc Natl Acad Sci U S A. 1998;95(15):8869–8873. doi: 10.1073/pnas.95.15.8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 7.Wong JK, Hezareh M, Gunthard HF, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 8.Persaud D, Pierson T, Ruff C, et al. A stable latent reservoir for HIV-1 in resting CD4(+) T lymphocytes in infected children. J Clin Invest. 2000;105(7):995–1003. doi: 10.1172/JCI9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monie D, Simmons RP, Nettles RE, et al. A novel assay allows genotyping of the latent reservoir for human immunodeficiency virus type 1 in the resting CD4+ T cells of viremic patients. J Virol. 2005;79(8):5185–5202. doi: 10.1128/JVI.79.8.5185-5202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med. 1995;1(12):1284–1290. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 11.Finzi D, Blankson J, Siliciano JD, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 12.Ho Y, Shan L, Hosmane NN, et al. Replication-Competent Noninduced Proviruses in the Latent Reservoir Increase Barrier to HIV-1 Cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laird GM, Eisele EE, Rabi SA, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brussel A, Sonigo P. Analysis of early human immunodeficiency virus type 1 DNA synthesis by use of a new sensitive assay for quantifying integrated provirus. J Virol. 2003;77(18):10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76(21):10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strain MC, Lada SM, Luong T, et al. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4):e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu JJ, Wu TL, Liszewski MK, et al. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology. 2008;379(1):78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriksson S, Graf EH, Dahl V, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun TW, Justement JS, Lempicki RA, et al. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proc Natl Acad Sci U S A. 2003;100(4):1908–1913. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasternak AO, Jurriaans S, Bakker M, Prins JM, Berkhout B, Lukashov VV. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12):e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Procopio FA, Fromentin R, Kulpa DA, et al. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine. 2015;2(8):872–881. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archin NM, Bateson R, Tripathy MK, et al. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210(5):728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, Margolis DM. Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses. 2009;25(2):207–212. doi: 10.1089/aid.2008.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barton KM, Archin NM, Keedy KS, et al. Selective HDAC inhibition for the disruption of latent HIV-1 infection. PLoS One. 2014;9(8):e102684. doi: 10.1371/journal.pone.0102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Contreras X, Schweneker M, Chen CS, et al. Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem. 2009;284(11):6782–6789. doi: 10.1074/jbc.M807898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83(10):4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archin NM, Eron JJ, Palmer S, et al. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS. 2008;22(10):1131–1135. doi: 10.1097/QAD.0b013e3282fd6df4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crooks AM, Bateson R, Cope AB, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archin NM, Vaidya NK, Kuruc JD, et al. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc Natl Acad Sci U S A. 2012;109(24):9523–9528. doi: 10.1073/pnas.1120248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis. 2015;2(4):ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou S, Jones C, Mieczkowski P, Swanstrom R. Primer ID Validates Template Sampling Depth and Greatly Reduces the Error Rate of Next-Generation Sequencing of HIV-1 Genomic RNA Populations. J Virol. 2015;89(16):8540–8555. doi: 10.1128/JVI.00522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douek DC, Brenchley JM, Betts MR, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorzer I, Guelly C, Trajanoski S, Puchhammer-Stockl E. The impact of PCR-generated recombination on diversity estimation of mixed viral populations by deep sequencing. J Virol Methods. 2010;169(1):248–252. doi: 10.1016/j.jviromet.2010.07.040. [DOI] [PubMed] [Google Scholar]
- 37.Meyerhans A, Vartanian JP, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18(7):1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodrich DW, Duesberg PH. Retroviral recombination during reverse transcription. Proc Natl Acad Sci U S A. 1990;87(6):2052–2056. doi: 10.1073/pnas.87.6.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cronn R, Cedroni M, Haselkorn T, Grover C, Wendel JF. PCR-mediated recombination in amplification products derived from polyploid cotton. Theor Appl Genet. 2002;104(2–3):482–489. doi: 10.1007/s001220100741. [DOI] [PubMed] [Google Scholar]
- 40.Di Giallonardo F, Zagordi O, Duport Y, et al. Next-generation sequencing of HIV-1 RNA genomes: determination of error rates and minimizing artificial recombination. PLoS One. 2013;8(9):e74249. doi: 10.1371/journal.pone.0074249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8(7):R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simonetti FR, Sobolewski MD, Fyne E, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilcher CD, Fiscus SA, Nguyen TQ, et al. Detection of acute infections during HIV testing in North Carolina. N Engl J Med. 2005;352(18):1873–1883. doi: 10.1056/NEJMoa042291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.