Abstract
More than 50 distinct amino acid transporter genes have been identified in the genome of Arabidopsis, indicating that transport of amino acids across membranes is a highly complex feature in plants. Based on sequence similarity, these transporters can be divided into two major superfamilies: the amino acid transporter family and the amino acid polyamine choline transporter family. Currently, mainly transporters of the amino acid transporter family have been characterized. Here, a molecular and functional characterization of amino acid polyamine choline transporters is presented, namely the cationic amino acid transporter (CAT) subfamily. CAT5 functions as a high-affinity, basic amino acid transporter at the plasma membrane. Uptake of toxic amino acid analogs implies that neutral or acidic amino acids are preferentially transported by CAT3, CAT6, and CAT8. The expression profiles suggest that CAT5 may function in reuptake of leaking amino acids at the leaf margin, while CAT8 is expressed in young and rapidly dividing tissues such as young leaves and root apical meristem. CAT2 is localized to the tonoplast in transformed Arabidopsis protoplasts and thus may encode the long-sought vacuolar amino acid transporter.
In addition to the indispensable role as protein constituents, amino acids play many other metabolic and signaling roles in plants (Coruzzi and Zhou, 2001). In plants, the primary organic compounds after assimilation of inorganic nitrogen are Gln and Glu, the most prominent amino acids in light-adapted Arabidopsis (Lam et al., 1995; Pilot et al., 2004). The other amino acids are found at lower concentrations in plants. The relative amount of each amino acid is similar in the phloem, leaf apoplasm, and cytosol, indicating that amino acid transport at plant plasma membranes does not discriminate much between individual amino acids (Lohaus et al., 1995). It was early recognized that amino acid transport systems are not highly specific for single amino acids, but transporters with some specificity for acidic or basic amino acids were identified in many species (Kinraide, 1981; Wyse and Komor, 1984; Bush, 1993).
Though some reports identified the bulk tissue amino acid content compartmented in vacuoles, it has been shown in many studies that amino acids are not strongly concentrated in vacuoles. Some studies found similar cytosolic and vacuolar amino acid concentrations (Dietz et al., 1990; Carroll et al., 1992; Farre et al., 2001), while others identified very low vacuolar amino acid concentrations, suggesting active or secondary active extrusion of amino acids from vacuoles (Winter et al., 1993, 1994). The molecular identity of vacuolar amino acid transporters is still unknown.
The genomic sequences of plant genomes provide an outline of the complexity of the amino acid transport in plants. Neglecting families of plastidic and mitochondrial transporters, the genomic sequence from Arabidopsis still contains more than 50 genes putatively involved in amino acid transport, which are classified into two major groups: the amino acid transporter family (ATF) superfamily and the amino acid polyamine choline (APC) transporters (Wipf et al., 2002). As in many other plants, the majority of putative Arabidopsis amino acid transporters belong to the ATF superfamily (approximately 46 members). Several of these members have been characterized as broad-specific transporters, transporting all neutral and cationic amino acids with high capacity when heterologously expressed in yeast (Saccharomyces cerevisiae) and Xenopus laevis oocytes (Fischer et al., 2002). The highly abundant acidic amino acids are also transported in their neutral form (Fischer et al., 2002). Some ATF transporters, however, are highly selective, e.g. for Pro (Rentsch et al., 1996). ATF-type amino acid transporters from plant species other than Arabidopsis have also been molecularly characterized (Tegeder et al., 2000; Miranda et al., 2001). Proteins of the ATF superfamily have so far not been identified in bacteria but are identified in animals and humans, where they are involved in plasma membrane, vesicular, and lysosomal amino acid transport. In yeast, seven ATF homologs are identified, and four were identified as vacuolar amino acid importers or exporters. All characterized ATF transporters, irrespective from which kingdom, are proton coupled either as proton symporters or antiporters and have a 9 to 11 transmembrane (TM) domain topology (Wipf et al., 2002).
APC transporters are functionally, but not structurally, related to ATF transporters. Members of the APC superfamily were identified in almost all organisms, including bacteria, suggesting that they represent the evolutionarily older group of amino acid transporters, but it remains unclear why two separate protein families have obviously evolved in parallel. In unicellular yeast, amino acid uptake across the plasma membrane is catalyzed by a coordinated array of 25 amino acid transporters, all belonging to the APC superfamily (Andre, 1995). While some of these transporters are selective for a single substrate, others transport many amino acids. All are thought to be proton symporters. In humans, more than 10 APC superfamily members have been identified and characterized (Verrey et al., 2004). Most of these display the typical topology with 12 putative TM spans and need heterodimer formation with an additional subunit for plasma membrane localization. They function as proton-independent, obligatory amino acid exchangers (Verrey et al., 2004). The human genome also contains sequencewise-related transporters with 14 TM spans that function as uniporters for basic amino acids. Within the complete Arabidopsis genome, 14 APC-type genes are identified. Nine APC proteins (cationic amino acid transporter [CAT]1–CAT9; see below) have a 14 TM topology, while the remaining 5, more distantly related members, have a 12 TM topology.
In plants, our molecular knowledge on amino acid transporters is almost exclusively restricted to transporters from the ATF superfamily, despite the fact that amino acid transport in yeast and humans is dominated by APC-type amino acid transporters. A single member of plant APC-type proteins has been functionally characterized and was initially named amino acid transporter 1 (AAT1; Frommer et al., 1995). Since the abbreviation AAT1 is also used for the Asp amino transferase, the transporter was renamed to CAT1 (Wipf et al., 2002), emphasizing its structural and functional similarity with mammalian CATs. The related genes and proteins of this family with a 14 TM domain topology will be characterized here and named CAT1 to CAT9.
RESULTS
Cloning and in Silico Analysis of APC-Type Amino Acid Transporters
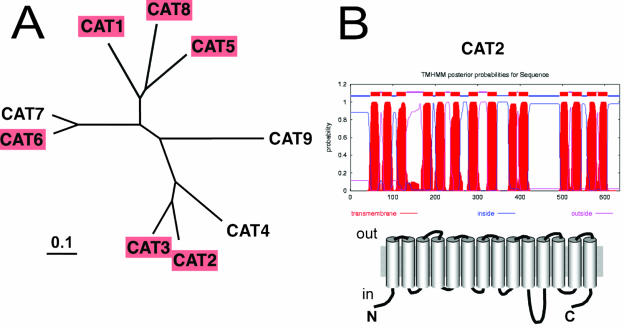
The locus code and nomenclatures for all CAT family members in Arabidopsis are given in “Materials and Methods.” The first member of this family, CAT1, has been characterized at the functional level (Frommer et al., 1995). CAT5 is the closest homolog of CAT1 and is 64% identical to CAT1 on the protein level (Fig. 1). CAT8 shows slightly lower identity to CAT1 (55%). CAT1 and CAT2 are only 34% identical at the protein level. Using TM prediction programs (THMM; http://www.CBS.dk) the proteins of this family are likely to have 14 TM domains with both termini on the cytosolic side (Fig. 1). The sequences can be phylogenetically grouped into small subgroups (Fig. 1). Genes from different subbranches differ markedly in their exon-intron structure (e.g. CAT5 and CAT8 contain no intron, while CAT3 contains 14 introns). The intron sizes and location of splice sites, however, are roughly conserved in genes in each subbranch.
Figure 1.
Computer-aided analysis of the similarities between CATs. A, Maximum parsimony analyses were performed using Phylogenetic Analysis Using Parsimony, version 4.0b 10 (Sinaur Associates, Sunderland, MA) with all DNA characters unweighted and gaps scored as missing characters. Heuristic tree searches were executed using 100 random sequence additions and the tree bisection-reconnection branch-swapping algorithm with random sequence analysis. Percentage bootstrap values of 1,000 replicates are given at each branch point. Branch lengths (drawn in the horizontal dimension only) are proportional to phylogenetic distance. The CAT proteins functionally investigated in this study are labeled in pink. B, Prediction of the TM domains of CAT2 using TMNMM (www.cbs.dtu.dk/services/TMHMM) and a cartoon showing the deduced two-dimensional structure.
Several CAT proteins contain putative transit peptides for targeting to plastids. Both CAT6 and CAT7 are predicted to contain chloroplast transit peptides; CAT3 contains putative mitochondrial targeting signals, while CAT9 is predicted to be targeted to chloroplast or mitochondria (see Aramemnon database at http://aramemnon.botanik.uni-koeln.de).
Expression Analysis of CAT Transporters
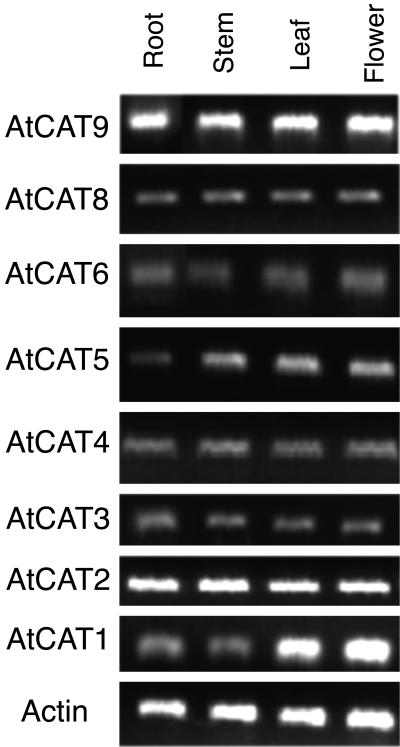
Expression of each CAT member is documented by numerous expressed sequence tags (ESTs) in public databases. The only exception is CAT7, for which no EST or cDNA could be identified. Public microarray data (e.g. available at John Ward's Arabidopsis membrane protein library at http://www.cbs.umn.edu/arabidopsis/) document expression in many tissues for most of the CATs. According to the EST abundance, CAT2 is the highest expressed at the transcriptional level. The tissue specificity of CAT2 was determined by RNA gel blots. CAT2 mRNA was similarly expressed in root, stem, flower, sink leaf, source leaf, and silique (data not shown). The same expression profile was obtained using reverse transcription (RT)-PCR (Fig. 2). Using ACTIN2 as control, expression of all CAT members was identified by RT-PCR in roots, stems, flowers, and leaves (Fig. 2), except for CAT7. Thus, most CAT members are broadly expressed, and more detailed analysis is necessary to assign a cellular function to these genes.
Figure 2.
Expression profile of CAT members detected by RT-PCR. Transcripts of each CAT could be amplified from all tissues tested, except for CAT7. The CATs are broadly expressed. Experiments were repeated for each member at least three times with similar results.
Functional Characterization of CAT Transporters in Yeast
Yeast has provided a genuine heterologous expression system for many plant nutrient and metabolite transporters. The yeast mutant 22Δ8AA, which lacks eight endogenous amino acid transport systems, was transformed with plasmids containing cDNAs of several CAT-family transporters. Yeast transformed with empty plasmid (pDR) was used as control. The yeast mutant 22Δ8AA is unable to efficiently utilize Arg, Asp, citrulline, γ-aminobutyric acid, Glu, or Pro as sole nitrogen sources.
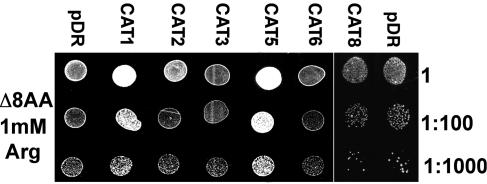
As expected from competition studies of His uptake (Frommer et al., 1995), expression of CAT1 in 22Δ8AA enhanced Arg, citrulline, and Glu import and supported growth on these substances as sole nitrogen sources. CAT5 behaved very similarly and complemented growth on Arg (Fig. 3), citrulline, and, to a lesser extent, Glu. Cells transformed with CAT2, CAT3, CAT6, or CAT8 showed no clear enhanced growth on media containing 1 mm Arg as sole nitrogen source (Fig. 3). Similar results were obtained on 1 mm citrulline and Glu (data not shown). Within 7 d, none of the CAT members mediated sufficient amino acid transport to enable growth on 3 mm Asp, Pro, or γ-aminobutyric acid. Growth of 22Δ6AAL (Fischer et al., 2002), a Lys auxotroph, was restored by expressing CAT1 or CAT5, but not by CAT2, CAT3, or CAT6 (data not shown).
Figure 3.
Complementation of yeast mutant strain 22Δ8AA by CATs. Comparison of the growth of the mutant yeast transformed with empty plasmid (pDR), or the plasmid containing the ORF of CAT1, CAT2, CAT3, CAT5, CAT6, or CAT8. Growth was assayed in three dilution steps on 1 mm Arg as sole nitrogen source.
The inability of several CATs to support growth on Arg, Asp, Pro, γ-aminobutyric acid, or Lys might be due to poor targeting to the yeast plasma membrane, lost functionality, or a different substrate spectrum than CAT1 and CAT5. As shown below, it is likely that an altered substrate spectrum is responsible for the poor support of yeast growth.
CAT5 Is a Basic Amino Acid Transporter
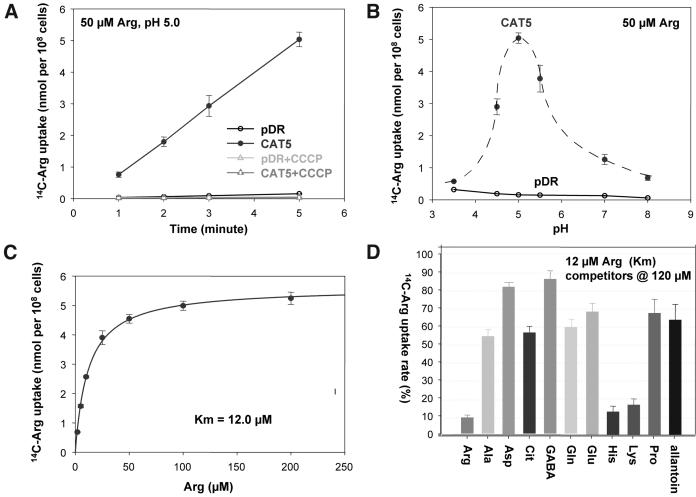
Direct l-[14C]Arg uptake was determined in 22Δ8AA carrying pDR-CAT5 and the same strain transformed with the empty plasmid. Uptake was linear over 20 min and was inhibited by the protonophore carbonyl cyanide 3-chlorophenylhydrazone (100 μm; Fig. 4A). Arg influx was pH dependent (Fig. 4B) with a bell-shaped optimum at pH 5, similar to other proton-coupled amino acid systems (Frommer et al., 1995; Wipf et al., 2002). Uptake was saturated at low Arg concentrations with a K0.5 = 12 μm (Fig. 4C). Glu was also taken up, but with a much lower affinity (K0.5 approximately 200 μm; data not shown). Arg transport was significantly inhibited only by basic amino acids, when supplied at 10-fold excess (Fig. 4D). Interestingly, some uncharged amino acids and even allantoin were also weak competitors. This may not necessarily indicate direct competition for the same binding site, but may be partially explained by unspecific lowering of the driving force for Arg. Allantoin and some amino acids are imported into the used yeast strain by H+-cotransport systems. The imported protons depolarize the yeast cells and reduce the driving force for the positively charged Arg and a possibly cotransported H+. In summary, the selectivity of CAT5 is thus similar to that of CAT1, both encoding high-affinity CATs.
Figure 4.
Functional analysis of CAT5 in yeast. A, Linear uptake by 50 μm Arg at pH 5 is blocked by carbonyl cyanide 3-chlorophenylhydrazone (100 μm). B, The pH optimum was determined at pH 5.0. C, Saturable Michaelis-Menten kinetics of Arg uptake. D, Competition of Arg uptake (12 μm) by 10-fold excess of Ala, Asp, citrulline, γ-amino butyric acid, Gln, Glu, His, Lys, or Pro. Allantoin, a nonamino acid heterocycle nitrogen compound, which is not expected as a substrate for amino acid transporters, was tested for noncompetition control. Arg uptake without competitor (control) was taken as 100%.
Expression of CAT1, CAT2, CAT3, and CAT5 Transporters in Xenopus Oocytes
As the activities of CAT2 and CAT3 were not successfully detected in yeast and to study the transport mechanism of CAT1 and CAT5 in more detail, several CAT transporters were expressed in Xenopus oocytes. Two unrelated amino acid transporters (AAP5, AAP6) that had been functionally expressed earlier in oocytes (Fischer et al., 2002) served as controls and were measured in parallel. While the AAPs generated large amino acid-dependent currents, no currents were observed with CATs (data not shown). This was somewhat surprising, as the experiments in the yeast system identified CAT1 and CAT5 as transporters for positively charged amino acids, suggesting that these substrates should generate substrate-induced currents. Direct l-[14C]Lys uptake measurements with oocytes expressing AAP5, CAT1, or CAT5 confirmed that only AAP5, but not CAT1 or CAT5, was functional at the plasma membrane of oocytes (data not shown).
Sensitivity to Toxic Amino Acids Is Enhanced by Expression of Amino Acid Transporters
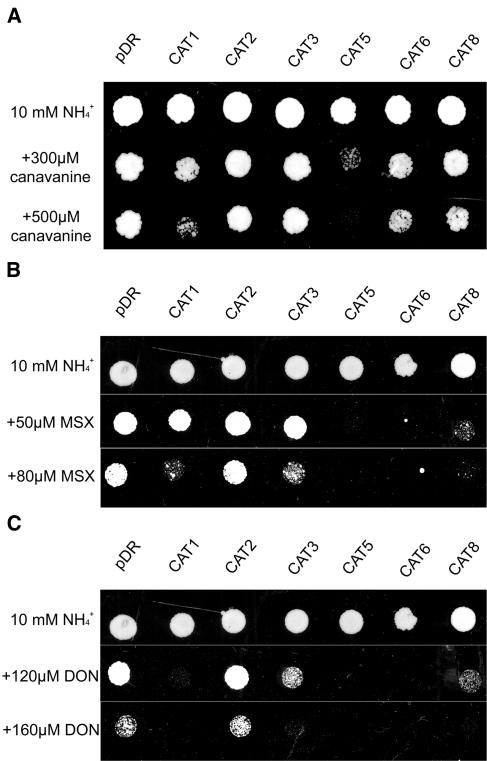
The yeast strain 22Δ8AA does not transport Arg, Asp, citrulline, γ-aminobutyric acid, Glu, or Pro, while other amino acids are transported efficiently. To test if other amino acids may be preferred or if CATs may be functional in detoxification of unwanted toxic amino acid analogs, a growth test on toxic amino acids was established. Yeast growth is sensitive to low concentrations of the Glu analog l-Met sulfoximine (MSX), an inhibitor of Gln synthetase. Yeast strains with increased amino acid import activity are more sensitive to MSX (Marek and Dickson, 1987). Similarly, the sensitivity to toxic (S)-2-amino-6-diazo-5-oxo-l-norleucine (DON), an analog of the neutral amino acid Gln, is expected to increase with higher uptake activity. DON inhibits Gln-requiring enzymes such as glucosamine-6-phosphate synthase (Milewski, 1993). The toxicity of the Arg analog canavanine has been used to identify basic amino acid transporters in yeast (Hoffmann, 1985). Sensitivity to toxic amino acids was found to be higher in the wild-type strain 23344c. Endogenous amino acid uptake is known to be strongly suppressed by high ammonium in wild-type yeast, largely due to suppression and lower uptake by general amino acid permease GAP1 (Springael and Andre, 1998). Due to the higher sensitivity, the wild-type strain was used in the following experiments, but highly similar results were also obtained from similar experiments conducted with the 22Δ8AA strain (data not shown).
The expression of functional amino acid transporters CAT1 and CAT5 increased the sensitivity of the mutant 22Δ8AA strain to canavanine, the toxic Arg analog, when supplemented to the medium containing 10 mm ammonium (Fig. 5). Thus, both transporters, which mediated efficient transport of Arg (Fig. 3), also transported its toxic analog, making yeast cells be more sensitive to this toxic drug. Expression of the other family members did not lead to increased sensitivity to canavanine, in accordance with the fact that they did not transport Arg in yeast growth assays.
Figure 5.
Growth inhibition of yeast cells by toxic amino acid analogs. A, Canavanine (analog of Arg). B, MSX, analog of Glu. C, DON, analog of Gln. Expression of CAT1, CAT3, CAT5, CAT6, or CAT8 increased sensitivity to toxic amino acids compared to the control (empty vector = pDR).
CAT1 and CAT5 both increased the sensitivity to MSX and DON (Fig. 5), which fits their ability to transport Glu and the fact that Gln weakly competes for Arg in uptake experiments (Fig. 4D). CAT3, CAT6, and CAT8 also conferred higher sensitivity to toxic MSX and DON (Fig. 5), indicating that they mediate at least some uptake of these toxic amino acids. It should be noted that some transporters did confer sensitivity to the Glu analog MSX, although growth on Glu was not observed. However, the ability to grow on a single amino acid as sole nitrogen source requires high-capacity uptake of that specific amino acid, since all other amino acids and nucleotides need to be synthesized from that amino acid. In contrast, only low import of a toxic amino acid analog may be sufficient to confer toxicity. The inability of CAT3, CAT6, and CAT8 to mediate efficient uptake of basic amino acids, but their transport of neutral and acidic amino acid analogs, may suggest that the substrate spectrum of these transporters is different from that of CAT1 and CAT5. In all experimental conditions CAT2-transformed yeast were indistinguishable from vector-transformed controls.
Promoter Analysis and Expression Pattern of CAT5
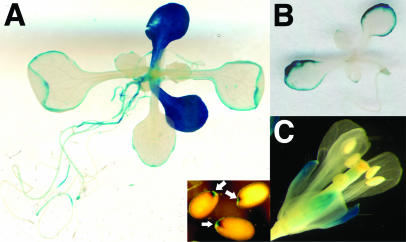
The expression pattern of the high-affinity/basic amino acid transporter CAT5 was analyzed by promoter activity analysis. As the neighboring gene is located only 0.5 kb upstream of the CAT5 open reading frame (ORF), Arabidopsis plants containing this fragment as promoter::β-glucuronidase (GUS) fusion were generated. When the transgenic Arabidopsis lines were examined with short histochemical staining periods (3–5 h), CAT5 promoter activity was visible only at the leaf rim (Fig. 6B), but intensive staining of cotyledons of developing seedlings was detected after long (24 h) incubation in staining solution (Fig. 6A). Promoter activity was identical in more than 10 lines and was not detected by short staining periods in older plants, with the exception of some staining in flowers (Fig. 6C), which explains the low abundance of transcript in roots using RT-PCR. Staining was also observed in seeds (Fig. 6A, inset). The expression pattern was compared to predictions of cis-acting regulatory elements using the PLACE database (Higo et al., 1999). A TGACGT motif that strongly drives α-amylase expression during seedling development (Yamauchi, 2001) is identified −177 before the start ATG in the CAT5 promoter, in accordance with expression during early seedling development.
Figure 6.
Expression of the GUS gene under the control of CAT5 promoter in Arabidopsis. A, Promoter activity in a 3-week-old seedling. Staining was visible at the leaf edges after brief staining periods. After long incubation in staining solution (>24 h) cotyledons were dark blue and a faint staining was visible in roots. The inset shows staining in seeds. B, Leaf edges were stained in a 10-d-old seedling after brief staining (3 h). C, Staining of sepals in flowers.
Expression Pattern of CAT8
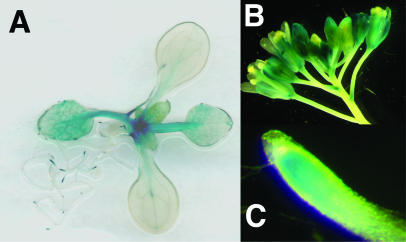
The expression pattern of the closest relative of CAT1 and CAT5 was also analyzed. GUS histochemical analysis was performed on transgenic Arabidopsis plants stably expressing a CAT8 promoter::GUS fusion, in which the 1.8-kb genomic sequence located upstream from the ATG, was driving GUS expression. Analysis of the CAT8 promoter::GUS plants identified expression in rapidly dividing cells of the shoot and root apical meristem and in young leaves and petioles during seedling development (Fig. 7A). At later developmental stages, staining was visible in flowers as well (Fig. 7B). In root tips, expression was absent from the columella and epidermis (Fig. 7C). Consistent with the expression pattern, an ACTTTA motif for apical meristem expression is identified at position −187 (Baumann et al., 1999).
Figure 7.
Expression of the GUS gene under the control of the CAT8 promoter in Arabidopsis. A, Expression pattern in a 3-week-old plant in flowers (B) and in root tips (C).
CAT5 and CAT8 Are Localized to the Plasma Membrane and CAT2 to the Tonoplast
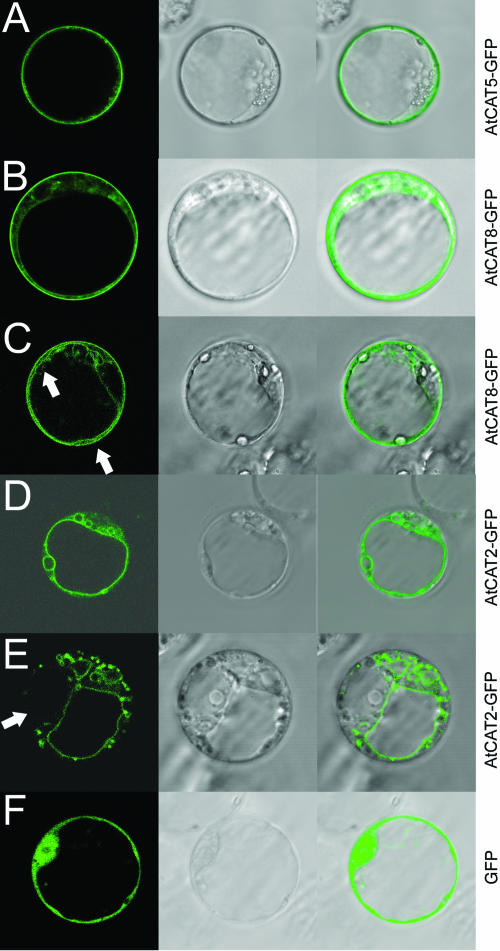
Fluorescence of CAT5-green fluorescent protein (GFP) in transiently transformed Arabidopsis protoplasts was restricted to the plasma membrane (Fig. 8A). CAT8-GFP also localized to the plasma membrane (Fig. 8B), but more than one-half of the protoplasts showed additional GFP fluorescence on intracellular membranes (Fig. 8C).
Figure 8.
Subcellular localization of CAT5-GFP, CAT8-GFP, and CAT2-GFP. A, Expression of CAT5-GFP at the plasma membrane in protoplasts from Arabidopsis suspension culture. B and C, Protoplasts expressing CAT8-GFP fusion showed staining of the plasma membrane and often staining of intracellular membranes (indicated by arrows). D and E, CAT2-GFP localized to the large central tonoplast membrane, while in some of the CAT2-GFP-expressing protoplasts the staining was observed in dot-like structures and in a subset of vacuoles. Staining was excluded from some vacuoles (arrow in lower section). F, Protoplasts expressing cytosolic and nuclear GFP5.
A possible explanation for the lack of function of CAT2 in heterologous systems is that the transporter might not be localized at the plasma membrane, but at intracellular membranes. To evaluate such a possibility, CAT2 was also fused with GFP at the carboxy terminus and transiently expressed in Arabidopsis protoplasts. Confocal images from fluorescent proteins showed that CAT2-GFP localized to the tonoplast, implying a function in amino acid storage or export from the vacuole (Fig. 8D). In a few CAT2-GFP-expressing cells a punctuate fluorescence was additionally observed, and only some vacuoles were fluorescent (Fig. 8E). As control, GFP was also expressed alone (Fig. 8F).
DISCUSSION
Broad Expression of CATs
RNA gel-blot and RT-PCR analyses suggest a broad expression pattern for most CAT genes. This agrees with public transcriptome analysis, e.g. from nitrate feeding experiments (Wang et al., 2003), where CAT members are detected in roots and shoots. The only exceptions are CAT6, the transcript of which is detected only in roots, and CAT7, which is lowly abundant in both tissues (Wang et al., 2003). CAT7 was also not detected by RT-PCR in our growth condition, and no ESTs are identified for this member, suggesting that CAT7 is lowly expressed under standard growth conditions. However, similar to the compatible solute transporter ProtT1, 80 mm sodium chloride treatment transiently, but specifically, enhances transcript levels of CAT7 (Maathuis et al., 2003), which may suggest a role for CAT7 as a compatible solute transporter. None of the transcripts of these amino acid transporters is rapidly regulated by short-time nitrate supply (Wang et al., 2003), although amino acid transporters are involved in transport of downstream products of nitrate assimilation. Promoter-reporter gene fusions revealed that the expression of both CAT5 and CAT8 is developmentally regulated. However, the distinct expression patterns and tissue specificities between the two closely related transporters may indicate that each fulfills a specific and nonredundant function. Public microarray data (available at http://www.cbs.umn.edu/arabidopsis/) suggest that CAT5 is expressed in seeds, where it may be involved in import of basic amino acids, especially the limiting essential amino acid Lys (Galili, 2002). The histochemical staining of the CAT5::GUS seeds may suggest a function in loading of basic amino acids from seed stalks (funiculi) into seeds. Each member of the many amino acid transporters thus seems to have a distinct tissue distribution and developmentally regulated expression; similar results have been reported for other plant amino acid permeases (Rentsch et al., 1996; Hirner et al., 1998; Okumoto et al., 2002).
Toward the Physiological Function
CAT1 and CAT5 function as specific, high-affinity basic amino acid transporters, while the structurally unrelated AAP3 and AAP5 transport a broad range of amino acids including basic and neutral amino acids. While AAP3 and AAP5 transport basic amino acids when expressed in hererologous expression systems, their natural substrates in planta are possibly neutral amino acids, which are more abundant in the apoplast (Fischer et al., 2002). Lys transport in AAP5, for example, can be efficiently blocked by other amino acids, including the neutral amino acid Gln (Fischer et al., 2002). It has been frequently observed that the pH dependence of APC-type amino acid transporters expressed in yeast has a bell-shaped form (Fig. 4B; Frommer et al., 1995; Wipf et al., 2002), while the pH dependence of structurally distinct ATF members was monophasic (Fischer et al., 1995). The pH dependence may be a general difference between amino acid transporters from these two different families.
Three kinetically distinct uptake systems for Lys have been identified in Arabidopsis roots: a high affinity, (roughly approximate to 2 μm), lower affinity (roughly approximate to 160 μm), and a very low or linear system (Heremans et al., 1997). The rlt11 and raec1 Arabidopsis mutants lack the lower-affinity, high-capacity amino acid transport systems, and the mutations were mapped to a region on chromosome 1 (Heremans et al., 1997). Neither an APC-type amino acid transporter, nor an ATF transporter maps to that particular region, which may imply the activity (or the targeting to the plasma membrane) of certain amino acid transporters may be controlled by regulatory components located in that chromosomal region.
The selectivity of CAT1 in growth assays is in agreement with previous studies showing that the basic amino acids are recognized and transported by the transporter. The functional properties of the CAT5 protein are similar to that of CAT1, which has been reported to be expressed in seeds and vascular tissue (Frommer et al., 1995). The apparent K0.5 of CAT5 fits well to the low apoplasmic concentrations of basic amino acids, suggesting that it also transports basic amino acids in planta.
Human APC-type amino acid transporters require a second subunit (4F2hc or rBAT) for functional expression at the plasma membrane (Verrey et al., 2004), but no homolog of such secondary subunit is identified in plants. It may be envisioned that plant APC-type amino acid transporters also need to associate with such an unidentified subunit. This could explain why the amino acid transporters failed to express amino acid uptake activity in Xenopus oocytes. It is also possible that the transport mechanism in some CAT amino acid transporters is different from proton cotransport, and some CATs may mediate electroneutral amino acid exchange, a mechanism of several related mammalian amino acid transporters (Verrey et al., 2004). Such a transport mechanism may be difficult to identify by yeast growth tests.
CAT5 and CAT8 expression is developmentally regulated and both appear to be involved in early seedling development. The transport of toxic Gln and Glu analogs by CAT8 suggests that this transporter may be involved in allocation of the highly abundant amino acids Gln and Glu to young and rapidly dividing tissues. It thus may supply the root and shoot meristem with precursors for other amino acids.
Amino Acid Transport across the Tonoplast
CAT5 and CAT8 GFP fusions were detected at the plasma membrane in transiently transformed plant protoplasts, in accordance with a transport function at the plasma membrane. In many protoplasts, CAT8-GFP also stained internal, and frequently vacuolar, membranes. It remains to be shown in future work if this behavior is artificially due to overexpression or if this variable subcellular localization is in fact part of a complex regulation of transporter function.
CAT2-GFP was consistently identified at the tonoplast of transiently transformed protoplasts. In some protoplasts CAT2-GFP was visible only in a subset of vacuoles, suggesting that tonoplast amino acid transport may be restricted to specific vacuole types. Tonoplast aquaporins also selectively label different vacuole types (Paris et al., 1996).
Amino acid transport at the tonoplast, probably by facilitated diffusion, has been identified in several species including carrot (Daucus carota) and barley (Hordeum vulgare; Dietz et al., 1990; Carroll et al., 1992), but it is controversial if plant vacuoles are storage compartments for amino acids. A combination of nonaqueous fractionation and subcellular volume determination suggested that, in barley and spinach (Spinacia oleracea) leaves, amino acids are exported from vacuoles, as vacuolar concentrations are 25 to 160 times lower than cytosolic concentrations (Winter et al., 1993, 1994). Similar experiments identified that vacuoles and cytosol contain similar concentrations of amino acids in potato (Solanum tuberosum) tubers (Farre et al., 2001). Though the selectivity and transport mechanism of CAT2 has not been identified, its subcellular localization suggests that it is a vacuolar amino acid transporter in Arabidopsis.
MATERIALS AND METHODS
Plant Material, Growth, Transformation, and Analysis
Arabidopsis L. ecotype Columbia (Col-0) was grown either in axenic culture on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 2% Suc or cultured in soil in the greenhouse. Arabidopsis plants were transformed using Agrobacterium tumefaciens strain GV3101 (pMP90) with the floral dipping method (Clough and Bent, 1998). Seeds were collected and germinated on Murashige and Skoog medium containing 50 μg/mL of kanamycin. Transformants were identified for the resistance to kanamycin and selected for further analyses.
DNA Cloning
ORFs for CAT1 (At4g21120), CAT3 (At5g36940), CAT5 (At2g34960), and CAT6 (At5g04770) were amplified by PCR from a cDNA library from Arabidopsis ecoptype Landsberg (Ler; Minet et al., 1992). CAT8 (At1g17120) was amplified from cDNA generated from seedling RNA from ecotype Col-0. CAT2 (At1g58030) was obtained as full cDNA clone containing 5′ and 3′ untranslated regions. Accession numbers of the other CATs are CAT4 (At3g03720), CAT7 (At3g10600), and CAT9 (At1g05940). The PCR primers contained appropriate sites for restriction enzymes to facilitate the subsequent cloning into expression vectors. The fidelity of PCR amplifications was verified by sequencing. cDNAs were inserted into the Xenopus laevis oocyte expression plasmid pOO2 (Ludewig et al., 2002) and the yeast expression plasmid pDR196 (Rentsch et al., 1995). Compared with the Col-0 sequence, the Ler CAT1 ORF has two silent basepair changes. CAT3 has 5-bp differences between the two ecotypes in the ORF, which lead to two amino acid exchanges in Ler (F13V and V563A). CAT5 was identical in both ecotypes. The Ler ORF of CAT6 has five silent differences compared to Col-0. Differences between ecotypes were verified by sequencing several independent PCR fragments from Ler genomic DNA. For C-terminal GFP fusion constructs the stop codons were removed by PCR and the respective ORFs were cloned in frame to the GFP gene driven by the cauliflower mosaic virus 35S promoter. The promoters of CAT5 and CAT8 were amplified by PCR using Arabidopsis L. ecotype Col-0 genomic DNA as template. The CAT5 promoter was chosen as the region upstream from the ATG up to the neighboring gene and was only 455-bp long. A 1.8-kb fragment preceding the ATG was chosen as the CAT8 promoter. The amplified fragments were fused with the uidA gene in the binary expression vector pPTkan (a gift from Dr. Schumacher, Zentrum für Molekularbiologie der Pflanzen) and the constructs were used for transformation of A. tumefaciens.
RT-PCR
Gene-specific primers were designed for amplification of roughly approximate to 200- to 500-bp fragments, spanning an intron if available. Total RNA was extracted according to Downing et al. (1992) from 5-week-old greenhouse plants. Five micrograms of total RNA was converted to cDNA using the RevertAid H-M-MuLV Reverse Transcriptase (Fermentas, St. Leon-Rot, Germany). PCR was performed at an annealing temperature of 55°C and 25 cycles were used for CAT2, 28 cycles for CAT4 and CAT5, and 30 cycles for the other constructs. A 300-bp cDNA fragment of the constitutively expressed ACTIN2 gene was amplified simultaneously (25 cycles) and used as a control. RT-PCR was repeated on plants grown under different conditions (agar plates, hydroponic culture, soil) supplied with variable nitrogen sources and yielded highly similar results. Primers used were (5′ to 3′):
Forward
ACT2 GTGGGAATGGAAGCTGCTGG
CAT1 CATCGCTATTAATATCTTCCTTC
CAT2 GGTTGCAAGTACCGCAGAGG
CAT3 GTGGATTGTTCCTTCTTGTCG
CAT4 CTGTGTTCTTAGCTCGACAAAC
CAT5 GTGAAGAGAGAGGATACTGG
CAT6 CATCGGACGGTCCAGAGTGGTC
CAT7 CGACCCGATAACCCGGAAAAC
CAT8 GCATTGCTTCCGAAATATCGTG
CAT9 GTCTCAATCTTAATCAGTATTG
Reverse
ACT2 GACCTGCCTCATCATACTCGG
CAT1 CAGAACAAGAATTATCAGCAAC
CAT2 GCTCATCTGGAGGAACATATCTT
CAT3 GCGACGAATCTCCTGGAGATG
CAT4 CGCTGCCCATTGCATCCCAC
CAT5 AGTCCAGGCTCTAGCAACAGC
CAT6 GGACGTAAGAGGGTGCGTCCAAC
CAT7 CGTTTAGGTCGGTGAAGAGGG
CAT8 GGCTGATGAGCTACATCATAGG
CAT9 CAGCTAAACCCAGAACCTAGAG
Histochemical Localization of GUS Activity
Histochemical assays for GUS activity were performed as described (Okumoto et al., 2002). GUS staining solution contained 100 mm sodium phosphate (pH 7), 10 mm EDTA, 3 mm K4(Fe[CN]6), 0.5 mm K3(Fe[CN]6), 0.1% (v/v) Triton X-100, 2 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcUA for 3 to 24 h at 37°C. Slight vacuum was applied to facilitate substrate infiltration. Tissues were cleared in 70% ethanol.
Yeast Growth, Transformation, and Selection
The yeast strain 22Δ8AA (Fischer et al., 2002) was transformed with pDR vector containing cDNAs of CAT1, CAT2, CAT3, CAT5, CAT6, CAT8, or empty vector pDR196 by heat shock (Dohmen et al., 1991). Transformants were selected on yeast nitrogen base media without uracil, supplemented with 0.5 g/L ammonium sulfate. Growth assays were performed in uracil-free media containing Asp (2 mm), Glu (1.5 mm), Pro (3 mm), Arg (1 mm), or citrulline (1 mm) as sole nitrogen source. Similar experiments were performed with Lys auxotroph strain 22Δ6AAL, but with media containing 100 μm Lys and 10 mm ammonium (Fischer et al., 2002). All experiments were repeated multiple times with independent colonies from different batches of transformants. For growth on toxic amino acid analogs, the EUROFAN (http://mips.gsf.de/proj/eurofan/) wild-type yeast strain 23344c (MATα, ura3) was used. Cells expressing different CATs were adjusted to the same density and spotted at several dilutions on yeast nitrogen base plates containing 10 mm ammonium sulfate and MSX or DON at the indicated concentrations. Toxic analogs and amino acids were obtained from Sigma (St. Louis).
Transient Expression in Protoplasts
The fusion proteins CAT-GFP were observed by confocal microscopy (Leica DMRE microscope equipped with a confocal head TCS SP; Leica, Wetzlar, Germany) 1 or 2 d following the transformation of protoplasts from an Arabidopsis suspension cell culture (Forreiter et al., 1997).
Transport Measurements in Yeast
Yeast cells were grown to logarithmic phase harvested at OD600 of 0.5, washed, and resuspended in ice-cold buffer (50 mm KPi, pH 5.0, 0.6 m sorbitol) to a final OD of 5. One hundred microliters of cells were preincubated with 10 μL of 1 m Glc (30°C, 5 min). Uptake was initiated by the addition of 110 μL of radioactive substrate mixture, containing l-[14C]Arg (11.2 GBq/mmol; Amersham Biosciences, Freiburg, Germany), and unlabeled Arg of the desired concentration was added. Linear uptake rates were determined over 5 min. Samples were removed after 1, 2, 3, and 5 min, transferred to 4 mL of ice-cold buffer, filtered on glass fiber filters, and washed twice with 4 mL of buffer. Radioactivity was determined by liquid scintillation spectrometry. Endogenous uptake activity of yeast transformed with empty vector pDR196 was subtracted as background activity. Transport measurements were repeated independently and represent the mean of two experiments. Competition experiments were performed with 12 μm of 14C-labeled Arg and a 10-fold excess of respective amino acids.
Expression in Oocytes
The protocol has been published earlier (Ludewig et al., 2002). Briefly, cRNA was synthesized and injected (50 nL) into individual oocytes. After 3 to 5 d of incubation in ND96, currents were recorded in solution containing 100 mm choline-Cl, 1 mm CaCl2, 1 mm MgCl2, and 5 mm MES, buffered to pH 5.0 with Tris, supplemented with variable concentrations of different amino acids. Functional amino acid transporters AAP5 and AAP6 were used as positive controls (Fischer et al., 2002).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers At4g21120, At5g36940, At2g34960, At5g04770, At1g17120, At1g58030, At3g03720, At3g10600, and At1g05940.
Acknowledgments
We thank C. Brancato for transformation of protoplasts, G. Gomella and M. Benjdia for discussion, K. Schumacher for plant transformation vectors, and G. Pilot for helpful comments on the manuscript.
This work was supported by Bundesministerium für Bildung und Forschung (in the framework of Genomanalyse im biologischen System Pflanze-GABI).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.045278.
References
- Andre B (1995) An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11: 1575–1611 [DOI] [PubMed] [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the Dof protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11: 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush DR (1993) Proton-coupled sugar and amino acid transporters in plants. Annu Rev Plant Physiol Plant Mol Biol 29: 513–542 [Google Scholar]
- Carroll AD, Stewart GR, Phillips R (1992) Dynamics of nitrogenous assimilate partitioning between cytoplasmic and vacuolar fractions in carrot cell suspension cultures. Plant Physiol 100: 1808–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects.’ Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Dietz K-J, Jäger R, Kaiser G, Martinoia E (1990) Amino acid transport across the tonoplast of vacuoles isolated from barley mesophyll protoplasts. Plant Physiol 92: 123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Strasser AW, Honer CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7: 691–692 [DOI] [PubMed] [Google Scholar]
- Downing WL, Mauxion F, Fauvarque MO, Reviron MP, de Vienne D, Vartanian N, Giraudat J (1992) A Brassica napus transcript encoding a protein related to the Kunitz protease inhibitor family accumulates upon water stress in leaves, not in seeds. Plant J 2: 685–693 [PubMed] [Google Scholar]
- Farre EM, Tiessen A, Roessner U, Geigenberger P, Trethewey RN, Willmitzer L (2001) Analysis of the compartmentation of glycolytic intermediates, nucleotides, sugars, organic acids, amino acids, and sugar alcohols in potato tubers using a nonaqueous fractionation method. Plant Physiol 127: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WN, Kwart M, Hummel S, Frommer WB (1995) Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis. J Biol Chem 270: 16315–16320 [DOI] [PubMed] [Google Scholar]
- Fischer WN, Loo DD, Koch W, Ludewig U, Boorer KJ, Tegeder M, Rentsch D, Wright EM, Frommer WB (2002) Low and high affinity amino acid H+-cotransporters for cellular import of neutral and charged amino acids. Plant J 29: 717–731 [DOI] [PubMed] [Google Scholar]
- Forreiter C, Kirschner M, Nover L (1997) Stable transformation of an Arabidopsis cell suspension culture with firefly luciferase providing a cellular system for analysis of chaperone activity in vivo. Plant Cell 9: 2171–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer WB, Hummel S, Unseld M, Ninnemann O (1995) Seed and vascular expression of a high-affinity transporter for cationic amino acids in Arabidopsis. Proc Natl Acad Sci USA 92: 12036–12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G (2002) New insights into the regulation and functional significance of lysine metabolism in plants. Annu Rev Plant Biol 53: 27–43 [DOI] [PubMed] [Google Scholar]
- Heremans B, Borstlap AC, Jacobs M (1997) The rlt11 and raec1 mutants of Arabidopsis thaliana lack the activity of a basic amino acid transporter. Planta 201: 219–226 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirner B, Fischer WN, Rentsch D, Kwart M, Frommer WB (1998) Developmental control of H+/amino acid permease gene expression during seed development of Arabidopsis. Plant J 14: 535–544 [DOI] [PubMed] [Google Scholar]
- Hoffmann W (1985) Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae: a transmembrane protein without N-terminal hydrophobic signal sequence. J Biol Chem 260: 11831–11837 [PubMed] [Google Scholar]
- Kinraide TB (1981) Interamino acid inhibition of transport in higher plants. Plant Physiol 68: 1327–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh MH, Corruzzi G (1995) Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohaus G, Winter H, Riens B, Heldt HW (1995) Further studies of the phloem loading process in leaves of barley and spinach: the comparison of metabolite concentrations in the apoplastic compartment with those in the cytosolic compartment and in the sieve tubes. Bot Acta 108: 270–275 [Google Scholar]
- Ludewig U, von Wirén N, Frommer WB (2002) Uniport of NH4+ by the root hair plasma membrane ammonium transporter LeAMT1;1. J Biol Chem 277: 13548–13555 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ, Filatov V, Herzyk PC, Krijger GB, Axelsen K, Chen S, Green BJ, Li Y, Madagan KL, Sanchez-Fernandez R, et al (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Marek ET, Dickson RC (1987) Cloning and characterization of Saccharomyces cerevisiae genes that confer L-methionine sulfoximine and tabtoxin resistance. J Bacteriol 169: 2440–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski S (1993) Chemical modification studies of the active site of glucosamine-6-phosphate synthase from baker's yeast. Biochim Biophys Acta 1161: 279–284 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417–422 [DOI] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Heim U, Sauer N, Wobus U, Weber H (2001) Amino acid permeases in developing seeds of Vicia faba L.: expression precedes storage protein synthesis and is regulated by amino acid supply. Plant J 28: 61–71 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–479 [Google Scholar]
- Okumoto S, Schmidt R, Tegeder M, Fischer WN, Rentsch D, Frommer WB, Koch W (2002) High affinity amino acid transporters specifically expressed in xylem parenchyma and developing seeds of Arabidopsis. J Biol Chem 277: 45338–45346 [DOI] [PubMed] [Google Scholar]
- Paris N, Stanley CM, Jones RL, Rogers JC (1996) Plant cells contain two functionally distinct vacuolar compartments. Cell 85: 563–572 [DOI] [PubMed] [Google Scholar]
- Pilot G, Stransky H, Bushey DF, Pratelli R, Ludewig U, Wingate VP, Frommer WB (2004) Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from hydathodes of Arabidopsis leaves. Plant Cell 16: 1827–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB (1996) Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8: 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370: 264–268 [DOI] [PubMed] [Google Scholar]
- Springael JY, Andre B (1998) Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell 9: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Offler CE, Frommer WB, Patrick JW (2000) Amino acid transporters are localized to transfer cells of developing pea seeds. Plant Physiol 122: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y (2004) CATs and HATs: the Park next to us or thereabouts, and our door is the one in front of where the cars are SLC7 family of amino acid transporters. Pflugers Arch 447: 532–542 [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1993) Subcellular volumes and metabolite concentrations in barley leaves. Planta 191: 180–190 [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1994) Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193: 530–535 [Google Scholar]
- Wipf D, Benjdia M, Tegeder M, Frommer WB (2002) Characterization of a general amino acid permease from Hebeloma cylindrosporum. FEBS Lett 528: 119–124 [DOI] [PubMed] [Google Scholar]
- Wipf D, Ludewig U, Tegeder M, Rentsch D, Koch W, Frommer WB (2002) Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem Sci 27: 139–147 [DOI] [PubMed] [Google Scholar]
- Wyse RE, Komor E (1984) Mechanism of amino acid uptake by sugar cane suspension cells. Plant Physiol 29: 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi D (2001) A TGACGT motif in the 5′-upstream region of alpha-Amylase gene from Vigna mungo is a cis-element for expression in cotyledons of germinated seeds. Plant Cell Physiol 42: 635–641 [DOI] [PubMed] [Google Scholar]