Abstract
Heat shock factors (HSFs) are transcriptional regulators of the heat shock response. The major target of HSFs are the genes encoding heat shock proteins (HSPs), which are known to have a protective function that counteracts cytotoxic effects. To identify other HSF target genes, which may be important determinants for the generation of stress tolerance in Arabidopsis, we screened a library enriched for genes that are up-regulated in HSF3 (AtHsfA1b)-overexpressing transgenic plants (TPs). Galactinol synthase1 (GolS1) is one of the genes that is heat-inducible in wild type, but shows constitutive mRNA levels in HSF3 TPs. The generation and analysis of TPs containing GolS1-promoter::β-glucuronidase-reporter gene constructs showed that, upon heat stress, the expression is transcriptionally controlled and occurs in all vegetative tissues. Functional consequences of GolS1 expression were investigated by the quantification of raffinose, stachyose, and galactinol contents in wild type, HSF3 TPs, and two different GolS1 knockout mutants (gols1-1 and gols1-2). This analysis demonstrates that (1) raffinose content in leaves increases upon heat stress in wild-type but not in the GolS1 mutant plants; and (2) the level of raffinose is enhanced and stachyose is present at normal temperature in HSF3 TPs. These data provide evidence that GolS1 is a novel HSF target gene, which is responsible for heat stress-dependent synthesis of raffinose, a member of the raffinose family oligosaccharides. The biological function of this osmoprotective substance and the role of HSF-dependent genes in this biochemical pathway are discussed.
Plants are sessile and hence cannot escape unfavorable environmental conditions such as heat, cold, drought, and salt stress. However, they have the potential to acclimate to these stresses by triggering a cascade of events that lead to changes in gene expression and, subsequently, to biochemical and physiological changes required for survival and growth during or after stress.
Plant acclimation to cold and drought induces to some extent common reactions, including signaling pathways, target gene expression, and biochemical/metabolic changes. One pathway that leads to the activation of a number of target genes is controlled by the transcriptional activator CBF/DREB (Stockinger et al., 1997; Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Shinozaki and Yamaguchi-Shinozaki, 2000; Haake et al., 2002). Multiple biochemical changes associated with cold acclimation in Arabidopsis are attributed to the function of CBF3, which upon transgenic overexpression leads to elevated levels of osmoprotective substances—Pro, Suc, raffinose, Glc, and Fru (Gilmour et al., 2000). Raffinose family oligosaccharides (RFOs), e.g. raffinose, stachyose, and verbascose, also seem to play a role in drought tolerance. RFOs are common in the plant kingdom, abundant in seeds of a large variety of plant species, and can also accumulate in other tissues during stresses. Galactinol synthase (GolS), a key enzyme in the pathway leading to RFOs, synthesizes galactinol (from UDP-Gal and myoinositol), which serves as a galactosyl donor to form raffinose, stachyose, and verbascose. Initial reports showed enhanced GolS activity in seeds of kidney bean upon exposure to cold (Liu et al., 1998). In tomato (Lycopersicon esculentum) seeds, the presence of GolS mRNA showed a correlation with desiccation tolerance; the mRNA declined during imbibition, but, in seedling leaf tissue of tomato, GolS RNA was induced by cold and desiccation (Downie et al., 2003).
In Arabidopsis, there are seven members belonging to the GolS gene family, of which GolS1 and GolS2 mRNAs were detected in mature seeds (Taji et al., 2002) and were also induced by drought and salt stress in leaf tissue; GolS3 was induced by cold stress (Taji et al., 2002). Experimental proof of a protective role of RFO came from the analysis of transgenic plants (TPs) that constitutively expressed GolS2 in vegetative tissue. These plants showed enhanced levels of galactinol and raffinose in leaves and exhibited improved drought tolerance (Taji et al., 2002).
In contrast to drought and cold, the acquisition of tolerance to heat stress (HS) is correlated with the induction of heat shock protein (HSP) expression. HSPs are molecular chaperones whose functions are thought to counteract proteotoxic effects of stress-dependent protein denaturation and thus help to maintain the metabolic and structural integrity of the cell. The expression of HSPs is regulated at the level of transcription by HS-dependent activation of heat shock transcription factors (HSFs), which recognize conserved heat shock element (HSE)-binding sequences in the promoter upstream regions of HS genes. In Arabidopsis, 21 different HSF genes have been identified (Nover et al., 2001). The high multiplicity of HSFs, evident for many plant species, suggests that the obvious genetic redundancy may be important for both backup and diversification of HSF function. By the analysis of gene knockout mutations, AtHSF1 (AtHsfA1a) and AtHSF3 (AtHsfA1b) were recently identified as fast response regulators that are responsible for the immediate early transcription of several HS genes tested (Lohmann et al., 2004). On the other hand, constitutive overexpression of AtHSF3 in transgenic Arabidopsis led to a low-level constitutive HSP synthesis at normal temperature and to increased basal thermotolerance (Prändl et al., 1998). This finding supported the protective function of HSPs for the acquisition of thermotolerance. Direct evidence for the importance of HSP100 in acquired thermotolerance was provided by mutant isolation and analysis of Arabidopsis. Based on a hypocotyl elongation screen, a number of mutants (hot mutants) have been isolated, which showed deficiencies in their ability to acquire thermotolerance at certain developmental stages (Hong et al., 2003). The hot1 mutant was identified to have a mutation in the Hsp101 gene (Hong and Vierling, 2000); however, the phenotypic, molecular, and physiological characterizations of hot2, hot3, and hot4 mutants provide evidence of yet unknown genes that are not related to the expression and function of HSPs (Hong et al., 2003). Another line of evidence for the involvement of other novel genes and metabolic pathways in the HS response was the identification of ascorbate peroxidase genes, Apx1 (Storozhenko et al., 1998) and Apx2 (Panchuk et al., 2002), the expression of which is induced by HS and controlled by HSFs. The importance of APX during HS was indicated by the appearance of a novel heat-stable APX isoform and higher levels of APX activity upon HS in TPs, which overexpressed AtHSF3 (Panchuk et al., 2002). Thus, HSFs may integrate the signaling and cross-talk between different environmental stresses by controlling the expression of key enzymes in biochemical pathways that contribute to common stress tolerance in plants.
The objective of this study was to identify other novel target genes of HSF regulation that may serve important functions in environmental stress responses of Arabidopsis. Taking advantage of AtHSF3 TPs, which show HSP synthesis at normal temperature, we were able to identify GolS1 mRNA in leaf tissue. Furthermore, by promoter::reporter gene expression and functional analysis of T-DNA insertion mutants, we clearly identify GolS1 as a novel HSF-dependent HS gene of Arabidopsis. The correlation between expression of GolS1 and the synthesis of RFOs in leaf tissue suggests an important role of this gene in stress-induced osmolyte synthesis in vegetative tissue and a common role of this pathway in environmental stress responses.
RESULTS
GolS1 Expression in Wild-Type and AtHSF3 TPs
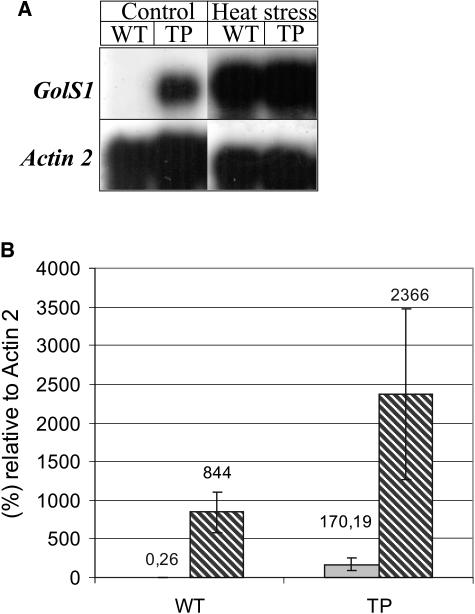
In an attempt to identify novel HSF target genes, we screened a differential suppression subtractive hybridization library (Diatchenko et al., 1996) that was enriched for genes that are overexpressed at normal temperature in leaf tissue of HSF3 TPs compared to wild-type Arabidopsis (C. Leisgen and F. Schöffl, unpublished data). DNA sequencing of preselected clones revealed the presence of HSP18.2 (12%), HSP17.6 (2%), HSP17.4 (4%), HSP21 (7%), and HSF3 (7%) sequences, which were known to be overexpressed in HSF3 TPs from previous investigation (Prändl et al., 1998). Among unconventional HSP genes present in this library, only GolS sequences were overrepresented (6%). To verify the enhanced levels of mRNAs, we used northern-blot hybridization (Fig. 1A) of wild-type and HSF3 TPs. It is shown that GolS1 mRNA is not present in wild type at normal temperature, but is strongly induced after HS. In HSF3 TPs, GolS1 mRNA is present at normal temperature, but its abundance is further enhanced after HS. The quantitative differences determined by real-time PCR (Fig. 1B) show that GolS1 mRNA is 650-fold overexpressed at normal temperature in HSF3 TPs, which is approximately 20% of the maximum level of GolS1 mRNA induced after HS in wild-type plants. After HS, the abundance of GolS1 mRNA was increased about 14-fold in HSF3 TPs. The maximum of heat-induced GolS1 mRNA is about 3 times higher in HSF3 TPs compared to wild type. These data demonstrate that the expression of the GolS1 gene is heat inducible in wild-type and HSF3 TPs. The constitutive mRNA levels in HSF3 TPs, which contain derepressed AtHSF3 (Prändl et al., 1998), indicate that GolS1 expression is controlled by HSF.
Figure 1.
GolS1 expression in wild-type and HSF3 TPs. Poly(A+) RNA was isolated from leaves of wild-type and HSF3 TPs that were subjected to HS (37°C, 2 h) or maintained at control temperature (25°C, 2 h). A, Northern-blot hybridization. One microgram of mRNA was electrophoresed on a 1% agarose gel; after blotting, the membrane was hybridized with the GolS1 probe that spans 13 bp in the 5′-UTR of the entire first exon and 95 bp of the second exon; to ensure equal loading, the membrane was stripped and reprobed with Actin 2. B, GolS1 mRNA quantification. mRNA was reverse transcribed into cDNA and used as a template for quantitative RT-PCR. Specific primers for GolS1 and Act 2 standard were designed toward the 3′-end. Relative amounts were normalized to Act 2 mRNA (=100%). The values are mean ± sd (n = 3).
GolS1 Promoter::β-Glucuronidase Expression in Transgenic Arabidopsis
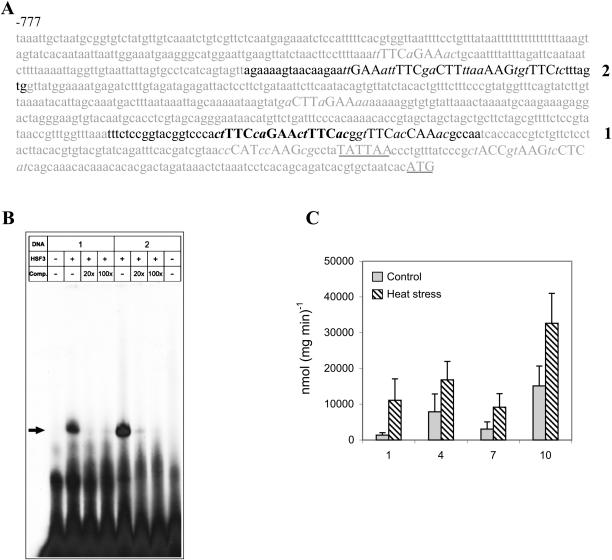
To determine whether GolS1 expression is in fact regulated at the transcriptional level, we generated chimeric promoter-reporter gene constructs for expression analysis in transgenic Arabidopsis plants. The inspection of the putative promoter region of GolS1 (http://intra.psb.ugent.be:8080/PlantCARE) revealed several perfect and imperfect HSE sequences with the consensus -GAA-/-TTC- (see Fig. 2A), the binding sites of HSF, upstream of a putative TATA box. The binding of AtHSF3 to two subfragments (1 and 2), which include the putative binding sites, was investigated by electrophoretic mobility shift analysis (EMSA) using recombinant protein expressed in Escherichia coli. The shifted bands indicate that HSF3 forms DNA-binding complexes with both fragments (Fig. 2B). The formation of the HSE-HSF3-binding complexes is completed by an excess of unlabeled probe, which proves the specificity of binding. It should be noted that recombinant Arabidopsis HSF proteins have the capacity to bind to HSEs in vitro without HS (Hübel and Schöffl, 1994; Lohmann et al., 2004).
Figure 2.
Binding of HSF3 to GolS1 promoter fragments and promoter::GUS activity. A, The DNA sequence of the promoter upstream region of GolS1. HSE sequences are indicated by capital letters; perfect elements with alternating -GAA-/-TTC- boxes are marked in bold. The putative TATA-box sequence and the ATG are underlined. The sequences of two DNA fragments (1, 2), which were used for EMSA, are in black. B, Binding of recombinant HSF3 protein to HSE-containing promoter fragments. Radioactively labeled promoter fragments 1 and 2 were incubated with purified recombinant HSF3 in the absence or presence of 20- or 100-fold excess of unlabeled competitor (Comp.) followed by EMSA. The arrowhead marks the position of HSF-DNA complexes. C, GolS1 promoter-driven GUS activity. A single leaf from individuals of GolS1 promoter::GUS TPs (T3 progeny) was incubated at control (C) at 25°C or HS at 37°C for 2 h. GUS activity was measured in nanomoles (min mg protein)−1. The values are mean ± sd (n = 7–10).
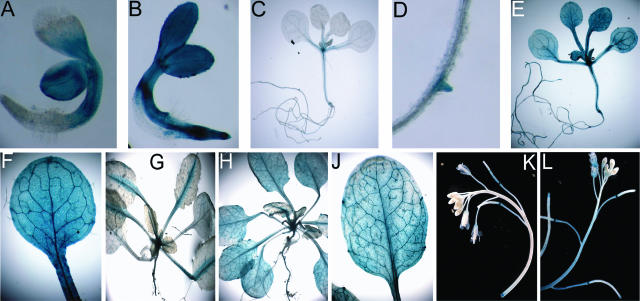
A fragment spanning 1,902 bp upstream from the start codon, including the HSEs containing regions 1 and 2, was inserted in front of the uidA reporter of the plant vector pGPTV-BAR-Asc (Überlacker and Werr, 1996). Following transformation of Arabidopsis, TPs were selected by means of BASTA resistance and subsequent generations (T2, T3) were analyzed for the expression of the reporter gene by assaying β-glucuronidase (GUS) activity in leaf tissue. Figure 2C exemplifies the results in the T3 generation: At normal temperature, there is low basal activity and, after HS, there is a significant increase in GUS activity. TPs of the T3 generation were chosen for determining the expression pattern of the GolS1 promoter by histochemical staining of GUS activity in different stages of development (Fig. 3). Without HS, 1-d-old seedlings show GUS activity in the axes and cotyledons (Fig. 3A), whereas after HS, GUS staining is observed throughout the seedling, with enhanced levels in the roots and vascular tissue (Fig. 3B). After 13 d of germination, the nonstressed plants show GUS staining in the shoot meristematic region, the vascular tissue of shoots, roots, and petioles (Fig. 3C), and in the emerging lateral roots as well (Fig. 3D). After HS, the staining of GUS activity spreads throughout the plant tissue (Fig. 3E), with enhanced staining of major veins in leaves (Fig. 3, E and F). In 4-week-old plants, the GUS activity patterns are very similar to that of plants at day 13 (Fig. 3, H and J). In addition, a developmental expression of GUS was observed in the abscission zones (Fig. 3K) and, after HS, in the basal part of the sepals/petals, expression was not observed in pollen, young siliques, mature siliques, or seeds (Fig. 3L). These data indicate that, at normal temperature, GolS1 expression is developmentally regulated but restricted to meristematic and vascular tissues. After HS, GolS1 expression is induced in almost all cells and tissues of the plant.
Figure 3.
Expression patterns of GolS1. Histochemical localization of GUS expression in GolS1 promoter::GUS TPs (T3 generation). Seeds were sown on Murashige and Skoog medium; seedlings were picked at different days after germination (dag) and tested for the effect of HS at 37°C for 2 h or control (C) at 25°C for 2 h of treatment. Plants investigated after 4 and 7 weeks were grown on soil. One dag, A (C), B (HS); 13 dag, C and D (C), E and F (HS); 4 weeks, G (C), H and J (HS); 7 weeks, K (C), L (HS).
Isolation of GolS1 T-DNA Insertion Mutants
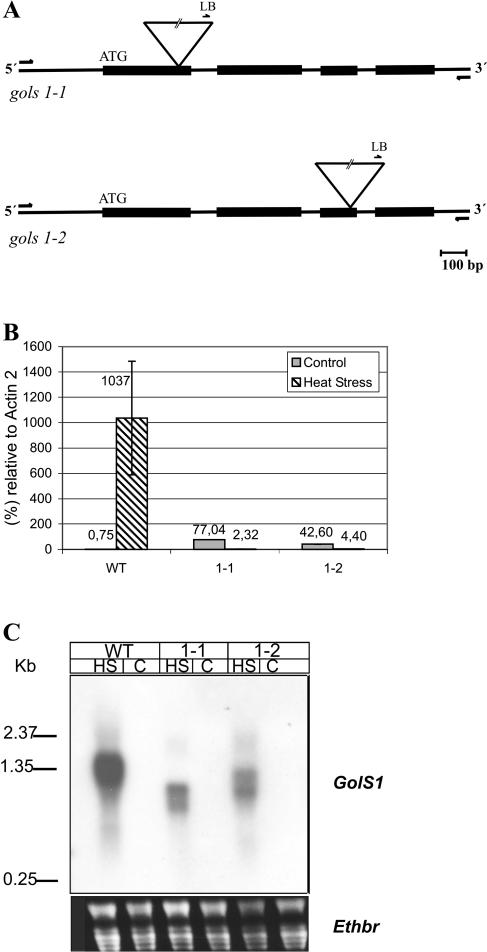
To determine the biological function of GolS1, the Wisconsin alpha population was screened for T-DNA insertions in GolS1. Two insertion mutants, gols1-1 and gols1-2, were obtained from the subpopulation CSH266 and CSH113, respectively. The T-DNA insertions were mapped by sequencing, in gols1-1 to the first exon (336 nucleotides downstream from the ATG start codon) and in gols1-2 to the third exon (1,000 nucleotides downstream from the ATG), as depicted in Figure 4A. The two homozygous mutants showed no obvious phenotypic variations compared to wild-type plants.
Figure 4.
Isolation of GolS1 knockout mutants. A, Localization of T-DNA insertion sites in GolS1. Insertions were determined by sequencing the PCR product obtained from genomic DNA of the tagged lines with the T-DNA left-border primer (LB) and 3′-gene-specific primer. Exons are represented by black boxes and introns by lines. T-DNA is depicted by an inverted triangle. T-DNA elements and primers are not drawn to scale. B, Quantification of GolS1 RNA levels in wild type and GolS1 mutants. Total RNA and, subsequently, poly(A+) RNA were isolated from leaves of wild type and GolS1 knockout mutants (gols1-1 [1-1] and gols1-2 [1-2]) that were subjected to HS (37°C, 2 h) or control temperature (25°C, 2 h). Poly(A+) RNA was reverse transcribed to cDNA and used as a template for quantitative RT-PCR. The primers are specific for GolS1 and Act 2 toward the 3′-end. Relative amounts were normalized to Act 2 mRNA (=100%). The values are mean ± sd (n = 3). Note: The primers for GolS1 are located downstream from the T-DNA insertion site. C, Northern-blot analysis of GolS1 T-DNA insertion mutants. Total RNA was isolated from both wild type and GolS1 mutants (gols1-1 [1-1] and gols1-2 [1-2]) that were subjected to HS at 37°C or kept at 25°C for 2 h as control (C). Fifteen micrograms of total RNA were separated on a 1% agarose gel. Prior to blotting, the RNA was stained with ethidium bromide and visualized under UV to ensure equal loading. After blotting, the membrane was hybridized with a GolS1 probe that spans 13 bp in the 5′-UTR, the entire first exon, and 95 bp of the second exon. The 0.24 to 9.5-kb RNA ladder was used as a standard for estimation of RNA sizes.
The effects of the T-DNA interruptions on GolS1 transcript levels were investigated by quantitative reverse transcription (RT)-PCR. The results show that the heat-induced expression of GolS1 mRNA is almost completely blocked (<1% of heat-induced wild type) in mutant lines (gols1-1 and gols1-2; Fig. 4B). Surprisingly, at normal temperature, low levels of GolS1 transcripts were detectable in the mutant lines but not in the wild type. It should be noted that the primers chosen for PCR quantification of mRNA levels map downstream of the T-DNA-insertions in the 3′-terminal part of the mRNA, spanning a region of 144 nucleotides, including the last exon and the 3′-untranslated sequences of GolS1. The absence of heat-inducible GolS1 mRNAs (in quantitative RT-PCR analysis) indicates that the T-DNA insertions cause gene knockout mutations. However, using an upstream probe (upstream to the T-DNA insertion) in northern-blot hybridizations (Fig. 4C), we were able to detect heat-induced but truncated transcripts of different sizes in gols1-1 (1.09 and 0.68 kb) and gols1-2 plants (1.22 and 0.89 kb). The low levels of constitutive GolS1 mRNA at normal temperature in gols1-1 and gols1-2 (77% and 42.6% relative to actin 2 standard; Fig. 4B) are probably the result of an initiation or read-through of transcription starting within the T-DNA. This background transcription, not observed in wild-type plants (only 0.9% relative to actin 2 mRNA), is heat sensitive as indicated by the strong reduction to 2.32% and 4.4%, respectively, relative to actin 2 mRNA.
Determination of RFOs in Wild-Type, AtHSF3 TPs, and GolS1 Mutant Lines
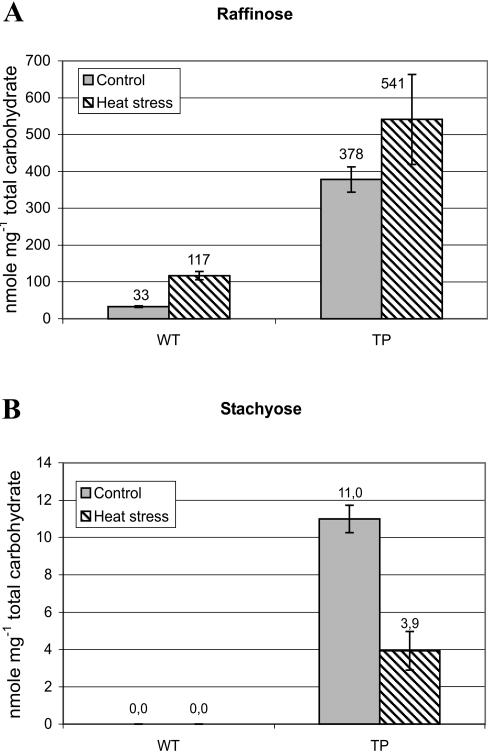
GolS is a key enzyme in the biosynthesis of RFOs: raffinose, stachyose, and verbascose. To determine the functional consequences of changes in GolS1 expression, we assessed the levels of raffinose and stachyose in leaves of HSF3 TPs and in wild-type plants. Leaf samples representing the same developmental stage were subjected to HS for 2 h or incubated at normal temperature as a control. Carbohydrates were extracted and samples were subjected to HPLC analyses. In wild-type (Arabidposis C-24) leaves, raffinose is present at control temperature conditions (33 nmol mg−1 carbohydrate); however, there is a 3.5-fold increase (to 117 nmol mg−1 carbohydrate) of the raffinose content after HS (Fig. 5A). HSF3 TPs show significantly higher raffinose content at control temperature (378 nmol mg−1 carbohydrate) and after HS (541 nmol mg−1 carbohydrate) compared to wild-type plants (Fig. 5A). Interestingly, stachyose was detected only in HSF3 TPs but not in wild type, and the level was significantly higher (approximately 3-fold) at control temperature compared to HS (Fig. 5B). After HS and at control temperature, the level of Suc did not show any significant change in wild-type and HSF3 TPs (data not shown). This indicates that the increase in RFO levels is a specific effect of HS.
Figure 5.
HPLC analysis for raffinose and stachyose content in wild-type and HSF3 TPs. Carbohydrates were isolated and HPLC analysis was performed on heat-treated (37°C, 2 h) and control (25°C, 2 h) wild-type and HSF3 TPs. Values are represented as mean ± sd (n = 3). A, Raffinose content. B, Stachyose content in wild type and TPs.
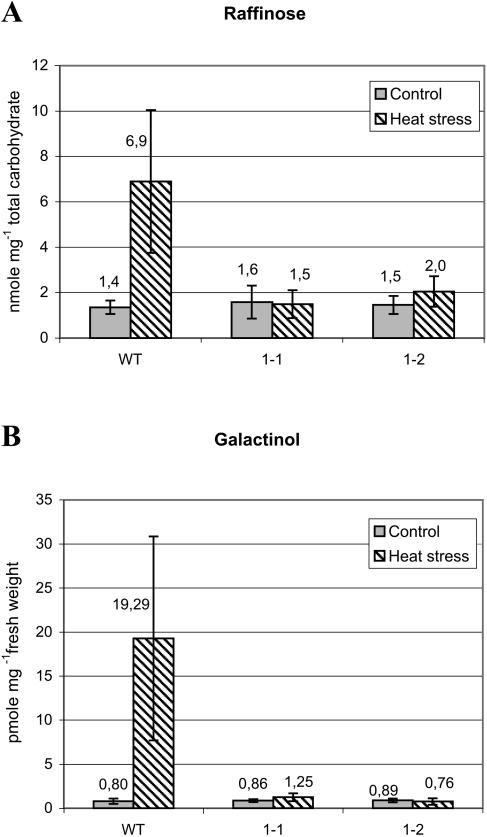
In wild-type Arabidopsis, ecotype Wassilewskija, there is also a strong increase in raffinose levels after HS but not in the knockout mutant lines, gols1-1 and gols1-2, which show only the basal raffinose contents present in wild type under nonstress conditions (Fig. 6A). In these lines we also examined the levels of galactinol, the primary product of GolS activity. Galactinol was detected and quantified in derivatized metabolite fractions after gas chromatography-mass spectrometry (GC-MS) analysis (Fig. 6B). In wild type, there is a strong (approximately 20-fold) increase in galactinol levels in leaves after HS. In gols1-1 and gols1-2 mutant lines, there is no increase after HS; they show only low basal galactinol contents as present in unstressed tissue.
Figure 6.
Raffinose and galactinol content in wild-type plants and GolS1 insertion mutants. Wild type and GolS1 knockout mutants (gols1-1 and gols1-2) were subjected to HS (37°C) or kept at 25°C (2 h) as a control. Metabolites were isolated from leaf tissue and analyzed for raffinose and galactinol content. Values are represented as mean ± sd (n = 3). A, Carbohydrates were isolated and raffinose content was analyzed by HPLC. B, Polar metabolites were isolated and derivatized and galactinol content was quantified after subjecting samples to GC-MS analysis.
These results indicate that there is a correlation between the induction of GolS1 mRNA (Figs. 1 and 4B) and an increase in raffinose-galactinol content after HS in wild-type plants. In HSF3 TPs, the largely enhanced levels of raffinose and the presence of stachyose correlate with the constitutive expression of GolS1 mRNA (Fig. 1).
Phenotypic Analysis of GolS1 Mutants
The GolS1 mutant lines, gols1-1 and gols1-2, show no obvious alterations in growth and morphology compared to wild-type plants. To test stress tolerance phenotypes, seedlings or grown plants were subjected to different HS regimes, which had been previously used for determination of thermotolerance of Arabidopsis (Lee et al., 1995; Prändl et al., 1998, Lohmann et al., 2004). There was no difference in thermotolerance between wild-type and mutant lines (data not shown). Therefore, we examined whether combined HS and dehydration (drought) had differential effects on the survival of plants. Soil-grown plants were subjected to HS for 8 h/d (37°C) for 7 d in a growth chamber under otherwise regular growth conditions. One set of plants of each line was watered regularly (HS only condition); the other set was not irrigated during the experiment (HS/drought condition). There was no significant difference in survival and morphology between the lines and stress condition tested. Data are exemplified in Supplemental Figure 1 (available at www.plantphysiol.org).
Even after using electrolyte leakage as a more quantitative assay for detecting subtle differences in stress tolerance (Lohmann et al., 2004), it was not possible to find significant differences between the lines. The levels of electrolyte leakage, tested for cut leaves of heat-stressed or heat-stressed/drought-stressed plants (from the above experiments), were very similar for all three lines and also not affected by drought stress (see Supplemental Fig. 2).
DISCUSSION
Heat-Induced GolS1 Expression
Our analysis showed that GolS1 is a novel heat-inducible, HSF-dependently expressed gene in Arabidopsis. At normal temperature, GolS1 mRNA is hardly detectable in the leaf tissue of wild-type plants, but its abundance is greatly enhanced by a factor of >3,000 after HS (Fig. 1). The kinetics of mRNA accumulation (data not shown) are very similar to that observed for most HSP genes, which are transiently expressed with a maximum after about 1 h HS (Lohmann et al., 2004). GolS1 also meets other criteria of a typical HS gene: (1) the presence of HSE sequences in the promoter upstream region (a perfect [nTTCnnGAAnnTTCn] present at position −192 bp from ATG and a number of imperfect sequences further upstream); and (2) an enhanced expression when an active HSF is present in the cell. The dependence on an active HSF is indicated by the greatly enhanced (650-fold) basal mRNA level in HSF3-overexpressing plants, which are derepressed for both HSF activity and the expression of HSP (Prändl et al., 1998) and also of Apx2 (Panchuk et al., 2002). Apx2 was the first non-HSP gene identified as HS- and HSF-dependent in this line. The conclusion that GolS1 is a true target of HSF regulation is confirmed by total transcriptome analysis of HSF knockout mutants (W. Busch and F. Schöffl, unpublished data) and by the investigation of TPs carrying GolS-promoter::GUS-reporter constructs (Fig. 2). The high basal levels of GolS1 mRNA in HSF3 TPs are attributed to the derepressed activity of HSF3 in this line, as indicated by a constitutive, HSF3 overexpression-dependent HSF DNA-binding complex in electrophoretic mobility shift experiments (Prändl et al., 1998). The higher abundance of GolS1 mRNA after HS in this line compared to wild type is also attributed to the approximately 10-fold higher levels of HSF3 in TPs. HSF3 and HSF1 have been identified as early response regulators required for immediate heat-induced expression of primary HSF target genes in Arabidopsis (Lohmann et al., 2004). As shown by EMSA, recombinant HSF3 binds to HSE-containing fragments of the GolS1-promoter upstream region. However, HSF3 is not the sole regulator of GolS1 induction in wild-type plants, since heat induction of the mRNA of this gene is not compromised in hsf3-tt1 knockout mutants (Lohmann et al., 2004; T.J. Panikulangara and F. Schöffl, unpublished data). Only in HSF1/HSF3 double knockout plants (hsf1-tt1/hsf3-tt3) did we observe a significant reduction (approximately 10-fold) of GolS1 mRNA levels after HS (data not shown). Hence, AtHSF1 and AtHSF3 are probably involved in the rapid up-regulation of GolS1 mRNA; both seem to be able to functionally replace each other in single knockout mutants. The fast induction and transient accumulation of GolS1 mRNA is the signature of HSF1/HSF3-dependent regulation (Lohmann et al., 2004; W. Busch and F. Schöffl, unpublished data). Therefore, it is unlikely that other HSFs, whose expression depends on the activity of HSF1/HSF3, are involved in the early heat-induced expression of GolS1. Such factors, as for example AtHSF4 and AtHSF7, are probably required for delayed regulatory functions in the HS response (Lohmann et al., 2004).
The HS-induced GUS activity also provides evidence that GolS1 expression is controlled at the transcriptional level (Fig. 2). Histochemical GUS staining is observed in almost all cells and tissues upon HS, with only little variation during plant development. The generalized HS induction of GolS1 expression in all cells is reminiscent of the global expression of HSPs, and it suggests that this gene/enzyme represents another important pathway that may be required for plant cell protection from deleterious effects of HS. In the absence of HS, the GUS-staining pattern indicates that GolS1 is expressed at a low level, but only in certain tissues, including meristematic and major vascular tissue of roots, shoots, and leaves. Neither basal nor heat-induced expression of GolS1::GUS showed a preference for minor veins in leaves, which would have been consistent with a role of this gene in phloem loading, as inferred from cucurbits (Turgeon, 1996) and from heterologous expression of a Cucumis melo GolS promoter in transgenic Arabidopsis (Haritatos et al., 2000). Hence, GolS1 expression does not seem to be involved in phloem loading. At present, the most likely mechanism of phloem loading in Arabidopsis is a carrier-mediated apoplastic transport of Suc (Haritatos et al., 2000).
What is the function of GolS1? Previously, it was shown that the expression of both GolS1 and GolS2 was induced by drought and high-salinity stress (Taji et al., 2002). Interestingly, the expression of both genes, in contrast to GolS3, seems to be insensitive to regulation by the transcriptional activator DREB1/CBF (Taji et al., 2002). In addition, there was no DREB/CBF recognition site detected within 1,000 bp of the promoter upstream region of GolS1. Our results show that GolS1 expression is clearly induced by HS in a HSF-dependent fashion. It seems possible that HSFs may be involved in drought- and salinity-induced GolS1/GolS2 expression, whereas the expression of cold-inducible GolS3 is regulated by DREB1/CBF. The examination of the putative promoter region for other cis-regulatory elements revealed a large number of different motifs, including several light-, abscisic acid-, and ethylene-responsive elements and a MYB recognition site. The presence of such sequences suggests that GolS1 may be a target of multiple signaling pathways for expression. There is ample evidence for cross-talk between different environmental stresses, leading to the expression of common stress genes (Patori and Foyer, 2002; Chinnusamy et al., 2004) and transcriptome analysis (Seki et al., 2001; Kreps et al., 2002). Under natural conditions, there is a high probability that plants subjected to HS will subsequently suffer dehydration stress.
GolS1 Expression and RFO Synthesis
In our analyses of wild-type, HSF3 TPs, and GolS1 knockout plants, there is a strong correlation between the expression levels of GolS1 mRNA and the levels of raffinose or galactinol in leaf tissue. Heat-induced raffinose levels are significantly enhanced in both wild-type and HSF3 plants. In HSF3 TPs, the effect is more pronounced under nonstress conditions (11-fold higher basal raffinose level compared to wild type) than after HS (only about 5-fold difference). It has to be taken into account that the HS-induced RFO content was assayed immediately after the 2-h treatment, representing only a snapshot of a short-term effect, whereas the constitutive overexpression of GolS1 at normal temperature in HSF3 TPs represents long-term steady-state levels. An interesting phenomenon was the presence of stachyose, however, solely in leaves of HSF3 TPs, not in wild type. In Arabidopsis, stachyose accumulation has been observed during seed maturation but could not be detected in rosette leaves, neither without nor after drought, cold, or salinity stress treatments, which resulted in the induction of raffinose only (Taji et al., 2002).
Why is stachyose formed only in HSF3 TPs? There are two observations that may explain this phenomenon: (1) the relatively high constitutive levels of raffinose; and (2) the down-regulation of stachyose levels after HS (by a factor of about 3). Raffinose is the substrate that, along with galactinol, is required for stachyose synthesis. In wild type, the levels of raffinose are 5 to 10 times lower than in HSF3 TPs. These low substrate levels may not be sufficient to cause conversion to detectable levels of stachyose. The down-regulation of stachyose after HS indicates that this step is thermosensitive, either at the expression or at the activity level of putative stachyose synthases. This may explain why wild-type leaves, which show increased raffinose levels after HS, are nevertheless unable to synthesize stachyose. The threshold levels for reaction and detection are unknown. The isolation and analysis of stachyose synthases will be required to verify this model. It cannot be excluded entirely that HS activates α-galactosidases that would specifically degrade stachyose but not raffinose. However, it has been shown that down-regulation of α-galactosidase expression has a positive effect on the accumulation of raffinose, stachyose, and other soluble sugars in petunia (Pennycooke et al., 2003), which indicates that the substrate specificity of this enzyme is not very high. Stachyose-specific degrading α-galactosidases have been described for stachyose-translocating plant species (Bachmann et al., 1994).
In the GolS1 knockout mutants (gols1-1 and gols1-2) the inability to induce GolS1 mRNA by HS correlates with a total loss of heat-inducible levels of galactinol and raffinose. Hence, GolS1 makes the major contribution to the heat-induced formation of these compounds. The low basal levels of raffinose, which are approximately the same in wild-type and knockout lines, may result from the combined basal activities of different GolS enzymes expressed in leaves of Arabidopsis.
RFOs accumulate to relatively high levels during seed maturation in Arabidopsis (Taji et al., 2002), soybean (Blackman et al., 1992), and maize (Brenac et al., 1997). Its role in desiccation tolerance and seed storability is still a matter of controversy (Minorsky, 2003). Experimental evidence for an osmoprotective role in vegetative tissue is provided by experiments that showed that, in TPs overexpressing GolS2, levels of galactinol and raffinose are enhanced, which correlates with improved drought tolerance of Arabidopsis (Taji et al., 2002). The HS-induced accumulation of raffinose may have the same functional and biological consequences. In our experiments, there was no negative effect of heat/drought stress tolerance detectable in GolS1-deficient plants. This suggests that GolS1 may not be essential for thermotolerance, and its function during combined heat and drought stress may be compensated by the activity of drought-inducible GolS2. However, it is possible that GolS1 contributes to environmental stress tolerance under long-term natural conditions. Further analysis of double mutants in GolS genes may be required to investigate the functional relevance of RFO in plants.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Heat Treatment
For experimental purposes, different ecotypes of Arabidopsis L. Heynh were used. In this study, Arabidopsis C-24 was the genetic background of HSF3-overexpressing TPs (Prändl et al., 1998), in which HSF-dependent expression of GolS1 was identified. The analysis of GolS1-promoter::GUS-reporter gene expression was carried out after transformation of Arabidopsis ecotype Columbia (Col-0). T-DNA-tagged Arabidopsis ecotype Wassilewskija lines were used for the identification of GolS1 knockout mutants.
Unless stated otherwise, plants were grown on soil with a 16-h-dark/8-h-light cycle at 20°C, with a light intensity of 3,900 lux for 4 to 5 weeks. Prior to analysis, wild-type Arabidopsis C-24 and HSF3-overexpressing plants were incubated for 3 d at 28°C (Panchuk et al., 2002). Constitutive synthesis of small HSPs (HSP17.6, cytoplasmic class I sHSP) served as a marker for derepression of the HS response in HSF3 TPs (Prändl et al., 1998).
For HS treatment, pools of fully expanded leaves from 4- to 5-week-old plants were cut and incubated at 37°C for 2 h in prewarmed section incubation buffer (1 mm potassium phosphate, pH 6.0, and 1% [w/v] Suc) placed in a water bath with 40 oscillations/min. As a control, the leaves were incubated under the same experimental conditions but at room temperature (approximately 25°C). After treating the leaves, excess buffer was rinsed off with water, blotted dry with a filter paper, and then immediately frozen in liquid nitrogen and stored at −70°C until further use.
Stress Tolerance Tests
Experiments with combined heat and dehydration (drought) stress were conducted in a growth chamber under standard conditions. Soil-grown plants were subjected to 8 h/d HS (37°C) for 7 d. Dehydration stress was applied to one set of heat-stressed plants by withdrawing water supply for 7 d; the other set was irrigated every third day during the experiment (receiving only HS). Control plants were grown at room temperature (20°C) and either subjected to dehydration or watered.
Electrolyte leakage assays were conducted after collecting leaves from stressed plants in 15 mL of deionized water. Ion leakage was measured using a Mettler-Toledo MC126 conductometer following a 1- to 3-h incubation at 20°C in a shaker. Samples were then frozen at −80°C, thawed, shaken for 1 h at 20°C, and total ion leakage measurements were taken. With the two measurements, percentage of electrolyte leakage was determined for each sample. In each experiment, three individual samples were taken from each treatment and analyzed in parallel.
Isolation of Poly (A+) RNA and Northern-Blot Analysis
Total RNA was isolated from leaf tissue using the RNeasy kit (Qiagen, Valencia, CA), and poly(A+) RNA was isolated directly from 1 g of frozen leaf tissues using oligo(dT)-Dynabeads (Dynal Biotech, Oslo, Norway), following the manufacturer's instructions.
One microgram of poly(A+) RNA or 15 μg of total RNA was electrophoresed on a 1% agarose gel containing formaldehyde (Maniatis et al., 1982), transferred on Hybond Nx membrane (Amersham-Pharmacia Biotech, Uppsala), and followed by UV cross-linking (0.125 J cm−2). As a hybridization probe for GolS1 mRNA, a PCR product obtained from a cDNA template using nested PCR primers (primer 1, 5′-TCGAGCGGCCGCCCGGGCAGGT-3′ and primer 2R, 5′-AGCGTGGTCGCGGCCGAGGT-3′; CLONTECH Laboratories, Palo Alto, CA) was used. The cDNA probe (450 bp) spans a region including 13 bp in the 5′-untranslated region (UTR), the entire first exon, and 95 bp of the second exon of GolS1 (AC002337; Taji et al., 2002). To ensure equal loading of mRNA samples, the membrane was stripped after the first hybridization by incubation at 80°C in 0.1% SDS for 1 h before it was reprobed with Act 2 (U41998) mRNA using a gene-specific radioactively labeled PCR product derived from Actin 2 cDNA (An et al., 1996).
The hybridization probes were labeled using the Rediprime DNA Labeling System (Amersham-Pharmacia Biotech) with [α32P]dCTP. Hybridization was performed at 68°C overnight, in a solution containing 0.25 mm NaH2PO4, 1 mm EDTA, 1% (w/v) BSA fraction 5 (pH 7.0), and 7% SDS. The membrane was then washed twice at 68°C for 30 min each in 2× SSC containing 0.1% SDS and once in 0.2× SSC containing 0.1% SDS.
Primer Design and PCR Specificity
For quantitative RT-PCR, GolS1 promoter::GUS studies, and isolation of T-DNA insertions in GolS1, gene-specific primers were designed using the Primer 3 program (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). Primers used for quantitative RT-PCR amplification of GolS1 (AC002337) were: F10-52 ic L (5′-TTCATCACCGCTCTTACTGAAG-3′) and F10-52 ic R (5′-AAGAGGTTGGGGATTATGTGTC-3′). The amplification product included 65 bp of the last exon and 79 bp of the 3′-UTR, respectively, to yield a gene-specific product of 144 bp. Actin 2 (U41998) primers used for quantitative RT-PCR were: left primer 5′-AAGCTGGGGTTTTATGAATGG-3′, right primer 5′-GACTACGTGAACACACACTGTT-3′, yielding a gene-specific product covering 118 bp of the 3′-terminal region of the mRNA. Gradient PCR was performed to determine suitable annealing and amplification conditions for generating a gene-specific product. The specificity of the performed reaction was indicated by a single band after agarose gel electrophoresis and was confirmed by DNA sequencing of the PCR product.
cDNA Synthesis and Quantification of GolS1 RNA by Quantitative RT-PCR
Total RNA was isolated from 100 to 200 mg of frozen leaf material using the RNeasy Plant kit (Qiagen), and mRNA was purified from total RNA using Oligotex kit (Qiagen); cDNA was synthesized from either 100 ng or one-fifth of mRNA using the Thermo Script RT-PCR system (Invitrogen, Carlsbad, CA) with oligo(dT)20 primers.
Quantitative RT-PCR was performed in triplicate using undiluted and 1:10 diluted cDNA as templates. The 50-μL reactions contained 50 mm KCl, 20 mm Tris-HCl (pH 8.4), 0.5 units platinum Taq polymerase (Invitrogen), 0.1× SYBR-Green (Molecular Probes, Eugene, OR), 1.5 mm MgCl2 (Invitrogen), 100 μm of each dNTPs (TaKaRa Shuzo, Shiga, Japan), and 0.5 μm GolS1 primers or Actin 2 primers. PCR was performed with the following program: initial polymerase activation at 95°C for 15 min, then 40 cycles at 95°C for 20 s, and 56°C for 1 min 10 s. The amplification was monitored via intercalation of SYBR-Green using the iCycler iQ system (Bio-Rad Laboratories, Hercules, CA). All experiments were repeated three times using independently prepared RNA/cDNA templates. Deviations of threshold values were less than 1 cycle for independent cDNA preparations and less than 0.5 cycles for replicates of the same cDNA. The values were normalized to an internal standard Act 2 mRNA that was set as 100%.
HSF3 Binding to GolS1 Promoter Fragments
Double-stranded DNA fragments spanning the two HSEs containing regions 1 and 2 (58 bp each) of the GolS1 promoter (Fig. 2A) were generated from synthesized complementary single strands (Sigma-Aldrich, St. Louis) by slow annealing starting at 85°C with a decline of 0.5°C min−1. The double-stranded fragments were purified by polyacrylamide (10%) gel electrophoresis and eluted from the gel. These fragments were used to test binding of recombinant HSF3.
For expression of N-terminal 6×-His-tagged full-length HSF3 (Lohmann et al., 2004), the coding sequence of HSF3 (Prändl et al., 1998) was used in the pQE-30 expression vector (Qiagen). Purified HSF3 protein was prepared from 100 mL Escherichia coli M15 expression culture using the Protino Ni 150 kit (Macherey-Nagel, Duren, Germany), according to the manufacturer's protocol, with protease inhibitor cocktail (Roche, Basel, Switzerland) supplied in the lysis buffer. The purified protein was concentrated with Vivaspin 500 columns (3,000 molecular weight cutoff; Vivascience, Hannover, Germany).
For binding reactions, 1.5 μg of purified protein were incubated for 20 min at room temperature in the presence of 1 μg of poly[d(I-C)] and 2 ng of radiolabeled promoter fragment in a total volume of 25 μL 0.5× Tris-borate/EDTA. In the competition experiment, 40 or 200 ng of unlabeled promoter fragment were included. After adding 5 μL of loading buffer (30% [v/v] glycerol, 0.02% [w/v] bromphenol blue), samples were resolved on a 5% polyacrylamide gel containing 3% (v/v) glycerol in 0.5× Tris-borate/EDTA. After electrophoresis at 350 V for 2.5 h at 4°C, the gel was dried on DE81 anion-exchange chromatography paper (Whatman Biometra, Clifton, NJ) and subjected to autoradiography.
Chimeric GolS1 Promoter::GUS Construction and Generation of TPs
Genomic DNA was isolated from Arabidopsis C-24 plants, according to Li and Chory (1998), and used as a template for PCR amplification of a 1.9-kb fragment upstream of the putative ATG start codon of GolS1 (AC002337) by the following program: 95°C for 1 min, 30 cycles at 94°C for 1 min, 68°C for 40 s, 72°C for 4 min, and 72°C for 5 min, with Pwo polymerase (Peqlab, Erlangen, Germany) with the following primer pairs: 10-52P5′ (5′-GACTGAAGCTTGCATGTTGCCTAAACTCGGACA-3′) and 10-52P3′ (5′-GACTGTCTAGATAGCACGTGATCTGCTGTGAGG-3′), which provided HindIII and XbaI restriction sites, respectively (underlined sequence). The PCR product was inserted into TOPO Blunt vector (Invitrogen), and the construction was verified by DNA sequencing. One clone was digested with HindIII and XbaI, and following elution from agarose gel (QIA quick gel extraction kit; Qiagen), the promoter fragment was cloned via HindIII and XbaI sites upstream of the GUS gene (uidA) into the plant binary vector pGPTV-BAR-Asc (Überlacker and Werr, 1996).
Arabidopsis (Col-0) was transformed, with the resulting promoter::GUS construct, according to Bechtold et al. (1993). Transformed plants were selected for their resistance to 0.1% BASTA (glufosinat ammonium; AgrEvo, Norfolk, UK). Reporter gene activity was investigated in subsequent generations.
Determination of GUS Activity
For GUS activity measurements, a single leaf located at an identical position within a rosette was collected from 7 to 10 individual T3-generation TPs (4–5 weeks old) and was subjected to HS or kept at control temperature. GUS activity was measured fluorometrically in total protein extracts from leaf tissue according to Jefferson (1987). Protein concentrations were determined by the Bradford (1976) dye-binding method. GUS activities are expressed in nanomoles of 4-methylumbelliferone produced per minute per milligram of protein.
Histochemical Localization of GUS
For histochemical localization of GUS, seeds of the T3 generation were germinated on solidified Murashige and Skoog medium. Seedlings and plants were heat stressed or kept at room temperature followed by vacuum infiltration (Jefferson et al., 1987) with 1 mm 5-bromo-4-chloro-3-indolyl-glucuronide in 100 mm sodium phosphate buffer (pH 7.4), 0.5 mm of potassium ferric cyanide, and 0.5 mm potassium ferrocyanide, and incubated overnight at 37°C in the same buffer. The reaction was stopped by rinsing the plant material in 70% (v/v) ethanol. The expression pattern was recorded microscopically using Leica MZFL III supplied with Axiocam (Zeiss, Jena, Germany) digital camera and AxioVision 3.0 software.
Isolation of GolS1 T-DNA Knockout Lines
T-DNA insertions in GolS1 were screened using the alpha populations consisting of 60,480 T-DNA-tagged lines (ecotype Wassilewskija; Krysan et al., 1999) at the Arabidopsis knockout facility at the University of Wisconsin, Madison. Suitable PCR primers were designed and tested according to the instructions provided at http://www.biotech.wisc.edu/NewServicesAndResearch/Arabidopsis/Guidelines.asp. The facility at Wisconsin performed PCR using T-DNA left-border primer JL-202: 5′-CATTTTATAATAACGCTGCGGACATCTAC-3′ in combination with GolS1/5′-ko primer, 5′-GTGTACAATGCACCTCGTAGCAGGGAATA-3′ and GolS1/3′-ko primer, 5′-GTAAAGCAAATCACATGTCAACGTCCAAA-3′. The PCR products obtained from Wisconsin were subsequently analyzed by Southern-blot hybridization using the PCR product of 1.8-kb genomic DNA as probe. The T-DNA insertions were mapped within GolS1 by DNA sequence analysis. After a second round of screening, seed pools obtained from Arabidopsis Biological Resource Center (ABRC; http://www.biosci.ohiostate.edu/plantbio/Facilities/abrc/abrchome.htm) were grown according to the ABRC instructions and individual knockout plants were isolated by PCR with the above primers and the genomic DNA, which was extracted as described by Li and Chory (1998). Homozygosity of knockout mutations was assessed by PCR using left-border T-DNA primer and both 3′- and 5′-gene-specific primers in one reaction. In the case of homozygotes, a single band was obtained with the 3′-gene-specific primer and left border T-DNA primer.
HPLC Analysis of Free Sugars
Leaves (150–200 mg) from identical positions within a rosette were pooled from different plants and subjected to incubation at control temperature or HS (37°C, 2 h) in buffer; subsequently leaves were washed with deionized water, briefly blotted on filter paper, and frozen in liquid nitrogen. Samples were crushed in liquid nitrogen with a prechilled mortar and pestle. Sugars were extracted by boiling in 80% methanol (800 μL, 2 min), followed by centrifugation (12,000g, 5 min) to sediment the insoluble fraction. The pellet was extracted again by boiling in 20% methanol (500 μL, 2 min), as mentioned above. The two supernatants were pooled and evaporated in a speedvac at room temperature overnight. Samples were resuspended in 0.5 mL of HPLC-grade water, centrifuged (12,000g, 3 min), and filtered (0.2 μm). HPLC separation was done using a high-pH anion-exchange column (Carbo Pac PA1; Dionex, Sunnyvale, CA) with 62 mm NaOH as eluent and pulsed amperometric detection (PED-300; Biometra, Göttingen, Germany). The equipment used was a modular HPLC system (pump 420, autosampler 460; Kontron, Eching, Germany) with KromaSystem 2000 software (Bio-Tek, Winooski, VT). Identity and amount of raffinose and stachyose were confirmed by cochromatography with corresponding authentic sugars (Sigma-Aldrich). The amounts of raffinose and stachyose were expressed in nanomoles (mg total carbohydrate)−1. Values obtained are mean ± sd (n = 3).
For the colorimetric determination of total carbohydrate, according to Dubois et al. (1956), 100 μL of extract (see above) was diluted with 100 μL of distilled water and used for analysis. Three parallel assays of 50 μL were performed.
GC-MS Analysis of Galactinol
Extraction of Polar Arabidopsis Metabolites
Arabidopsis leaves of approximately 300 mg fresh weight were frozen in liquid nitrogen and crushed in a Retsch ball-mill (2-mL Eppendorf tubes, 1 steel ball of 5 mm diameter, 1 min at 30 strokes s−1). Immediately 1 mL methanol was added and the mixture was vortexed thoroughly. Fifty microliters of a ribitol stock solution (0.2 mg mL−1 water) were added as internal standard. Extraction was performed at 70°C for 15 min. After addition of 0.8 mL of water, the mixture was vortexed again, and centrifuged at 14,000g for 3 min. The supernatant was reduced to dryness in vacuo.
Derivatization (Methoxymation and Trimethylsilylation)
The residue was redissolved and derivatized for 90 min at 30°C with continuous shaking in 80 μL of methoxyamine hydrochloride (20 mg mL−1 pyridine) followed by a 30-min treatment at 37°C with 80 μL of MSTFA (N-methyl-N-[trimethylsilyl]trifluoroacetamide). Samples of 1 mL were injected with a split ratio of 25:1 after a period of 2 h at room temperature.
GC-MS
The GC-MS system used was composed of an autosampler (AS 7683), a gas chromatograph (GC 6890), and a quadruple mass spectrometer (MSD 5973; Agilent, Palo Alto, CA). GC was performed on a 30 m × 0.25 mm Supelco SPB50 column with 0.25-μm film thickness.
Conditions
The injection temperature at 260°C, MSD interface at 250°C, ion source at 230°C, helium flow mL min−1, solvent delay 5 min at 70°C, oven temperature is increased at 5°C min−1 to 280°C, 1 min isocratic, cool down to 70°C, followed by an additional 5-min delay (modified from Roessner et al., 2000).
Mass spectra of the putative galactinol peak from Arabidopsis leaves were matched with spectra obtained from pure galactinol provided by Fluka (code 79544; Buchs, Switzerland).
Supplementary Material
Acknowledgments
We thank Christine Leisgen of our laboratory for providing the SSH library, Marta Hoffman for excellent assistance in DNA sequencing and screening, Bettina Stadelhofer, Karl Wurster (Central Services, ZMBP-Tübingen) for technical assistance with HPLC and GC-MS, and Steffi Nagel (FG Seitz, ZMBP-Plant Physiology) for assistance with carbohydrate measurements.
This work was supported by Deutsche Forschungsgemeinschaft SFB 446, project A2.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.042606.
References
- An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S, Meagher RB (1996) Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J 10: 107–121 [DOI] [PubMed] [Google Scholar]
- Bachmann M, Matile P, Keller F (1994) Metabolism of the raffinose family oligosaccharides in leaves of Ajuga reptans, cold acclimation, translocation, and sink to source transition: discovery of chain elongation enzyme. Plant Physiol 105: 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellid J, Pelletier G (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Life Sci 316: 1194–1199 [Google Scholar]
- Blackman SA, Obendorf RL, Leopold AC (1992) Maturation proteins and sugars in desiccation tolerance of developing soybean seeds. Plant Physiol 100: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for quantification of microgram quantities of protein utilising the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brenac P, Horbowicz M, Downer SM, Dickerman AM, Smith ME, Obendorf RL (1997) Raffinose accumulation related to desiccation tolerance during maize (Zea mays L) seed development and maturation. J Plant Physiol 150: 481–488 [Google Scholar]
- Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K (1996) Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA 93: 6025–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie B, Gurusinghe S, Dahal P, Thacker RR, Synder JC, Nonogaki H, Yim K, Fukanaga K, Alvarado V, Bradford KJ (2003) Expression of a Galactinol synthase gene in tomato seeds is up-regulated before maturation desiccation and again after imbibition whenever radicle protrusion is prevented. Plant Physiol 131: 1347–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Over-expression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Haake V, Cook D, Reichmann JL, Pineda O, Thomashow MF, Zhang JZ (2002) Transcriptional factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haritatos E, Ayre BG, Turgeon R (2000) Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol 123: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Lee U, Vierling E (2003) Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol 132: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübel A, Schöffl F (1994) Arabidopsis heat shock factor: isolation and characterization of the gene and the recombinant protein. Plant Mol Biol 26: 353–362 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 over expression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper J (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11: 2283–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hubel A, Schöffl F (1995) Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J 4: 603–612 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J (1998) Methods in molecular biology. In JM Martinez-Zapater, J Salinas, eds, Arabidopsis Protocols. Humana Press, Totowa, NJ, pp 55–60
- Liu J-JJ, Krenz DC, Galvez AF, deLumen BD (1998) Galactinol synthase (GS): increased enzyme activity and levels of mRNA due to cold and desiccation. Plant Sci 134: 11–20 [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Gen Genet 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Minorsky PV (2003) The hot and classic. Plant Physiol 131: 1159–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchuk II, Volkov RA, Schöffl F (2002) Heat stress and heat shock transcription factor dependent expression and activity of ascorbate peroxidase in Arabidopsis. Plant Physiol 129: 838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patori GM, Foyer CH (2002) Common components, networks and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol 129: 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennycooke JC, Jones ML, Stushnoff C (2003) Down-regulating α-galactosidase enhances freezing tolerance in transgenic Petunia. Plant Physiol 133: 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prändl R, Hinderhofer K, Eggers-Schumacher, Schöffl F (1998) HSF3, a new heat shock factor from Arabidopsis thaliana, derepresses the heat shock response and confers thermo tolerance when over expressed in transgenic plants. Mol Gen Genetics 258: 269–278 [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K (2001) Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA micro array. Plant Cell 13: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3: 217–223 [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MJ (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/ DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S, De Pauw P, Van Montague M, Inzè D, Kushnir S (1998) The heat shock element is a functional component of the Arabidopsis APX1 gene promoter. Plant Physiol 118: 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Turgeon R (1996) Phloem loading and plasmodesmata. Trends Plant Sci 1: 418–423 [Google Scholar]
- Überlacker B, Werr W (1996) Vectors with rare-cutter restriction enzyme sites for expression of open reading frames in transgenic plants. Mol Breed 2: 293–295 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.