Abstract
Clear cell renal cell carcinoma (ccRCC) is the most lethal neoplasm of the urologic system. Clinical therapeutic effect varies greatly between individual ccRCC patients, so there is an urgent need to develop prognostic molecular biomarkers to help clinicians identify patients in need of early aggressive management. In this study, samples from primary ccRCC tumor and their corresponding nontumor adjacent tissues (n = 18) were analyzed by quantitative proteomic assay. Proteins downregulated in tumors were studied by GO and KEGG pathways enrichment analyses. Six proteins were found both downregulated and annotated with cell proliferation in ccRCC patients. Of these proteins, PDZK1 and FABP1 were also involved in the lipid metabolism pathway. The downregulation of PDZK1 was further validated in TCGA_KIRC dataset (n = 532) and independent set (n = 202). PDZK1 could discriminate recurrence, metastasis and prognosis between ccRCC patients. Low level of PDZK1 in both mRNA and protein was associated with reduced overall survival (OS) and disease–free survival (DFS) in two independent sets. In univariate and multivariate analyses, PDZK1 was defined as an independent prognostic factor for both OS and DFS. These findings indicated that low level of PDZK1 could predict poor clinical outcome in patients with ccRCC.
Abbreviations: ccRCC, clear cell renal cell carcinoma; PDZK1, PDZ domain containing 1; CAP70, cystic fibrosis transmembrane conductance regulator associated protein of 70 kDa; DEPs, differentially expressed proteins; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; WB, Western Blotting; IHC, immunohistochemistry; iTRAQ, isobaric tags for relative and absolute quantitaion; FABP1, fatty acid binding protein 1; TCGA_KIRC, The Cancer Genome Atlas kidney renal clear cell carcinoma; NCCN, national comprehensive cancer network; TNM, The tumor-node-metastasis; TMA, tissue microarray; OS, overall survival; AJCC, American Joint Committee on Cancer; UPLC, the Ultra Performance Liquid Chromatography; FDR, false discovery rate; DAVID, The Database for Annotation, Visualization, and Integrated Discovery; DFS, disease–free survival; STRING, The Search Tool for the Retrieval of Interacting Genes/Proteins; GSEA, The gene set enrichment analysis; ES, Enrichment Score; ROC, Receiver operator characteristic; AUC, area under the curve; KM, Kaplan–Meier; IMP3, insulin-like growth factor mRNA binding protein 3; CA9, carbonic anhydrase IX; LDHA, lactate dehydrogenase A; FSCN2, fascin actin-bundling protein 2; BIRC5, baculoviral IAP repeat containing 5; NHERF, Na+/H+ exchanger regulatory factor; SR-BI, scavenger receptor class B type I
Keywords: Renal cancer, Proteomics, Prognostic markers, PDZ, CAP70, NHERF3, CLAMP
Highlights
-
•
PDZK1, which was involved in cell proliferation and lipid metabolism pathway, was significantly downregulated in ccRCC.
-
•
PDZK1 was defined as an independent prognostic factor for OS and DFS in univariate and multivariate cox analyses.
-
•
Low level of PDZK1 could predict poor clinical outcome in patients with ccRCC.
Clinical therapeutic effect varies greatly between individuals of clear cell renal cell carcinoma (ccRCC). Development of prognostic molecular biomarkers can help clinicians identify patients in need of early aggressive management. Differentially expressed proteins between tumor and adjacent normal tissues were analyzed by proteomics. Subsequently, bioinformatics assay was combined to screen for prognostic markers in ccRCC. PDZ domain containing 1 (PDZK1), involved in cell proliferation and lipid metabolism pathway, was significantly downregulated in ccRCC and defined as an independent prognostic factor for poor clinical outcome in ccRCC patients. Our findings will facilitate patient counseling and individualize management of patients with ccRCC.
1. Introduction
Renal cell carcinoma (RCC) is the most common and lethal cancer of the adult kidney. Clear cell RCC (ccRCC) accounts for approximately 70% to 80% of all RCC. Patients with ccRCC are normally treated by the standard surgical resection. However, after undergoing a nephrectomy, the outcome of ccRCC patients greatly varies. Organ-confined disease confers the best prognosis, only 3% to 29% of patients passed away after five years of nephrectomy (Frank et al., 2005). For patients with locally advanced tumors, 47% to 80% of them passed away after five years of nephrectomy (Frank et al., 2005). In addition, 10% to 28% of ccRCC patients recurred or distantly metastasized (Levy et al., 1998, Figlin, 1999), leading to their poor outcome. The median survival of recurrent and metastatic ccRCC patients is 21 and 13 months, and the 5-year survival rates are reported as 30.5% and < 10%, respectively (Eggener et al., 2006, Minasian et al., 1993). These patients would benefit from a more aggressive treatment strategy and a more active monitoring (NCCN Guidelines®). Hence, there is an urgent need to develop prognostic molecular biomarkers to help clinicians identify patients in need of early aggressive management.
The tumor-node-metastasis (TNM) staging system is viewed as a predominant prognostic factor for ccRCC patients. However, the clinical outcomes of patients with ccRCC may vary considerably even within the same tumor stage, suggesting that further clues other than TNM staging system is needed for more accurate assessment of prognosis. Moreover, the TNM cancer staging systems predict survival on the basis of anatomic and histological extent of the tumor rather than on molecular changes. The molecular basis of this diversity in clinical behavior is due to large variations of molecular pathogenesis which originates from dysregulation of different gene or protein expression within the same TNM stage (Tsui et al., 2000, Frank et al., 2005, Veeratterapillay et al., 2012). Prognosis stratification by molecular markers, such as expression of specific genes could improve accuracy of outcome prediction (Tamayo et al., 2011). Therefore, new molecular prognostic markers which could precisely stratify patients into different risk categories are clearly warranted.
In the present study, isobaric tags for relative and absolute quantitation (iTRAQ)-based proteomics was used to screen for differentially expressed proteins (DEPs) between tumor and adjacent normal tissues in each of four stages. Gene ontology (GO) cell proliferation annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of these DEPs were analyzed to identify a predictor of patient outcome. PDZ domain containing 1 (PDZK1, also called cystic fibrosis transmembrane conductance regulator associated protein of 70 kDa, CAP70) was downregulated in ccRCC tissues and correlated with lipid metabolism pathway. Further, we validated the prognostic significance of PDZK1 by using multiple approaches and independent sets.
2. Materials and Methods
2.1. Patients and Study Design
We obtained 112 pairs of primary ccRCC and adjacent normal kidney tissues (Table 1) from nephrectomies conducted at the Beijing Friendship Hospital between 2013 and 2014. These tissues were cut into two parts: one part was immediately frozen in liquid nitrogen and stored at − 80 °C for further use in paired iTRAQ and western blotting (WB) analyses; the other part was formalin-fixed and paraffin-embedded for use in unpaired IHC analysis. We also obtained 90 pairs of samples (81 of 90 samples had follow-up data from 2 to 83 months with a median of 70 months) collected from 2006 to 2008 (Table 1), formalin-fixed and paraffin-embedded samples for tissue microarray (TMA) construction. TMA was used for paired IHC and overall survival (OS) analyses. The tumor specimens were classified according to the 2010 American Joint Committee on Cancer (AJCC) staging system. Before surgery, none of the patients has received chemotherapy or radiotherapy. All specimens were histologically confirmed by uro-pathologists. Patients with a histological diagnosis other than ccRCC were subsequently excluded. The study was approved by the Research Ethics Boards of both Capital Medical University and Beijing Friendship Hospital. All subjects included in the protocol signed a declaration of informed consent.
Table 1.
Summary of clinicopathological features of ccRCC patients.
| Discovery set (18 pairs of ccRCC and adjacent normal tissues for iTRAQ analysis) | Independent validation set (532 ccRCC and 72 adjacent normal tissues in TCGA_KIRC dataset for mRNA level, OS and DFS analyses) | Expanded validation set (38 pairs of ccRCC and adjacent normal tissues for WB analysis) | Expanded validation set (112 ccRCC and 19 adjacent normal tissues for unpaired IHC analysis) | Independent validation set (90 pairs of ccRCC and adjacent normal tissues for TMA construction and paired IHC and OS analyses) | |
|---|---|---|---|---|---|
| Characteristic | |||||
| Age (years mean (range) | 63.4(46–76) | 60(26–90) | 61.7(32–79) | 61(28–84) | 59(29–82) |
| Gender (no.) male, female, unknown | 18, 0 | 334, 181, 17 | 26, 12 | 68, 44 | 51, 39 |
| Pathological T stage no. (%) | |||||
| T1 | 9(50%) | 259(48.6%) | 23(60.53%) | 63(56.25%) | 63(70%) |
| T2 | 7(38.87%) | 68(12.8%) | 8(21.05%) | 26(23.2%) | 18(20.1%) |
| T3 | 1(5.57%) | 177(33.3%) | 5(13.16%) | 19(16.95%) | 5(5.5%) |
| T4 | 1(5.57%) | 11(2.1%) | 2(5.26%) | 4(3.6%) | |
| Unknown | 17(3.2%) | 4(4.4%) | |||
| Pathological N stage no. (%) | |||||
| NX | 261(49.1%) | ||||
| N0 | 17(94.4%) | 237(44.5%) | 36(94.74%) | 106(94.6%) | 87(96.7%) |
| N1 | 17(3.2%) | 1(2.63%) | 3(2.7%) | 1(1.1%) | |
| N2 | 1(0.9%) | ||||
| N3 | 1(5.6%) | 1(2.63%) | 2(1.8%) | ||
| Unknown | 17(3.2%) | 2(2.2%) | |||
| Pathological M stage no. (%) | |||||
| MX | 14(2.7%) | ||||
| M0 | 17(94.4%) | 421(79.1%) | 36(94.74%) | 110(98.2%) | 88(97.8%) |
| M1 | 1(5.6%) | 79(14.8%) | 2(5.26%) | 2(1.8%) | 2(2.2%) |
| Unknown | 18(3.4%) | ||||
| Pathological grade no. (%) | |||||
| G1 | 2(11.1%) | 13(2.5%) | 8(21.1%) | 18(16.1%) | 33(36.7%) |
| G2 | 14(77.8%) | 229(43.0%) | 26(68.4%) | 82(73.2%) | 42(46.7%) |
| G3 | 2(11.1%) | 205(38.5%) | 2(5.25%) | 7(6.2%) | 14(15.5%) |
| G4 | 77(14.5%) | 1(1.1%) | |||
| Unknown | 8(1.5%) | 2(5.25%) | 5(4.5%) | ||
| AJCC TNM stage, no. (%) | |||||
| I | 8(44.4%) | 255(47.9%) | 23(60.5%) | 61(54.5%) | 60(66.7%) |
| II | 7(38.9%) | 56(10.5%) | 8(21.1%) | 26(23.2%) | 18(20%) |
| III | 1(5.6%) | 127(23.9%) | 4(10.5%) | 19(17.0%) | 4(4.4%) |
| IV | 2(11.1%) | 81(15.2%) | 3(7.9%) | 6(5.3%) | 2(2.2%) |
| Unknown | 13(2.5%) | 6(6.7%) | |||
In addition, mRNA data and clinical information for patients in The Cancer Genome Atlas (TCGA_KIRC, Table 1) dataset were used for differential mRNA expression analysis.
2.2. Protein Sample Preparation and iTRAQ-based Proteomics
To identify DEPs between ccRCC and paired adjacent normal tissues, the protein samples from 18 ccRCC patients which were distributed in four AJCC TNM stages were extracted separately. Then according to stage and tissue types, eight groups (four tumor tissue groups I-IV and four paired adjacent normal tissue groups I-IV) were divided. Equal amounts of protein in stage III (n = 1) and equally pooled proteins in stage I (n = 8), stage II (n = 7) and in stage IV (n = 2), respectively were analyzed by iTRAQ-based proteomics as previously reported (Uhlen et al., 2015).
Peptides were labeled with the 8-plex iTRAQ kit (AB SCIEX, Darmstadt, Germany) and subjected to a nanospray ionization source followed by tandem mass spectrometry (MS/MS) in Q Exactive™ Plus (Thermo Scientific, Waltham, MA) coupled online to the Ultra Performance Liquid Chromatography (UPLC). For MS scans, the m/z scan range was 350 to 1600. The fixed first mass was set as 100 m/z.
2.3. Database Search
The MS/MS data were processed using the Mascot search engine (v.2.3.0, Matrixscience, London, UK) and searched against SwissProt_human database concatenated with reverse decoy database. Trypsin was specified as the cleavage enzyme allowing up to two missing cleavages. Mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. The false discovery rate (FDR) < 0.01 was considered as statistically significant.
2.4. GO and KEGG Analyses
To explore the functional annotation and involved pathways of genes, the GO and the KEGG analyses were executed by online analysis tools–Database for Annotation, Visualization, and Integrated Discovery (DAVID) The (http://david.abcc.ncifcrf.gov/) and the WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/option.php), respectively.
2.5. The Western Blotting
Western blotting (WB) was performed as previously described (Zheng et al., 2010a, Zheng et al., 2010b). Anti-PDZK1 and anti-β-actin antibodies were purchased from BD Biosciences (Cat# 612660, RRID: AB_399904, San Jose, CA) and Sigma–Aldrich (Cat# A5441, RRID: AB_476744, St. Louis, MO), respectively. HRP-conjugated secondary antibody was purchased from Amersham Biosciences (Cat# NA931 RRID: AB_772210, Little Chalfont, UK). The blots were quantified using NIH Image 1.62 program. The protein level was normalized with β-actin.
2.6. Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously reported (Ma et al., 2015). Sections were incubated at the optimal conditions with rabbit monoclonal anti-PDZK1 antibody (GeneTex, CA, Cat# GTX114628 RRID: AB_10620236, 1:1000). Image–Pro plus 6.0 (MediaCybernetics Inc., SilverSpring, MD) was used to analyze optical densitometry.
2.7. Bioinformatics Analysis
mRNA (RNA Seq v2) levels of genes in ccRCC patients, normal kidney tissues and clinical information about recurrence, metastasis, OS, disease–free survival (DFS) of patients in TCGA_KIRC dataset were obtained from https://www.synapse.org and http://www.cbioportal.org/public-portal, respectively. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/) was used to screen for interacting protein with a combined score of C0.4.
The gene set enrichment analysis (GSEA) was performed as previously described (Peng et al., 2016) to assess whether genes from pre-defined gene-sets are enriched among the highest- (or lowest-) ranked genes by calculating a pathway Enrichment Score (ES).
2.8. Statistical Analysis
The results of the iTRAQ, WB and IHC of paired samples were analyzed by paired sample t-test. The results of RNA seq v2 and IHC of unpaired samples were analyzed by independent sample t-test. Receiver operator characteristic (ROC) curve and area under the curve (AUC) analyses were applied to detect the optimal cutoff point that yielded the highest total accuracy with respect to discriminating different clinical classifications [good (≥ 5 years, living) and poor (≤ 2 years, die) prognosis]. The log-rank test for the generated Kaplan–Meier (KM) curve was conducted to evaluate the association between the expression level of PDZK1 and the survival rate. Univariate and multivariate Cox proportional hazard regression analyses were used to estimate the prognostic significance of PDZK1 in ccRCC. Statistical significance was set at two-tailed P values < 0.05. All statistical analyses were performed using SPSS Statistics 19.0 (IBM SPSS, Chicago, IL).
3. Results
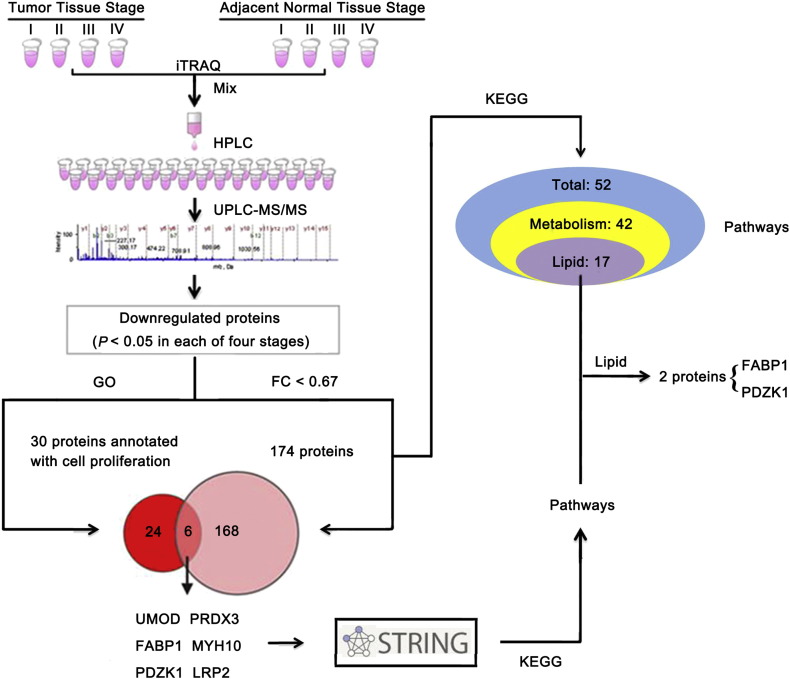
3.1. Proteomic Analysis Identifies 212 Differentially Expressed Proteins in ccRCC
To assess DEPs between ccRCC and normal tissues, a total of 3816 proteins were identified by using LC-MS/MS. Of these proteins, 2985 proteins were quantitatively analyzed. Thirty-eight upregulated proteins (> 1.5 fold over normal, P < 0.05) and 174 downregulated proteins (< 0.67 fold over normal, P < 0.05) were found in each of four stages in tumor tissues (Table S1).
3.2. Six Proteins are both Downregulated and Related to Cell Proliferation in ccRCC
To search the proteins correlated with cell proliferation in ccRCC, GO annotation was used to analyze the downregulated proteins, and six proteins were identified as follows: UMOD, FABP1, PDZK1, PRDX3, MYH10 and LRP2 (Table S2).
3.3. Downregulated Proteins in ccRCC are Mainly Involved in the Lipid Metabolism Pathway
To investigate the relevant pathways of the downregulated proteins, we performed a KEGG pathway enrichment analysis and found that these proteins participated in 52 KEGG pathways, including metabolic processes (42 pathways, 80.8%) and sodium reabsorption, etc. (10 pathways). Of metabolic processes, 17 pathways (32.7%) were related with lipid metabolism (Fig. 1). Results of GSEA study further verified that lipid metabolism pathway was dysregulated in ccRCC tissues (data not shown), which is consistent with the previous reports (Wettersten et al., 2015).
Fig. 1.
PDZK1 and FABP1, which are involved in both cell proliferation and lipid metabolism pathways, are downregulated in ccRCC tissues.
The proteins of paired ccRCC and its adjacent normal tissues from 18 ccRCC patients were extracted respectively. Protein lysates from the same TNM stage (stage I, n = 8; stage II, n = 7; stage III, n = 1; stage IV, n = 2) were combined as a group, respectively (Tumor Tissue Stage I, II, III, IV; Adjacent Normal Tissue Stage I, II, III, IV) in equal amounts and analyzed by iTRAQ quantitative proteomics. The downregulated proteins were performed with the GO and KEGG pathway analysis. Results revealed that 174 proteins were both downregulated and participated in 52 KEGG pathways, mainly lipid metabolism pathways. Six proteins were both downregulated and annotated with cell proliferation. Two proteins annotated with cell proliferation (FABP1 and PDZK1) participated in lipid metabolism pathways.
3.4. Cell Proliferation Associated Protein FABP1 and PDZK1 Are Involved in Lipid Metabolism Pathway
To explore whether the six proteins annotated with cell proliferation (UMOD, FABP1, PDZK1, PRDX3, MYH10 and LRP2) also participated in these metabolic pathways, especially lipid metabolism pathway, we first investigated their binding partners by using STRING database as most proteins usually exerted functions coordinately with their binding partners (Zheng et al., 2010b). A number of possible interacting proteins were identified. As shown in Table S3, UMOD, FABP1, PDZK1, PRDX3, MYH10 and LRP2 interacted with 10, 12, 16, 12, 14 and 13 proteins, respectively. Further KEGG pathway enrichment analysis demonstrated that UMOD, PRDX3, MYH10, LRP2 and their respective partners were not involved in metabolic pathways. FABP1 and its binding partners participated in two lipid metabolism pathways, PDZK1 and its binding partners participated in four metabolism-related pathways, with three of which involved in lipid metabolism (Fig. 1, Table S4). Thus, we further studied the clinical significance of FABP1 and PDZK1 in ccRCC.
3.5. mRNA Levels of FABP1 and PDZK1 Are Downregulated and Correlate With Lipid Metabolism Pathway
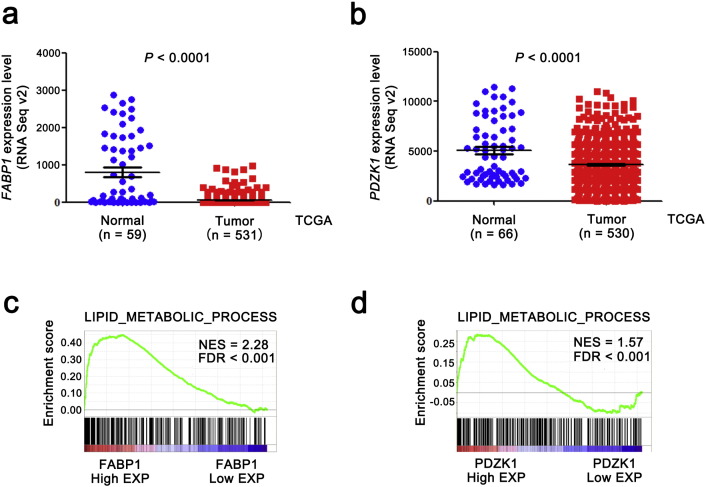
To confirm the results of our proteomics studies, we further analyzed the mRNA levels of FABP1 and PDZK1 from TCGA_KIRC dataset, and found that mRNA levels of both FABP1 and PDZK1 were significantly decreased in tumor tissues compared with normal tissues (Fig. 2a,b, P < 0.0001).
Fig. 2.
The levels of both PDZK1 and FABP1 mRNA are downregulated in ccRCC tissues and correlated with lipid metabolism in ccRCC.
(a,b) The mRNA levels of both FABP1 and PDZK1 in ccRCC by RNA sequencing were obtained from the TCGA_KIRC dataset. Seventy-two normal tissues and 532 ccRCC tissues were sequenced. Outlier values (> mean ± 3 SD) were removed. (c,d) The correlation between lipid metabolism process gene set and mRNA level of FABP1 or PDZK1 was analyzed by GSEA assay. FDR < 0.001 was considered as statistically significant.
Intracellular storage of lipid in the neoplastic cells of ccRCC leads to its distinctive pale and glassy cytoplasm (Sundelin et al., 2012). To confirm whether the FABP1 and PDZK1 were involved in lipid metabolism, GSEA was performed. The results showed that the mRNA levels of both FABP1 and PDZK1 positively correlated with the lipid metabolism process in renal tissues (FDR < 0.001, Fig. 2c,d).
3.6. Low Level of PDZK1 mRNA Predicts Poor Prognosis of ccRCC Patients
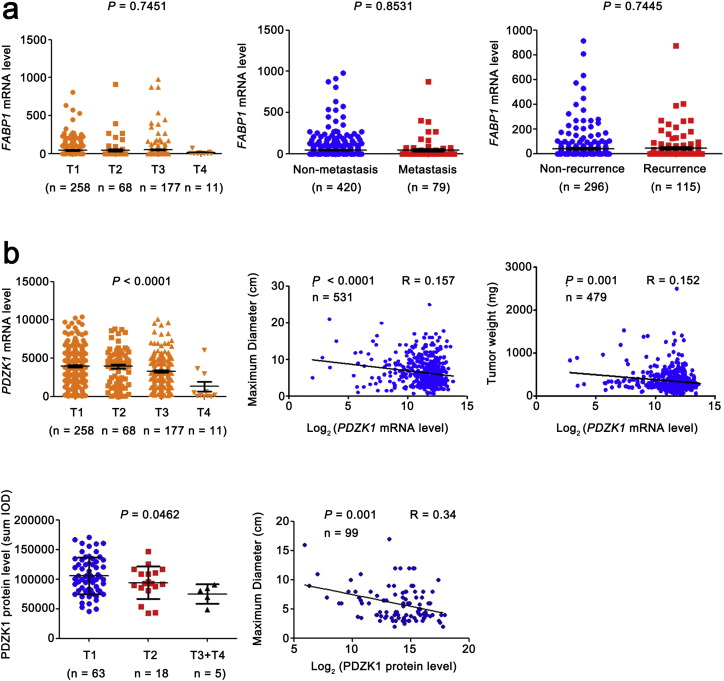
We further analyzed the correlation of the mRNA level of FABP1 and PDZK1 with TNM stage in ccRCC patients. The change of FABP1 mRNA level had no correlation with the T stages, and failed to distinguish between N0 and N1 (without and with lymph node metastasis, data not shown), non-distant metastasis and metastasis, non-recurrence and recurrence (Fig. S1a). These results suggest that FABP1 is not a potential prognostic biomarker as least in ccRCC.
Fig. S1.
The correlation between the mRNA levels of FABP1, PDZK1 and T, M stage, recurrence, tumor maximum diameter and weight.
(a) FABP1 mRNA level was obtained from TCGA_KIRC dataset and the trend of expression level with the increase of T stage was analyzed by ANOVA. The differences of FABP1 mRNA level between ccRCC patients without and with distant metastasis, recurrence were analyzed by independent sample t-test. (b) PDZK1 mRNA level was obtained from TCGA_KIRC dataset and the trend of expression level with the increase of T stage was analyzed by ANOVA. The correlations of PDZK1 mRNA level with tumor volume, maximum diameter and weight were analyzed by Pearson correlation analysis. PDZK1 protein level was obtained from IHC of 112 cases. The correlations of PDZK1 protein level with tumor maximum diameter was analyzed by Pearson correlation analysis.
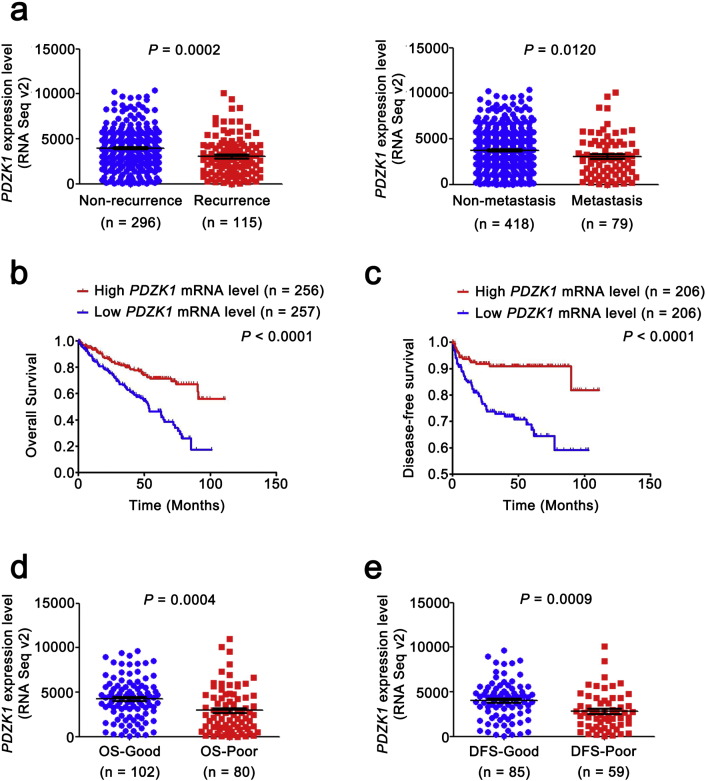
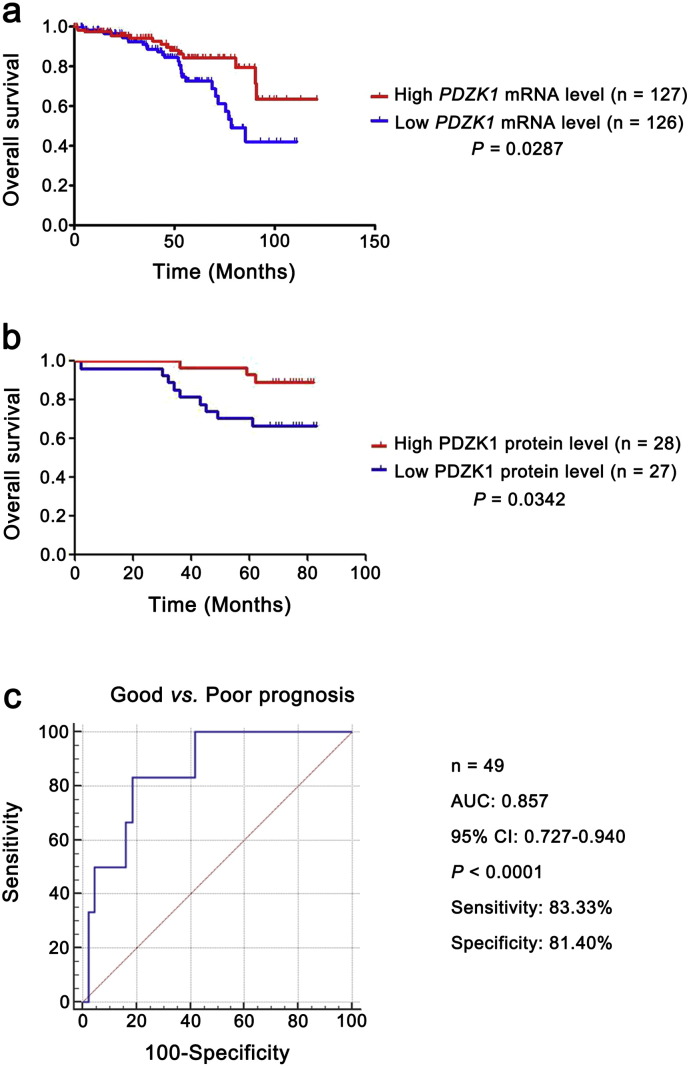
The PDZK1 mRNA level was decreased as T stage progressed and negatively correlated with the size and weight of tumors (Fig. S1b). It also exhibited differences between the patients with non-metastasis and metastasis (Fig. 3a), indicating that PDZK1 may be a potential prognostic biomarker for ccRCC. To investigate the prognostic significance of PDZK1, we compared PDZK1 mRNA level between patients with and without recurrence. PDZK1 mRNA expression was in significantly lower level with recurrent ccRCC (Fig. 3a).
Fig. 3.
Low level of PDZK1 mRNA predicts poor prognosis of ccRCC patients.
(a) The mRNA level of PDZK1 between ccRCC patients with and without recurrence, metastasis was compared by independent sample t-test. (b, c) Patients were divided into high and low groups by median value of PDZK1 mRNA level. Kaplan-Meier (KM) curves for OS and DFS of patients were performed. (d, e) The mRNA level of PDZK1 between ccRCC patients with good and poor prognosis for OS and DFS was compared by independent sample t-test.
To further evaluate the association of PDZK1 mRNA expression level with the survival time of ccRCC patients, KM survival curves were plotted. Patients were divided into ‘low’ and ‘high’ groups based on the median values of PDZK1 RNA seq quantification results. Patients with low PDZK1 mRNA level had shorter OS and DFS time (Fig. 3b, c, P < 0.0001), even for patients in early stage of ccRCC (Fig. S2a, P < 0.05). Moreover, the mRNA level of PDZK1 was able to classify ccRCC patients with a good or poor prognosis (Fig. 3d, e, P < 0.001).
Fig. S2.
The prognostic significance of PDZK1 expression levels for stage I ccRCC patients.
(a) KM curve for OS of patients in stage I with high and low PDZK1 mRNA level by median value. (b) KM curve for OS of patients in stage I with high and low PDZK1 protein level by median value. (c) That PDZK1 protein level stratified stage I ccRCC patients with good and poor prognosis for OS was analyzed by ROC curve.
3.7. Low Level of PDZK1 Protein in ccRCC Tissues is Validated by WB and IHC
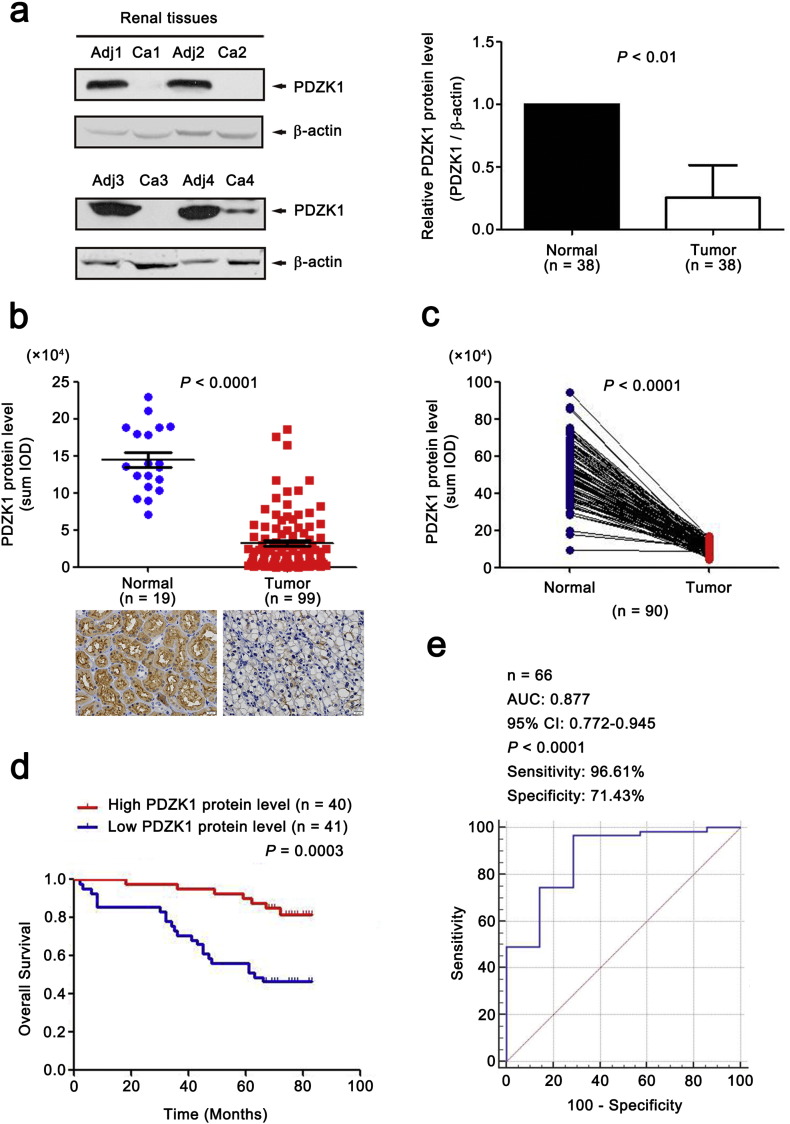
To further confirm our results, both WB and IHC assays were used on an expanded cohort of patients (n = 38 and 112, respectively). IHC was also performed on an independent set of 90 paired tissues. All results showed that PDZK1 protein level from ccRCC tissues was significantly lower than adjacent normal tissues (Fig. 4a,b,c, P < 0.01), and was gradually decreased with the increase of AJCC stage (data not shown).
Fig. 4.
Low level of PDZK1 protein predicts poor clinical outcome in patients with ccRCC.
(a) The protein level of PDZK1 in paired biopsies of tumor (Ca) and normal (Adj) tissues from 38 ccRCC patients were analyzed by WB analysis. Representative blots of PDZK1 protein level in normal and ccRCC tissues were shown. β-actin was used as a loading control. (b) IHC was performed with anti-PDZK1 antibody in 19 normal tissues and 112 tumor tissues. Outlier values (> Mean ± 3 SD) were removed. Representative IHC images (20 × magnification, all panels) were shown. (c) The protein level of PDZK1 in 90 paired ccRCC samples was detected with IHC. (d) Patients were divided into high and low groups by median value of PDZK1 protein level. KM survival curve for OS was performed. (e) ROC curve for PDZK1 protein level in classifying ccRCC patients with good and poor prognosis for OS. The area under the receiver operating characteristic curve (AUC) was 0.877 (95% CI, 0.772–0.945).
3.8. Low PDZK1 Protein Level Predicts Poor Prognosis of ccRCC Patients
To investigate the clinical relevance of PDZK1 protein level with prognosis in ccRCC patients, we further evaluated the OS time of ccRCC patients from the follow-up data via KM survival analysis. The patients were divided into ‘low’ and ‘high’ groups according to the median values of PDZK1 intensity/area. Low PDZK1 protein level was correlated with shorter OS time of ccRCC patients (Fig. 4d, P < 0.001). Moreover, PDZK1 protein level of ccRCC patients discriminated between good or poor OS prognosis with the AUC of 0.877 (Fig. 4e). Interestingly, the similar results were observed in the early stage of ccRCC patients (Figs. S2b,2c, P < 0.05, AUC 0.857). Consistent with the results of mRNA, the PDZK1 protein level was gradually decreased with the increase of the T stage (Fig. S1b).
3.9. PDZK1 is an Independent Prognostic Marker for ccRCC
The association between PDZK1 mRNA (Table 2), protein level (Table 3) and OS or DFS in the cohort of ccRCC patients was studied using univariate and multivariate analyses. Specimens were grouped into low and high PDZK1 expression categories according to the median value. Univariate analysis indicated that the patients with low PDZK1 level exhibited a shorter OS and DFS [PDZK1 mRNA OS: hazard ratio (HR) 0.415, P < 0.0001 and DFS HR 0.298, P < 0.0001; PDZK1 protein OS HR 0.236, P < 0.0001]. When controlling for other variables in the multivariate analysis, low PDZK1 mRNA and protein level retained its clinical significance as a marker of shorter survival (mRNA OS: HR 0.577, P = 0.002 and DFS HR 0.425, P = 0.002; protein OS HR 0.287, P = 0.003). Taken together, these data suggest that low PDZK1 expression level is an independent predictor of poor prognosis for ccRCC patients.
Table 2.
Univariate and multivariate analyses of PDZK1 mRNA level and patient survival.
| Variable | Univariate analysis |
Multivariate analysisc |
||||
|---|---|---|---|---|---|---|
| HRa | 95% CIb | P | HR | 95% CI | P | |
| Overall survival (n = 499) | ||||||
| Age (years) | ||||||
| ≤ 50 (n = 108) > 50 (n = 391) |
0.468 | 0.293–0.748 | 0.001 | 0.578 | 0.358–0.934 | 0.025 |
| Gender | ||||||
| Female (n = 175) Male (n = 324) |
1.028 | 0.747–1.414 | 0.866 | |||
| T stage | ||||||
| T1 or T2 (n = 316) T3 or T4 (n = 183) |
0.286 | 0.209–0.393 | 0.000 | 0.564 | 0.386–0.825 | 0.003 |
| N stage | ||||||
| N0 or NX (n = 483) N1 (n = 16) |
0.341 | 0.179–0.647 | 0.001 | 0.569 | 0.293–1.107 | 0.097 |
| M stage | ||||||
| M0 or MX (n = 421) M1 (n = 78) |
0.217 | 0.158–0.299 | 0.000 | 0.426 | (0.292–0.621 | 0.000 |
| G grade | ||||||
| G1 or G2 (n = 223) G3 or G4 (n = 276) |
0.360 | 0.252–0.514 | 0.000 | 0.566 | 0.385–0.831 | 0.004 |
| PDZK1 | ||||||
| High (n = 250) Low (n = 249) |
0.415 | 0.299–0.577 | 0.000 | 0.577 | 0.410–0.811 | 0.002 |
| Disease–free survival (n = 399) | ||||||
| Age (years) | ||||||
| ≤ 50 (n = 99) > 50 (n = 300) |
0.664 | 0.371–1.190 | 0.169 | |||
| Gender | ||||||
| Female (n = 133) Male (n = 266) |
0.911 | 0.556–1.494 | 0.713 | |||
| T stage | ||||||
| T1 or T2 (n = 262) T3 or T4 (n = 137) |
0.150 | 0.090–0.252 | 0.000 | 0.394 | 0.218–0.712 | 0.002 |
| N stage | ||||||
| N0 or NX (n = 385) N1 (n = 14) |
0.156 | 0.074–0.331 | 0.000 | 0.376 | 0.175–0.811 | 0.013 |
| M stage | ||||||
| M0 or MX (n = 347) M1 (n = 52) |
0.087 | 0.055–0.140 | 0.000 | 0.181 | 0.106–0.307 | 0.000 |
| G grade | ||||||
| G1 or G2 (n = 188) G3 or G4 (n = 211) |
0.197 | 0.106–0.365 | 0.000 | 0.354 | 0.185–0.675 | 0.002 |
| PDZK1 | ||||||
| High (n = 200) Low (n = 199) |
0.298 | 0.175–0.508 | 0.000 | 0.425 | 0.246–0.735 | 0.002 |
Hazard ratio, estimated from Cox proportional hazard regression model.
Confidence interval of the estimated HR.
Multivariate models were adjusted for T, N, M classification, age and gender.
Table 3.
Univariate and multivariate analyses of PDZK1 protein level and patient OS.
| Variable | Univariate analysis |
Multivariate analysisc |
||||
|---|---|---|---|---|---|---|
| HRa | 95% CIb | P | HR | 95% CI | P | |
| Overall survival (n = 77) | ||||||
| Age (years) | ||||||
| ≤ 60 (n = 40) > 60 (n = 37) |
0.467 | 0.215–1.012 | 0.054 | |||
| Gender | ||||||
| Female (n = 31) Male (n = 46) |
1.107 | 0.524–2.340 | 0.791 | |||
| Grade | ||||||
| G1 or G2 (n = 67) G3 or G4 (n = 10) |
0.157 | 0.070–0.351 | 0.000 | 0.547 | 0.202–1.477 | 0.234 |
| AJCC stage | ||||||
| I or II (n = 55) III or IV (n = 22) |
0.214 | 0.101–0.454 | 0.000 | 0.330 | 0.135–0.806 | 0.015 |
| PDZK1 | ||||||
| High (n = 47) Low (n = 30) |
0.236 | 0.108–0.513 | 0.000 | 0.287 | 0.125–0.657 | 0.003 |
Hazard ratio, estimated from Cox proportional hazard regression model.
Confidence interval of the estimated HR.
Multivariate models were adjusted for T, N, M classification, age and gender.
3.10. External Comparison of Biomarkers
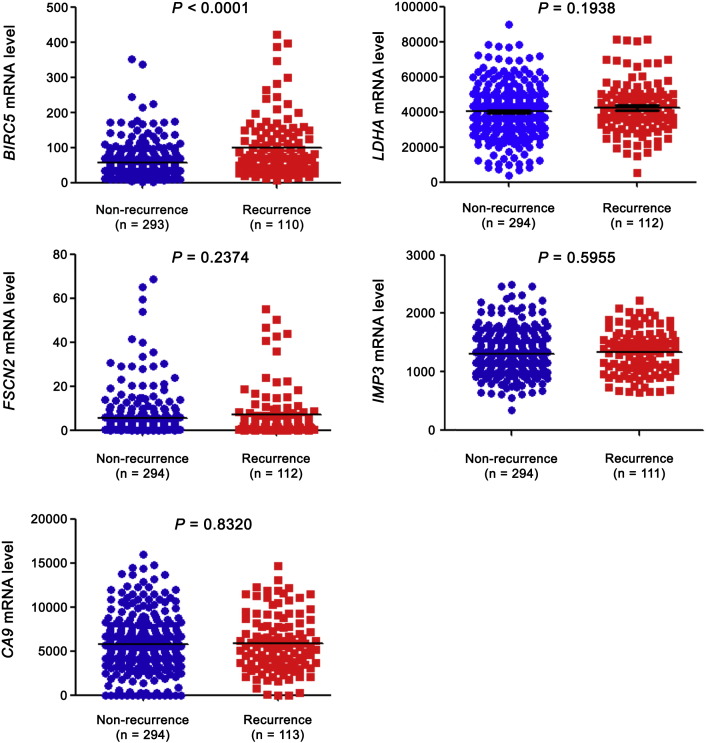
For external validation of PDZK1 as a prognostic biomarker in ccRCC, we compared it with five previously reported biomarkers. Fascin (FSCN2) (Jin et al., 2006), insulin-like growth factor mRNA binding protein 3 (IMP3) (Jiang et al., 2006) and carbonic anhydrase IX (CA9) (Zhang et al., 2013), etc. were the reported prognostic markers for ccRCC. However, these proteins failed to distinguish patients with good and poor prognosis in TCGA_KIRC dataset (Fig. 5). Although lactate dehydrogenase A (LDHA) (Girgis et al., 2014) could distinguish patients with good and poor prognosis for OS, it was not significant in identifying patients with poor prognosis for DFS or with recurrence (Fig. 5, Fig. 6). Survivin (BIRC5) (Parker et al., 2006) was able to discriminate between good and poor prognosis patients for both OS and DFS (Fig. 5).
Fig. 5.
External comparison of biomarkers-OS and DFS.
mRNA levels of five previously reported prognostic markers (BIRC5, LDHA, FSCN2, IMP3 and CA9) were extracted from TCGA_KIRC dataset. mRNA levels between patients with good and poor prognosis were compared by independent sample t-test. (a) for OS and (b) for DFS.
Fig. 6.
External comparison of biomarkers-recurrence.
mRNA levels of five previously reported prognostic markers (BIRC5, LDHA, FSCN2, IMP3 and CA9) were extracted from TCGA_KIRC dataset. mRNA levels between patients with and without recurrence were compared by independent sample t-test.
3.11. Comparison of DEP Identification Results Between Separate and Pooled Four Stage Samples
By comparison of ccRCC and their corresponding adjacent normal tissues separately in each of four stages, we generated the DEPs, which were consistently differentially expressed in each of four stages in this study. Meanwhile, we analyzed virtually pooled four stage samples, i.e., mixtures of biological samples of all four stages ccRCC tissues in one group and mixtures of all corresponding adjacent normal tissues in the other group. Some of DEPs found in virtually pooled groups were not differentially expressed in each of four stages, for example, aldose 1-epimerase was downregulated in pooled ccRCC group compared with pooled normal tissues group (Tumor/Normal: fold change (FC) 0.506, P < 0.0001), but not in ccRCC patient of stage IV (Tumor/Normal for stage I: FC 0.433, P < 0.0001; stage II: FC 0.424, P < 0.0001; stage III: FC 0.638, P < 0.0001; stage IV: no fold change, P = 0.174). Thus, analysis of consistent DEPs in each stage for separate four-stage samples could identify less DEPs than mixtures of biology samples of all four stages ccRCC tissues (n = 212 vs 593).
4. Discussion
In this study, PDZK1 was found to be downregulated in ccRCC tissues. The low PDZK1 level was strongly associated with a poor clinical outcome, especially in the early-stage of ccRCC patients.
Cell proliferation plays an important role in cancer development and progression (Feitelson et al., 2015). Proliferation-related proteins were able to predict a poor survival outcome in cancer patients (Yang et al., 2015). Downregulated proteins in ccRCC identified in this study are mainly involved in the lipid metabolism pathway, so we combined both GO cell proliferation annotation and lipid metabolism pathway to screen for prognostic markers in ccRCC and identified PDZK1 as a promising prognostic marker. PDZK1 could discriminate recurrence, metastasis and prognosis between ccRCC patients (Fig. 3, Fig. 4). In contrast, ccRCC prognostic markers proposed by other researchers failed to distinguish patients with good and poor prognosis (Fig. 5). The significance of LDHA in identifying patients with poor prognosis for DFS or with recurrence was insufficient. Survivin-BIRC5 was able to discriminate between good and poor prognosis patients for both OS and DFS, but its lower expression level made it not so easy to be detected using conventional method. Interestingly, both LDHA and BIRC5 were involved in lipid metabolism as revealed by KEGG pathway analysis and GSEA. These findings were consistent with previous reports–prognostic markers are usually metabolism pathway-related (Zaravinos et al., 2014), or pathway-derived metabolic products (Arsanious et al., 2009). This result further suggests that lipid metabolism pathway is important in screening prognostic markers for ccRCC. Indeed, we found that the downregulated expression level of PDZK1 is correlated with dysregulated lipid metabolism in ccRCC and PDZK1 may be a prognostic marker for ccRCC.
In this study, we identified FABP1 and PDZK1 as potential biomarkers by exploring the downregulated proteins involved in cell proliferation and the lipid metabolism pathway. Although the change of FABP1 mRNA level was not significant in prognosis prediction for ccRCC, it could distinguish ccRCC patients from normal individuals with AUC of 0.820 (95% CI 0.787–0.850). In addition, PDZK1 protein level could also discriminate ccRCC from normal tissues with AUC of 0.944 (95% CI, 0.906–0.982). Thus, the results of this study also verified the important role of cell proliferation and lipid metabolism-related phenotype in identifying biomarkers in ccRCC.
Compared with the strategy of analysis from pooled four stage samples, analysis of DEPs from separate four-stage ccRCC samples identified much less DEPs. Some DEPs found in pooled sample were not identified as differentially expressed in each of four stages, suggesting that not all DEPs found in pooled samples were appropriate as markers. Thus, it is our successful strategy to screen for biomarkers from DEPs which were consistently differentially expressed in each of four stages, and the previously reported markers, such as LDHA and FABP7 (Girgis et al., 2014, Zhou et al., 2015), were also in the list of identified DEPs in this study from the upregulated proteins (Table S1).
PDZK1 was expressed at the apical membrane of the renal proximal tubule cells, from which RCC originates. PDZK1 belong to PDZ proteins (Kocher et al., 1998), which play important roles in cell growth control, tumorigenesis and development (Yao et al., 2012). PDZK1 is also a member of Na+/H+ exchanger regulatory factor (NHERF) family. NHERFs were associated with malignant cell transformation (Zheng et al., 2010b, Yao et al., 2012), reminding that PDZK1 was correlated with ccRCC malignancy. PDZK1, as scavenger receptor class B type I (SR-BI)-binding protein, was also closely involved in the regulation of lipid metabolism (Kocher et al., 2008). Metabolism determined the biologically malignant behavior of multiple types of cancer (Ogawa et al., 2015). The disorder of metabolism related proteins, especially the abnormality of lipid metabolism related proteins was correlated with progression of RCC and could predict the prognosis of ccRCC patients (Yu et al., 2013). PDZK1 is most highly expressed in the kidney (Kocher et al., 1998), and significantly downregulated in ccRCC. PDZK1 could discriminate recurrence, metastasis and prognosis between ccRCC patients. Its role in ccRCC remains unknown. Further studies are required to clarify the role of PDZK1 in ccRCC tumorigenesis and progression. However, lines of evidence reported oncogenic activity for PDZK1 in breast cancer (Kim et al., 2013, OLeary et al., 2013). Thus, PDZK1 has been shown to play important and diverse roles in different tissues.
The drawback of this study was the lack of enough specimens with N1 or M1 stage or recurrence in the independent validation set of 81 samples, so we were unable to know if PDZK1 protein level could discriminate patients with lymph node involvement, distant metastasis or recurrence so far. In addition, ignorance of data collection for DFS time led to absence of KM-curve regarding PDZK1 protein level for DFS. These issues need to be further investigated in the future study.
In conclusion, PDZK1 is identified as an independent predictor for prognosis in ccRCC patients. These findings will facilitate patient counseling and individualize the management of patients with ccRCC.
The following are the supplementary data related to this article.
List of proteins differentially expressed in each of four stages for ccRCC samples
Supplementary tables
Funding Sources
This work was supported by the National Natural Science Foundation of the People's Republic of China (Nos. 81272887, 81372739, 81672521). These funding sources had no role in the study design; in the collection, analysis and interpretation of data; in the writing of the manuscript; or in the decision to submit the paper for publication.
Conflicts of Interest
The authors declare no conflicts of interest.
Author contributions
Zheng JF, Peng ZQ and He JQ designed the study. Zheng JF and He JQ analysed the data and wrote the report. All authors collected data, interpreted data, and approved the final report.
Acknowledgement
We highly appreciated the language editing of Dr. Qiong Qin.
References
- Arsanious A., Bjarnason G.A., Yousef G.M. From bench to bedside: current and future applications of molecular profiling in renal cell carcinoma. Mol. Cancer. 2009;8:20. doi: 10.1186/1476-4598-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggener S.E., Yossepowitch O., Pettus J.A., Snyder M.E., Motzer R.J., Russo P. Renal cell carcinoma recurrence after nephrectomy for localized disease: predicting survival from time of recurrence. J. Clin. Oncol. 2006;24(19):3101–3106. doi: 10.1200/JCO.2005.04.8280. [DOI] [PubMed] [Google Scholar]
- Feitelson M.A., Arzumanyan A., Kulathinal R.J., Blain S.W., Holcombe R.F., Mahajna J., Marino M., Martinez-Chantar M.L., Nawroth R., Sanchez-Garcia I., Sharma D., Saxena N.K., Singh N., Vlachostergios P.J., Guo S., Honoki K., Fujii H., Georgakilas A.G., Bilsland A., Amedei A., Niccolai E., Amin A., Ashraf S.S., Boosani C.S., Guha G., Ciriolo M.R., Aquilano K., Chen S., Mohammed S.I., Azmi A.S., Bhakta D., Halicka D., Keith W.N., Nowsheen S. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015;35:S25–S54. doi: 10.1016/j.semcancer.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlin R.A. Renal cell carcinoma: management of advanced disease. J. Urol. 1999;161(2):381–386. doi: 10.1016/s0022-5347(01)61897-4. [DOI] [PubMed] [Google Scholar]
- Frank I., Blute M.L., Leibovich B.C., Cheville J.C., Lohse C.M., Zincke H. Independent validation of the 2002 American Joint Committee on cancer primary tumor classification for renal cell carcinoma using a large, single institution cohort. J. Urol. 2005;173(6):1889–1892. doi: 10.1097/01.ju.0000158043.94525.d6. [DOI] [PubMed] [Google Scholar]
- Girgis H., Masui O., White N.M., Scorilas A., Rotondo F., Seivwright A., Gabril M., Filter E.R., Girgis A.H., Bjarnason G.A., Jewett M.A., Evans A., Al-Haddad S., Siu K.M., Yousef G.M. Lactate dehydrogenase A is a potential prognostic marker in clear cell renal cell carcinoma. Mol. Cancer. 2014;13:101. doi: 10.1186/1476-4598-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Chu P.G., Woda B.A., Rock K.L., Liu Q., Hsieh C.C., Li C., Chen W., Duan H.O., McDougal S., Wu C.L. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. Lancet Oncol. 2006;7(7):556–564. doi: 10.1016/S1470-2045(06)70732-X. [DOI] [PubMed] [Google Scholar]
- Jin J.S., Yu C.P., Sun G.H., Lin Y.F., Chiang H., Chao T.K., Tsai W.C., Sheu L.F. Increasing expression of fascin in renal cell carcinoma associated with clinicopathological parameters of aggressiveness. Histol. Histopathol. 2006;21(12):1287–1293. doi: 10.14670/HH-21.1287. [DOI] [PubMed] [Google Scholar]
- Kim H., Abd Elmageed Z.Y., Ju J., Naura A.S., Abdel-Mageed A.B., Varughese S., Paul D., Alahari S., Catling A., Kim J.G., Boulares A.H. PDZK1 is a novel factor in breast cancer that is indirectly regulated by estrogen through IGF-1R and promotes estrogen-mediated growth. Mol. Med. 2013;19(2):253–262. doi: 10.2119/molmed.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher O., Comella N., Tognazzi K., Brown L.F. Identification and partial characterization of PDZK1: a novel protein containing PDZ interaction domains. Lab. Investig. 1998;78(1):117–125. [PubMed] [Google Scholar]
- Kocher O., Yesilaltay A., Shen C.H., Zhang S., Daniels K., Pal R., Chen J., Krieger M. Influence of PDZK1 on lipoprotein metabolism and atherosclerosis. Biochim. Biophys. Acta Mol. basis Dis. 2008;1782(5):310–316. doi: 10.1016/j.bbadis.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.A., Slaton J.W., Swanson D.A., Dinney C.P.N. Stage specific guidelines for surveillance after radical nephrectomy for local renal cell carcinoma. J. Urol. 1998;159(4):1163–1167. [PubMed] [Google Scholar]
- Ma Q., Yang Y., Feng D., Zheng S., Meng R., Fa P., Zhao C., Liu H., Song R., Tao T., Yang L., Dai J., Wang S., Jiang W.G., He J. MAGI3 negatively regulates Wnt/beta-catenin signaling and suppresses malignant phenotypes of glioma cells. Oncotarget. 2015;6(34):35851–35865. doi: 10.18632/oncotarget.5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasian L.M., Motzer R.J., Gluck L., Mazumdar M., Vlamis V., Krown S.E. Interferon alfa-2a in advanced renal cell carcinoma: treatment results and survival in 159 patients with long-term follow-up. J. Clin. Oncol. 1993;11(7):1368–1375. doi: 10.1200/JCO.1993.11.7.1368. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Nagano H., Konno M., Eguchi H., Koseki J., Kawamoto K., Nishida N., Colvin H., Tomokuni A., Tomimaru Y., Hama N., Wada H., Marubashi S., Kobayashi S., Mori M., Doki Y., Ishii H. The combination of the expression of hexokinase 2 and pyruvate kinase M2 is a prognostic marker in patients with pancreatic cancer. Mol. Clin. Oncol. 2015;3(3):563–571. doi: 10.3892/mco.2015.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLeary P.C., Penny S.A., Dolan R.T., Kelly C.M., Madden S.F., Rexhepaj E., Brennan D.J., McCann A.H., Pontén F., Uhlén M., Zagozdzon R., Duffy M.J., Kell M.R., Jirström K., Gallagher W.M. Systematic antibody generation and validation via tissue microarray technology leading to identification of a novel protein prognostic panel in breast cancer. BMC Cancer. 2013;13:175. doi: 10.1186/1471-2407-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A.S., Kosari F., Lohse C.M., Thompson R.H., Kwon E.D., Murphy L., Riehle D.L., Blute M.L., Leibovich B.C., Vasmatzis G., Cheville J.C. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer. 2006;107(1):37–45. doi: 10.1002/cncr.21952. [DOI] [PubMed] [Google Scholar]
- Peng Z.Q., Wang Q.Q., Zhang Y., He J.Q., Zheng J.F. EBP50 interacts with EGFR and regulates EGFR signaling to affect the prognosis of cervical cancer (CC) patients. Int. J. Oncol. 2016;49(4):1737–1745. doi: 10.3892/ijo.2016.3655. [DOI] [PubMed] [Google Scholar]
- Sundelin J.P., Ståhlman M., Lundqvist A., Levin M., Parini P., Johansson M.E., Borén J. Increased expression of the very low-density lipoprotein receptor mediates lipid accumulation in clear-cell renal cell carcinoma. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo P., Cho Y.J., Tsherniak A., Greulich H., Ambrogio L., Meeteren N.S.V., Zhou T., Buxton A., Kool M., Meyerson M., Pomeroy S.L., Mesirov J.P. Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. J. Clin. Oncol. 2011;29(11):1415–1423. doi: 10.1200/JCO.2010.28.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui K.H., Shvarts O., Smith R.B., Figlin R.A., DeKernion J.B., Belldegrun A. Prognostic indicators for renal cell carcinoma: a multivariate analysis of 643 patients using the revised 1997 TNM staging criteria. J. Urol. 2000;163(4):1090–1095. doi: 10.1016/s0022-5347(05)67699-9. (quiz 1295) [DOI] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A.-K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Veeratterapillay R., Simren R., El-Sherif A., Johnson M.I., Soomro N., Heer R. Accuracy of the revised 2010 TNM classification in predicting the prognosis of patients treated for renal cell cancer in the north east of England. J. Clin. Pathol. 2012;65(4):367–371. doi: 10.1136/jclinpath-2011-200468. [DOI] [PubMed] [Google Scholar]
- Wettersten H.I., Hakimi A.A., Morin D., Bianchi C., Johnstone M.E., Donohoe D.R., Trott J.F., Abu Aboud O., Stirdivant S., Neri B., Wolfert R., Stewart B., Perego R., Hsieh J.J., Weiss R.H. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. 2015;75(12):2541–2552. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Lin H., Wu S., Lei F., Zhu X., Song L., Hong M., Guo L. Prostate tumor overexpressed 1 (PTOV1) is a novel prognostic marker for nasopharyngeal carcinoma progression and poor survival outcomes. PLoS One. 2015;10(8):1–13. doi: 10.1371/journal.pone.0136448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Feng D., Bian W., Yang L., Li Y., Yang Z., Xiong Y., Zheng J., Zhai R., He J. EBP50 inhibits EGF-induced breast cancer cell proliferation by blocking EGFR phosphorylation. Amino Acids. 2012;43(5):2027–2035. doi: 10.1007/s00726-012-1277-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Wang H., Zhao J., Yuan Y., Wang C., Li J., Zhang L., Zhang L., Li Q., Ye J. Expression of CIDE proteins in clear cell renal cell carcinoma and their prognostic significance. Mol. Cell. Biochem. 2013;378(1–2):145–151. doi: 10.1007/s11010-013-1605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaravinos A., Pieri M., Mourmouras N., Anastasiadou N., Zouvani I., Delakas D., Deltas C. Altered metabolic pathways in clear cell renal cell carcinoma: a meta-analysis and validation study focused on the deregulated genes and their associated networks. Oncoscience. 2014;1(2):117–131. doi: 10.18632/oncoscience.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.Y., Thompson R.H., Lohse C.M., Dronca R.S., Cheville J.C., Kwon E.D., Leibovich B.C. Carbonic anhydrase IX (CAIX) is not an independent predictor of outcome in patients with clear cell renal cell carcinoma (ccRCC) after long-term follow-up. BJU Int. 2013;111(7):1046–1053. doi: 10.1111/bju.12075. [DOI] [PubMed] [Google Scholar]
- Zheng J., Shen H., Xiong Y., Yang X., He J. The beta1-adrenergic receptor mediates extracellular signal-regulated kinase activation via Galphas. Amino Acids. 2010;38(1):75–84. doi: 10.1007/s00726-008-0207-6. [DOI] [PubMed] [Google Scholar]
- Zheng J., Sun L., Liu H., Huang Y., Li Y., He J. EBP50 exerts tumor suppressor activity by promoting cell apoptosis and retarding extracellular signal-regulated kinase activity. Amino Acids. 2010;38(4):1261–1268. doi: 10.1007/s00726-009-0437-2. [DOI] [PubMed] [Google Scholar]
- Zhou J., Deng Z., Chen Y., Gao Y., Wu D., Zhu G., Li L., Song W., Wang X., Wu K., He D. Overexpression of FABP7 promotes cell growth and predicts poor prognosis of clear cell renal cell carcinoma. Urol. Oncol. 2015;33(3):113.e9–113.e17. doi: 10.1016/j.urolonc.2014.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of proteins differentially expressed in each of four stages for ccRCC samples
Supplementary tables