Abstract
A joint analysis of all-atom molecular dynamics (MD) calculations and picosecond time-resolved x-ray structures was performed to gain single-molecule insights into mechanisms of protein function. Ensemble-averaged MD simulations of the L29F mutant of myoglobin after ligand dissociation reproduce the direction, amplitude, and time scales of crystallographically determined structural changes. This close agreement with experiments at comparable resolution in space and time validates the individual MD trajectories. From 1,700 single-molecule trajectories, we identified and structurally characterized a conformational switch that directs dissociated ligands to one of two nearby protein cavities. Subsequent ligand migration proceeds through a network of transiently interconnected internal cavities, with passage between them involving correlated protein–ligand motions. The simulations also suggest how picosecond protein motions modulate the functional dissociation of oxygen and suppress the geminate recombination of toxic carbon monoxide.
Beginning with the pioneering work of Frauenfelder and coworkers (1), myoglobin (Mb), an oxygen-storage protein found in muscle tissue, has served as a model system for probing the relations between protein structure, dynamics, and function (1–30). A ferrous heme located in the hydrophobic interior of this protein reversibly binds small gaseous ligands such as O2, CO, and NO. The photosensitive Fe–ligand bond has allowed transient intermediates to be studied by flash photolysis methods ranging from spectroscopy (1, 5–7) to x-ray crystallography (9–16). Since the seminal work of Case and Karplus (17), Mb has also been the target of molecular dynamics (MD) simulations (17–27). This combination of experimental and theoretical investigations has made Mb one of the most thoroughly studied proteins.
Ligand migration between the active site and the surrounding solvent is facile; yet, the static structure of Mb reveals no obvious channel large enough to facilitate ligand passage (1, 17, 31). Evidently, ligand migration requires conformational fluctuations that open and close channels through which the ligand can pass (17). Most of the protein side chains circumscribing the active site in mammalian Mb are highly conserved, suggesting that the structure and conformational flexibility of these residues is tuned to optimize the reversible binding of O2. The L29F mutant of Mb demonstrates how relatively small structural changes near the active site can have large functional effects. Replacing the highly conserved leucine in the 29 position with phenylalanine decreases the volume of the primary docking site, a cavity where ligands become transiently trapped after dissociating from the heme. This mutation leads to accelerated ligand-migration dynamics (15) and an unusually high O2-binding affinity (4). A structural explanation for the rapid ligand-migration dynamics was implicated by a series of picosecond time-resolved crystal structures of photolyzed L29F MbCO (Mb with carbon monoxide bound to the heme) (15). These structures, which represent an ensemble average of the ≈5 × 1013 protein molecules contained within the pump-illuminated volume of the protein crystal, provide a wealth of experimental data on protein structural changes in real time at atomic resolution (10, 15). However, a mechanistic understanding of protein function requires analysis of correlated motions in individual molecules, as can be obtained from MD simulations (2, 3). Here, we employ MD simulations to provide this single-molecule view.
Materials and Methods
Time-Resolved Crystallography. Diffraction data were acquired on the ID09B beamline at the European Synchrotron and Radiation Facility (ESRF) by using the “pump-probe” technique (15, 32). Briefly, a picosecond laser pulse (pump) that was focused onto a protein crystal (L29F MbCO, P6 space group) triggered ligand dissociation, and a time-delayed polychromatic x-ray pulse (probe) passing through the pump-illuminated volume produced a time-resolved Laue diffraction pattern that was recorded on a marCCD x-ray detector (marresearch, Norderstedt, Germany). The dissociated CO rebound to the Mb within a few milliseconds, allowing the pump–probe sequence to be repeated thousands of times. To increase the dynamic range of the diffraction images, the CCD was read after integrating 32 repeats of the pump–probe sequence. Images were acquired at 31 equally spaced orientations spanning a 30° range of crystal rotation. Each image contained ≈3,000 spots, the intensities of which measured Fourier components (structure factors) of the electron density within the unit cell. The complete set of images provided ≈5-fold redundant data for ≈20,000 structure factors; these data were sufficient to obtain, by means of inverse Fourier transformation, electron density maps with 1.8-Å resolution. The time resolution of the structural determination is limited by the x-ray pulse duration, which was ≈150 ps full width at half maximum (FWHM).
MD Simulation Protocol. The MD simulations were performed with the AMBER code (University of California, San Francisco) and parm94 (33) force fields, constant temperature (300 K) and pressure (1 bar), periodic boundary conditions, particle-mesh Ewald summation (34), and a 1-fs time step. We used the AMBER all-atom force field of Giammona and Case (35) for the CO-bound state of the heme and adapted their united-atom potential for the deoxy state. The crystal structure of the L29F MbCO (Protein Data Bank ID code 2SPL) (36) was solvated with 3,516 TIP3P water molecules (37) together with six Na+ and seven Cl- ions (38). During the final nanosecond of the 1.58-ns MbCO simulation, the time-averaged structure remained close to the crystal structure (α-carbon rms distance = 0.55 Å). Moreover, the conformations sampled by side chains in the vicinity of the heme pocket were symmetrically distributed about the crystal structure. To create starting structures for 17 photolyzed trajectories, we saved configurations from the equilibrium simulation of MbCO at 50-ps intervals, starting at 630 ps. Photolysis was simulated by instantaneously changing the heme potential from carboxy to deoxy (18, 21, 23) in 17 MD runs of 1.1-ns duration. To calculate the time-dependent branching probabilities that photolyzed CO appears in the primary docking site or in the Phe-29 site, we initiated 1,700 short (2 ps) photolysis trajectories (simulated at constant energy and volume) with starting structures sampled every 0.5 ps from the equilibrium MbCO trajectory. Each trajectory led to a single site, and the resulting binary probabilities for each site were time-averaged over a 50-ps moving window. For comparison, the branching probability was also calculated for 34 starting structures separated by 25 ps along the equilibrium trajectory by initiating 20 photolysis trajectories with random (Maxwell–Boltzmann) velocities assigned to each degree of freedom. The two methods gave consistent results.
To explore the effect of CO kinetic energy on the branching ratio, we created “cool” CO by energy-minimizing (19) the Fe position immediately after switching the heme potential from carboxy to deoxy (as compared with the “hot” CO, obtained without the energy-minimization step). We found that CO ejected with reduced kinetic energy samples the same internal cavities but in modestly different proportions.
To study the effect of the Fe out-of-plane motion and the resulting heme doming on the ligand dynamics, we performed 17 additional simulations of 1.1-ns duration for each of two modified deoxy-heme force fields: one with partial heme doming and the other without heme doming. As the heme doming decreased, the proportion of ligands that ventured into the Xe4 site increased and the number of ligands that escaped to the solvent decreased, but the qualitative aspects of ligand migration were largely unchanged. For comparison, we also performed a 1.95-ns equilibrium simulation of WT MbCO, from which 34 CO dissociation simulations of 1.1-ns duration were initiated.
Mapping of Internal Cavities. Internal cavities were identified by computing the time-averaged probability that a 1-Å spherical probe does not overlap with any protein atom. Rendering the spatial dependence of this probability produces a 3D map of sites where ligands might become transiently trapped.
Protocol for Converting MD Structures to Electron Density. Structures sampled every 0.5 ps from all 17 MD trajectories were aligned to the Cα backbone of the 2SPL model and then converted into electron-density distributions with the sfall program (ccp4 package, Daresbury Laboratories, Cheshire, U.K.). For this conversion, the 5-Gaussian approximation for the atomic electron density distribution was used. Time-dependent electron-density maps were constructed as a weighted sum of structures whose occupancies were assigned according to a normalized Gaussian function (75-ps standard deviation) centered at the experimental pump–probe delay time (100 ps, 316 ps, and 1 ns).
Nonlinear Mapping of Electron Density. A visually intuitive image-rendering scheme (39) was used to compare MD simulations with time-resolved x-ray structures. Briefly, the experimental and calculated electron densities were mapped nonlinearly to image brightness with the unphotolyzed and photolyzed states color-coded magenta and green, respectively. The nonlinear mapping of electron density preserves weak features (e.g., ligand-docking sites with low occupancy) that would otherwise disappear when rendered with conventional contour levels. The contrasting colors blend to white where they overlap, whereas atomic displacements are manifested as magenta-to-green color gradients.
Results and Discussion
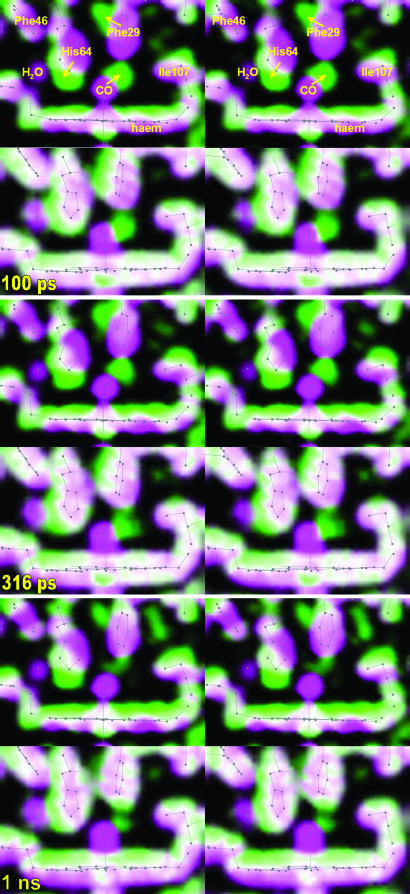
Photolysis-Induced Conformational Changes. To understand how the highly conserved side chains circumscribing the heme in Mb mediate diffusive ligand transport to and from the surrounding tissues, it is crucial to characterize the correlated motion of the ligand and protein. This information can be extracted from experimental and simulated time-resolved structures of photolyzed L29F MbCO, which are compared in stereo at three different time delays in Fig. 1. The magenta-to-green color gradients establish that the ensemble-averaged simulations reproduce the direction, amplitude, and time scale of atomic motion with high fidelity. [The sharper contrast of the experimental maps may stem from the use of a static, homogeneous structural model to phase the experimental structure factors (15)]. In the 100-ps structures, CO has moved from the binding site to the primary docking site located below Phe-29. The collision between unbound CO and Phe-29 causes that side chain to move toward His-64, which, in turn, is pushed away from the CO-binding site. Remarkably, the simulations even reproduce the concomitant loss of bound water near His-64, illustrating how protein motion can couple to the surrounding solvent. The strain induced by CO on the residues that confine it within the primary docking site expedites its departure. As CO migrates to other nearby sites, that strain is relieved, and the Phe-29 and His-64 side chains swing back toward their starting conformation. Whereas CO translocation triggers significant structural rearrangement on the distal side of the heme, downward displacement of the heme iron triggers rearrangement in both distal and proximal cavities: As the iron pushes His-93 downward, the heme recoils upward, dragging with it Ile-107 and the G-helix to which it is connected.
Fig. 1.
Stereoviews of protein structural changes recorded 100 ps, 316 ps, and 1 ns after photolysis of L29F MbCO. The two stereo panels at each time point depict electron densities determined by time-resolved x-ray crystallography (Upper) and MD simulations (Lower). The electron density of MbCO is colored magenta, and that of the photolyzed protein is colored green. Where both densities overlap, the two colors blend to white. The direction of molecular motion follows the magenta-to-green color gradient. The gray stick models correspond to the MbCO crystal structure (Upper) or the averaged simulated structure (Lower) and are included to guide the eye. Movies 1 and 2, which are published as supporting information on the PNAS web site, compare the time dependence of the experimental and simulated electron densities.
Ligand-Migration Pathways. Where does CO migrate after departing from the primary docking site? To answer this question, we first turn to results from MD simulations, as summarized in Fig. 2. Upon simulated photolysis, CO appears promptly in the primary docking site and, to a lesser degree, in a site located to the left of Phe-29. This site has not been observed in WT Mb (40) but has been seen in simulations of NO rebinding to L29F Mb, as reported by Li, Elber, and Straub (41). As CO escapes from the primary docking site, the population of CO in Xe4 and Phe-29 increases. Transient electron density in the time-resolved x-ray structures (Fig. 1) crossvalidates the presence of CO in both sites, although the assignment of the Phe-29 feature requires a careful analysis. For example, MD simulations show that Phe-29 is much more mobile while CO is localized in the primary docking site, with 17 ring flips occurring during 8.6 ns of simulations with CO in the primary docking site, and no ring flips occurring in 2.7 ns of simulations with CO either bound to the heme or free in solution. These frequent ring flips not only accelerate CO migration from the primary docking site to the Phe-29 site (see Fig. 3) but also contribute directly to the green electron density observed in the Phe-29 site. Persistence of that feature beyond the time required for Phe-29 to relax back toward its starting position (15) suggests that it is dominated by CO at 1 ns. Therefore, it is likely that the experimentally determined electron density in the Phe-29 site at 100 ps has contributions from both CO and rotated Phe-29. This assignment could not have been made unambiguously from experimental structures alone.
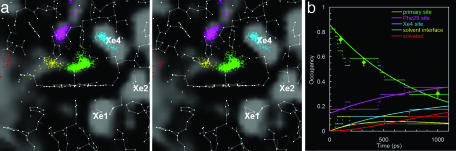
Fig. 2.
Stereoview of internal cavities (gray) and their time-dependent occupancies. (a) As CO migrates through the protein, it becomes transiently trapped in various internal cavities. Three of the cavities found in the MD simulations (labeled Xe1, Xe2, and Xe4) overlap with previously identified xenon docking sites (39). Snapshots of the CO location were uniformly sampled from 17 1.1-ns trajectories and are color-coded according to the site (see legend in b). (b) The time-dependent occupancies are well modeled (solid curves) by coupled first-order kinetic equations. The primary docking-site occupancy decays with a rate of (0.8 ns)-1. For comparison, the chart includes the experimental electron density in the primary docking site. Green diamonds with error bars, data normalized to 0–0.8.
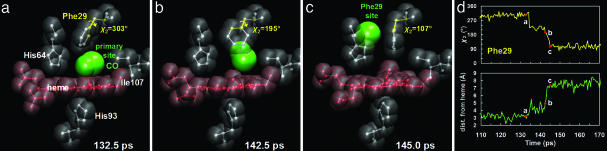
Fig. 3.
Simulated protein conformations before (a), during (b), and after (c) a CO migration event to the Phe-29 site. Rotation of the Phe-29 ring by Δχ2 ≈ 90° (d Upper) precedes CO migration to the Phe-29 site, as measured by the distance of the CO from the heme (d Lower). Movie 3, which is published as supporting information on the PNAS web site, depicts this correlated motion.
From the time-averaged probability that a small spherical probe does not overlap with any protein atom (Fig. 2a), we characterize a network of internal cavities where migrating ligands can become transiently trapped. The time-dependent occupancies in several of these sites have been extracted from the MD simulations, as shown in Fig. 2b. The (0.8 ns)-1 decay of the primary site population is in excellent agreement with the decay of the locally integrated electron density in the experimental maps (Fig. 2b). These rates are somewhat slower than the (0.14 ns)-1 rate found with femtosecond time-resolved IR spectroscopy (15); however, that determination was based on a spectroscopic change that may be sensitive to conformational changes as well as ligand migration. The sites labeled Xe1 and Xe2 in Fig. 2 have zero population, because the time scale for migration to these sites is well beyond the 1.1-ns duration of the MD simulation. The picture that emerges from the simulations is a network of transiently interconnected internal cavities through which a ligand can migrate, with its translocation slaved to conformational fluctuations that open and close interconnecting channels.
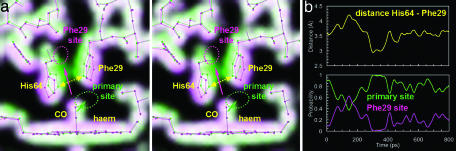
Conformational Gating of Ligand Migration. The CO trajectories that immediately follow simulated photolysis lead either to the primary docking site or to the Phe-29 site, with a 4:1 time-averaged ratio. The single-molecule simulations show that the partitioning between the two sites is time-dependent (Fig. 4b). To visualize the structural changes that control the ligand-migration pathway, time-averaged structures were constructed and color-coded according to the probability that the trajectory leads to the primary docking site (green) or to the Phe-29 site (magenta). The magenta-to-green color gradient unveils the conformational changes that favor the primary docking site: a narrowing of the channel between the bound CO and the Phe-29 docking site and a downward displacement of the heme. The time dependence of the partitioning is highly correlated with the His-64–Phe-29 separation (Fig. 4b; correlation coefficient of 0.88). This observation illustrates how modest (≈1-Å) protein-structure fluctuations occurring on a variety of time scales modulate a conformational switch that controls ligand-migration pathways.
Fig. 4.
Stereoview and time dependence of the conformational switch that gates the CO migration pathway. (a) The colored stick models and their associated electron density represent the average of all MbCO structures from which the dissociated CO ends up in the primary docking site (green) or the Phe-29 site (magenta). The green-to-magenta color gradient unveils the conformational motion responsible for biasing CO toward the Phe-29 site. (b) Time dependence of the nearest atom separation between His-64 and Phe-29 (Upper), and the time-dependent probability (Lower) that the CO ends up in the primary docking site (green) or the Phe-29 site (magenta); the curves represent a 50-ps moving average. When time-averaged over the MbCO equilibrium trajectory, the partitioning probabilities reproduce the occupancies at t = 0 shown in Fig. 2b.
Paths leading to the Xe4 and Phe-29 sites dominate the first stage of CO migration; however, CO manages to slip around His-64 to the solvent interface in 3 of 17 trajectories. It has been suggested that an out-of-pocket His-64 conformation (42) might open a pathway for ligand escape into the surrounding solvent (31). Although the His-64 gate is not “open” in Fig. 1, its outward motion may leave the gate slightly ajar. Only after CO departs from the primary docking site does His-64 move toward the heme-binding site in the L29F mutant (see Movie 3 and x-ray movie s1 in the supporting online material of ref. 15) and block the shortcut to the solvent interface. Indeed, in WT Mb, where this conformation is adopted promptly after photolysis (39), we have not observed a single CO escape into the surrounding solvent in any of 34 trajectories of 1.1-ns duration.
Suppression of Geminate Recombination. Both WT and L29F Mb suppress geminate recombination of toxic CO, but the mechanism used by L29F Mb is more effective. In WT Mb, His-64 moves promptly to block access to the binding site, whereas in the L29F mutant, the Phe-29 residue sweeps CO away from the docking site, and then His-64 moves into position to block access to the binding site. It is remarkable how such a modest mutation can alter so dramatically both the structural changes that ensue and the mechanistic consequences of those structural changes.
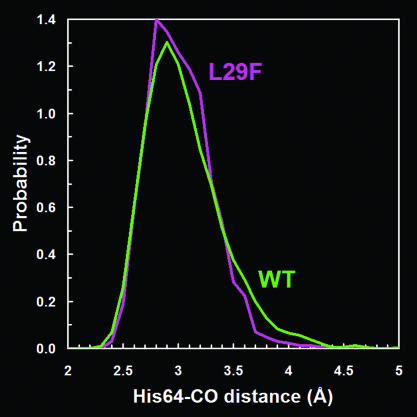
Modulation of O2-Binding Affinity. The L29F mutation improves 10-fold its discrimination against toxic CO (4); however, this improvement arises primarily from a 14-fold increase in the O2-binding affinity (4), which slows the release of O2 to the extent that it becomes detrimental to the protein's function. The increased O2-binding affinity in L29F Mb is largely accounted for by a 10-fold decrease in its “off” rate (4). It has been suggested that the O2-binding affinity is enhanced by a stabilizing H bond with the Nε-H tautomer of His-64 (43, 44). Conformational fluctuations that displace His-64 out of H-bonding distance would lower the thermal barrier to dissociation and would speed up the off rate. According to simulations, the probability distribution for the His-64–CO distance has a tail toward longer distances that is significantly larger in WT than in L29F MbCO (Fig. 5). Evidently, the side-chain packing in the L29F mutant suppresses His-64 excursions out of H-bonding distance with the bound ligand, thereby keeping the gate (1, 8) for thermal dissociation closed and slowing the ligand off rate. This result must be interpreted with caution, because the distance distributions were computed from single trajectories of MbCO. Moreover, the lack of an explicit H-bonding potential between CO and His-64 likely enhanced the probability of His-64 excursions in both WT and L29F MbCO.
Fig. 5.
Probability distribution of CO–His-64 distances computed from MD simulations of WT and L29F MbCO. The distance refers to the separation between the O atom and the nearest heavy atom of the His-64 ring.
Conclusion
The fidelity of the observed agreement between time-resolved x-ray structures and MD simulation is remarkable in both space and time, demonstrating the level of accuracy achievable in current simulations. By translating ensemble-averaged MD trajectories into electron-density maps, weak features in the experimental maps can be assigned with confidence to functional protein motions and/or ligand migration. Because individual MD simulations track structural changes at the single-molecule level, they provide detailed pictures of transient intermediates and the transitions between them. This information allowed us to characterize the structure of a conformational gate that directs the ligand-migration pathway at the time of dissociation. Without the picosecond time-resolved x-ray structures, insights gained from the simulations would not be nearly so compelling. Without the simulations, weak features in the experimental structures would be far more difficult to interpret. This unique combination of simulation and experiment unveils functional protein motions and illustrates at an atomic level the relationship among protein structure, dynamics, and function.
Supplementary Material
Acknowledgments
We thank W. A. Eaton and A. Szabo for helpful discussions, J. S. Olson and J. Soman (both of Rice University, Houston) for providing L29F MbCO crystals, and M. Wulff for supporting our efforts to develop the method of picosecond time-resolved x-ray crystallography.
Author contributions: G.H., F.S., and P.A.A. designed research; G.H., F.S., and P.A.A. performed research; G.H., F.S., and P.A.A. analyzed data; and G.H. and P.A.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mb, myoglobin; MbCO, Mb with carbon monoxide bound to the heme; MD, molecular dynamics.
References
- 1.Austin, R. H., Beeson, K. W., Eisenstein, L., Frauenfelder, H. & Gunsalus, I. C. (1975) Biochemistry 14, 5355-5373. [DOI] [PubMed] [Google Scholar]
- 2.Brooks, C. L., III, Karplus, M. & Pettitt, B. M. (1988) Proteins: A Theoretical Perspective of Dynamics, Structure, and Thermodynamics (Wiley, New York).
- 3.McCammon, J. A. & Harvey, S. C. (1987) Dynamics of Proteins and Nucleic Acids (Cambridge Univ. Press, New York).
- 4.Carver, T. E., Brantley, R. E., Jr., Singleton, E. W., Arduini, R. M., Quillin, M. L., Phillips, G. N., Jr., & Olson, J. S. (1992) J. Biol. Chem. 267, 14443-14450. [PubMed] [Google Scholar]
- 5.Lim, M., Jackson, T. A. & Anfinrud, P. A. (1995) Science 269, 962-966. [DOI] [PubMed] [Google Scholar]
- 6.Lim, M., Jackson, T. A. & Anfinrud, P. A. (1997) Nat. Struct. Biol. 4, 209-214. [DOI] [PubMed] [Google Scholar]
- 7.Merchant, K. A., Noid, W. G., Akiyama, R., Finkelstein, I. J., Goun, A., McClain, B. L., Loring, R. F. & Fayer, M. D. (2003) J. Am. Chem. Soc. 125, 13804-13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frauenfelder, H., Sligar, S. G. & Wolynes, P. G. (1991) Science 254, 1598-1603. [DOI] [PubMed] [Google Scholar]
- 9.Schlichting, I., Berendzen, J., Phillips, G. N., Jr., & Sweet, R. M. (1994) Nature 371, 808-812. [DOI] [PubMed] [Google Scholar]
- 10.Srajer, V., Teng, T., Ursby, T., Pradervand, C., Ren, Z., Adachi, S., Schildkamp, W., Bourgeois, D., Wulff, M. & Moffat, K. (1996) Science 274, 1726-1729. [DOI] [PubMed] [Google Scholar]
- 11.Chu, K., Vojtchovsky, J., McMahon, B. H., Sweet, R. M., Berendzen, J. & Schlichting, I. (2000) Nature 403, 921-923. [DOI] [PubMed] [Google Scholar]
- 12.Ostermann, A., Waschipky, R., Parak, F. G. & Nienhaus, G. U. (2000) Nature 404, 205-208. [DOI] [PubMed] [Google Scholar]
- 13.Brunori, M., Vallone, B., Cutruzzola, F., Travaglini-Allocatelli, C., Berendzen, J., Chu, K., Sweet, R. M. & Schlichting, I. (2000) Proc. Natl. Acad. Sci. USA 97, 2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srajer, V., Ren, Z., Teng, T. Y., Schmidt, M., Ursby, T., Bourgeois, D., Pradervand, C., Schildkamp, W., Wulff, M. & Moffat, K. (2001) Biochemistry 40, 13802-13815. [DOI] [PubMed] [Google Scholar]
- 15.Schotte, F., Lim, M., Jackson, T. A., Smirnov, A. V., Soman, J., Olson, J. S., Phillips, G. N., Jr., Wulff, M. & Anfinrud, P. A. (2003) Science 300, 1944-1947. [DOI] [PubMed] [Google Scholar]
- 16.Bourgeois, D., Vallone, B., Schotte, F., Arcovito, A., Miele, A. E., Sciara, G., Wulff, M., Anfinrud, P. & Brunori, M. (2003) Proc. Natl. Acad. Sci. USA 100, 8704-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Case, D. A. & Karplus, M. (1979) J. Mol. Biol. 132, 343-368. [DOI] [PubMed] [Google Scholar]
- 18.Henry, E. R., Levitt, M. & Eaton, W. A. (1985) Proc. Natl. Acad. Sci. USA 82, 2034-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elber, R. & Karplus, M. (1990) J. Am. Chem. Soc. 112, 9161-9175. [Google Scholar]
- 20.Straub, J. E. & Karplus, M. (1991) Chem. Phys. 158, 221-248. [Google Scholar]
- 21.Schaad, O., Zhou, H. X., Szabo, A., Eaton, W. A. & Henry, E. R. (1993) Proc. Natl. Acad. Sci. USA 90, 9547-9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma, J., Huo, S. & Straub, J. E. (1997) J. Am. Chem. Soc. 119, 2541-2551. [Google Scholar]
- 23.Vitkup, D., Petsko, G. A. & Karplus, M. (1997) Nat. Struct. Biol. 4, 202-208. [DOI] [PubMed] [Google Scholar]
- 24.Meller, J. & Elber, R. (1998) Biophys. J. 74, 789-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nutt, D. R. & Meuwly, M. (2003) Biophys. J. 85, 3612-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nutt, D. R. & Meuwly, M. (2004) Proc. Natl. Acad. Sci. USA 101, 5998-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossa, C., Anselmi, M., Roccatano, D., Amadei, A., Vallone, B., Brunori, M. & Di Nola, A. (2004) Biophys. J. 86, 3855-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinbach, P. J. & Brooks, B. R. (1993) Proc. Natl. Acad. Sci. USA 90, 9135-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kachalova, G. S., Popov, A. N. & Bartunik, H. D. (1999) Science 284, 473-476. [DOI] [PubMed] [Google Scholar]
- 30.Spiro, T. G. & Kozlowski, P. M. (2001) Acc. Chem. Res. 34, 137-144. [DOI] [PubMed] [Google Scholar]
- 31.Perutz, M. F. & Mathews, F. S. (1966) J. Mol. Biol. 21, 199-202. [DOI] [PubMed] [Google Scholar]
- 32.Bourgeois, D., Ursby, T., Wulff, M., Pradervand, C., Legrand, A., Schildkamp, W., Laboure, S., Srajer, S., Teng, T. Y., Roth, M. & Moffat, K. (1996) J. Synchrotron Radiat. 3, 65-74. [DOI] [PubMed] [Google Scholar]
- 33.Cornell, W. D., Cieplak, P., Bayly, C. I., Gould, I. R., Merz, K. M., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179-5197. [Google Scholar]
- 34.Darden, T., York, D. & Pedersen, L. (1993) J. Chem. Phys. 98, 10089-10092. [Google Scholar]
- 35.Giammona, D. A. (1984) Ph.D. thesis (Univ. of California, Davis).
- 36.Quillin, M. L., Arduini, R. M., Olson, J. S. & Phillips, G. N., Jr. (1993) J. Mol. Biol. 234, 140-155. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926-935. [Google Scholar]
- 38.Straatsma, T. P. & Berendsen, H. J. C. (1988) J. Chem. Phys. 89, 5876-5886. [Google Scholar]
- 39.Schotte, F., Soman, J., Olson, J. S. & Anfinrud, P. A. (2004) J. Struct. Biol. 127, 235-246. [DOI] [PubMed] [Google Scholar]
- 40.Tilton, R. F., Jr., Kuntz, I. D., Jr., & Petsko, G. A. (1984) Biochemistry 23, 2849-2857. [DOI] [PubMed] [Google Scholar]
- 41.Li, H., Elber, R. & Straub, J. E. (1993) J. Biol. Chem. 268, 17908-17916. [PubMed] [Google Scholar]
- 42.Yang, F. & Phillips, G. N., Jr. (1996) J. Mol. Biol. 256, 762-774. [DOI] [PubMed] [Google Scholar]
- 43.Pauling, L. (1964) Nature 203, 182-183.14207238 [Google Scholar]
- 44.Springer, B. A., Sligar, S. G., Olson, J. S. & Phillips, G. N., Jr. (1994) Chem. Rev. 94, 699-714. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.