Abstract
Transcriptome studies of psoriasis have identified robust changes in mRNA expression through large-scale analysis of patient cohorts. These studies, however, have analyzed all mRNA changes in aggregate, without distinguishing between disease-specific and non-specific differentially expressed genes (DEGs). In this study, RNA-seq meta-analysis was used to identify (1) psoriasis-specific DEGs altered in few diseases besides psoriasis and (2) non-specific DEGs similarly altered in many other skin conditions. We show that few cutaneous DEGs are psoriasis-specific and that the two DEG classes differ in their cell type and cytokine associations. Psoriasis-specific DEGs are expressed by keratinocytes and induced by IL-17A, whereas non-specific DEGs are expressed by inflammatory cells and induced by IFN-gamma and TNF. PBMC-derived DEGs were more psoriasis-specific than cutaneous DEGs. Nonetheless, PBMC DEGs associated with MHC class I and NK cells were commonly downregulated in psoriasis and other autoimmune diseases (e.g., multiple sclerosis, sarcoidosis and juvenile rheumatoid arthritis). These findings demonstrate “cross-disease” transcriptomics as an approach to gain insights into the cutaneous and non-cutaneous psoriasis transcriptomes. This highlighted unique contributions of IL-17A to the cytokine network and uncovered a blood-based gene signature that links psoriasis to other diseases of autoimmunity.
Keywords: RNA-seq, interferon, IL-17A, natural killer cells, PBMC
Introduction
Psoriasis is a cytokine-driven skin disease in which lesions develop due to abnormal epidermal thickening and keratinocyte (KC) hyperproliferation. Transcriptome studies of psoriasis have been performed using large patient cohorts to understand how gene expression is altered in lesional as compared to macroscopically normal skin (Li et al., 2014; Swindell et al., 2015b; Tian et al., 2012). These studies have been instrumental for identifying genes (Tian et al., 2012), pathways and transcription factors altered in lesional skin (Swindell et al., 2015b), treatment response biomarkers (Swindell et al., 2013a), and for developing new bioinformatic methods (Swindell et al., 2013a). Nearly all prior psoriasis transcriptome studies, however, have focused exclusively on psoriasis, or psoriasis in combination with one or two other inflammatory conditions (e.g., atopic dermatitis) (Choy et al., 2012; Dhingra et al., 2013; Nomura et al., 2003). Broader investigations seeking to establish connections between psoriasis and more distantly related skin conditions, such as skin cancers, have been less common (Haider et al., 2006). Transcriptomic features of psoriasis, however, overlap strongly with other skin conditions (e.g., squamous cell carcinoma, atopic dermatitis, acne), even though mechanisms governing the etiology of psoriasis are distinct (Swindell et al., 2014a). This raises the question of whether aggregate analysis of psoriasis DEGs will inform our understanding of disease etiology, since such DEGs may reflect generic cutaneous responses that occur in many skin diseases in the absence of shared causal mechanisms (D'Erme et al., 2015; Swindell et al., 2015a).
Disease-specific and non-specific signals in transcriptome data can be identified by “cross disease transcriptomics” (Swindell et al., 2015a). This approach emphasizes comparison of expression data across disease states and draws upon the wealth of transcriptome data archived in public databases (e.g., GEO) (Rung and Brazma, 2013). Large-scale analyses of such data have not focused on psoriasis, but have nonetheless shed light on functional aspects of transcriptome overlap between psoriasis and other skin diseases (Inkeles et al., 2015; Wong et al., 2012). Wong et al. (2012) identified genes differentially expressed between dermatomyositis and normal skin, and compared these to expression profiles from other skin conditions (e.g., lupus, atopic dermatitis). This showed that genes with similarly altered expression in psoriasis and other diseases are associated with epidermal barrier, interferon, and lipid metabolism (Wong et al., 2012). More recently, Inkeles et al. (2015) compared microarray gene expression profiles from normal skin and 16 human skin diseases. Interestingly, cluster analysis grouped psoriasis expression profiles with neoplastic skin diseases (e.g., squamous cell carcinoma and basal cell carcinoma) rather than prototypical inflammatory conditions (e.g., atopic dermatitis) (Inkeles et al., 2015). These findings highlight proliferative and inflammatory processes shared between psoriasis and other diseases, which together contribute to transcriptome overlap and convolution of disease-specific and non-specific signals (Haider et al., 2006).
This study identifies psoriasis DEGs using RNA-seq meta-analysis and applies cross-disease transcriptomics to stratify DEGs based on psoriasis-specificity. RNA-seq analysis of psoriasis lesions has been reported previously (Jabbari et al., 2012; Keermann et al., 2015; Li et al., 2014; Swindell et al., 2014b; Tsoi et al., 2015), but these data have not been integrated or compared systematically. We here integrate these data and identify protein-coding psoriasis DEGs based upon the meta-dataset (n = 44 patients). We then discriminate between psoriasis-specific and non-specific DEGs and characterize their respective functional properties and cell type associations. With this approach, we illustrate a generally applicable strategy for skin disease transcriptome analysis. This disentangles disease-specific expression patterns from those generically observed in all skin conditions, leads to functional insights not discernable from aggregate analysis of all psoriasis DEGs, and represents one useful in silico pathway towards large-scale integration of gene expression data in dermatology.
Results
RNA-seq meta-analysis of lesional (PP) and uninvolved (PN) skin
Meta-analysis was performed using RNA-seq reads generated from cDNA of paired lesional (PP) and uninvolved (PN) skin biopsies (GSE41745, GSE54456/GSE63979 and GSE66511; n = 44 patients) (Jabbari et al., 2012; Li et al., 2014; Swindell et al., 2015a; Tsoi et al., 2015). 15643 protein-coding genes satisfied our criteria as expressed features, with detectable expression in at least 25% of samples (see Methods). Samples from the three datasets differed with respect to the first FPKM principal component axis (Figure S1A), but PP versus PN differences showed good agreement across data sources (Figures S1B and S1C). Expected trends were observed among genes known to be altered in psoriasis lesions from previous work (Figure S1B) (Swindell et al., 2015b).
RNA-seq meta-analysis FC estimates (n = 44 patients) were compared to those from a prior microarray data meta-analysis (n = 237 patients) (Swindell et al., 2015b). RNA-seq meta-analysis FC estimates were highly correlated with those obtained by microarray (rs = 0.837; Figure S2). For each RNA-seq dataset individually, however, FC estimates were less strongly correlated (0.678 ≤ rs ≤ 0.798; Figure S2). Meta-analysis of RNA-seq data thus improved correspondence to microarray findings.
RNA-seq meta-analysis identifies 2113 differentially expressed protein-coding genes in psoriasis lesions (PP) as compared to uninvolved skin (PN)
The 15643 expressed genes were analyzed to assess evidence for differential expression (PP vs. PN skin), leading to the identification of 2113 differentially expressed genes (DEGs), including 954 PP-increased DEGs (FC > 2.0 with FDR < 0.05) and 1159 PP-decreased DEGs (FC < 0.50 with FDR < 0.05). Most meta-analysis DEGs could be identified from analysis of at least one individual RNA-seq dataset, although some were uniquely identified by meta-analysis (e.g., PP-increased: CXCR5, BATF, PNMA5; PP-decreased: KRT13, FSTL5, GPC4; Figure S3). Consistent with prior work (Swindell et al., 2014b), expression of long genes tended to be decreased in PP skin (Figure S4). There was no indication of GC content bias among DEGs identified from the pooled meta-dataset (Figure S5), even though this was observed among DEGs from one individual dataset (i.e., GSE54456/GSE63979; Figure S4). RNA-seq meta-analysis thus attenuated GC content bias.
Genes with weak expression were more frequently identified as PP-decreased DEGs (Figure S4C). For instance, among genes with mean FPKM < 0.20 (PP and PN samples), 21.1% were PP-decreased DEGs; in contrast, among genes with mean FPKM > 100, only 6.1% were PP-decreased DEGs (Figure S4C). PP-decreased DEGs also showed weaker overlap with microarray findings as compared to PP-increased DEGs (Figure S4D), with PP-decreased DEGs identified specifically by RNA-seq having relatively low expression (FPKM < 1.30 on average; Figure S4E). Comparison of FC estimates between RNA-seq and microarray also supported weaker correspondence among low-expressed DEGs (Figure S4F).
Genes differentially expressed in psoriasis lesions overlap strongly with those altered in neoplastic and other inflammatory skin diseases
We expected that many psoriasis DEGs would be non-specific and similarly altered in other skin diseases. We thus screened 98 gene lists derived from microarray comparisons between diseased and normal skin (Supplemental Data File 2), with the goal of identifying diseases mirroring psoriasis (i.e., elevation of PP-increased DEGs; repression of PP-decreased DEGs). This revealed strong transcriptome-level correspondence between psoriasis and diverse skin conditions (P < 10−200), including Mediterranean spotted fever eschars, acne, infection, eczema, wounding, atopic dermatitis and squamous cell carcinoma (SCC) (Figure 1A).
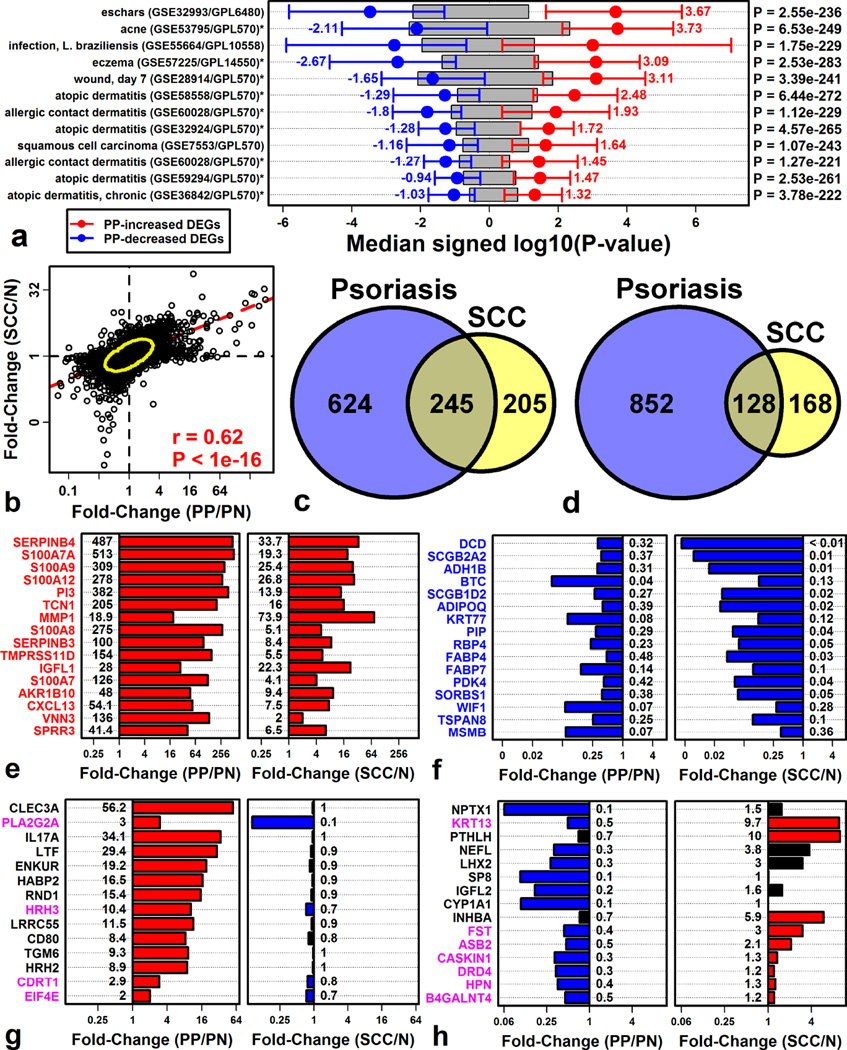
Figure 1. Gene expression changes in psoriasis lesions overlap significantly with those in squamous cell carcinoma (SCC).
(A) Skin diseases with psoriasis-like gene expression changes. Shifts in gene expression were quantified using signed log10-transformed p-values (Log10P; involved skin vs. control). Log10P was compared between PP-increased and PP-decreased DEGs (right margin, Wilcoxon rank sum test). Red and blue bars span the middle 50% of Log10P values among PP-increased and PP-decreased DEGs, respectively (grey boxes: non-DEGs). (B) FC comparison between SCC (GSE7553) and psoriasis (RNA-seq meta-analysis). (C) Overlap between PP-increased and SCC-increased DEGs. (D) Overlap between PP-decreased and SCC-decreased DEGs. Parts (E) – (H) show genes similarly (E and F) and oppositely (G and H) altered in each disease (red/blue bars: FDR < 0.05).
SCC, for example, is etiologically distinct from psoriasis (Arlette and Trotter, 2004), but despite this, PP-increased DEGs were biased towards SCC-increased expression, and likewise, PP-decreased DEGs were biased towards SCC-decreased expression (Figure 1A). Genome-wide FCs also correlated between the two diseases (rs = 0.62; PP/N versus SCC/N; Figure 1B). Genes elevated in both conditions included SERPINB4, S100A7A, and S100A9 (Figure 1E), while genes decreased in both conditions included DCD, SCGB2A2, and ADH1B (Figure 1F). Notably, IL17A was included among genes altered in psoriasis but not similarly altered in SCC (Figures 1G and 1H).
A psoriasis specificity index (PSI) discriminates between psoriasis-specific and non-specific DEGs to highlight distinct gene set functional associations
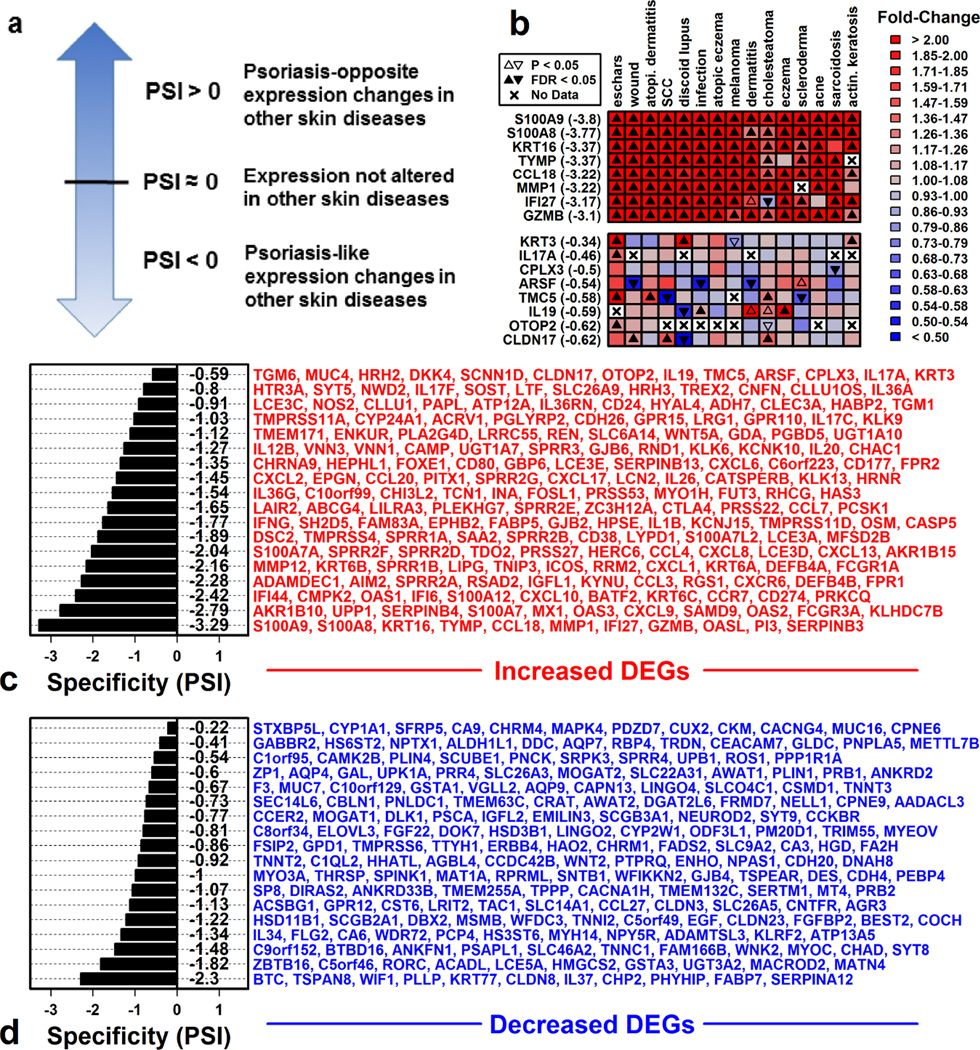
We next asked whether psoriasis-specific DEGs differ functionally from non-specific DEGs. To address this, we developed a psoriasis specificity index (PSI) for DEG stratification (Figure 2), with lower values (PSI < 0) indicating relatively non-specific DEGs and higher values (PSI ≈ 0 or PSI > 0) indicating relatively greater psoriasis-specificity (Figure 2A). Applying this index indicated that most psoriasis DEGs are non-specific (PSI < 0). Average PSI among PP-increased and PP-decreased DEGs, for example, was −1.44 (± 0.023) and −0.86 (± 0.014), respectively (Figure S6). Highly non-specific DEGs included S100A9 (PSI = −3.80), KRT16 (PSI = −3.37), BTC (PSI = −2.67) and TSPAN8 (PSI = −2.66) (Figures 2 and S7). Although few DEGs were strongly psoriasis-specific (PSI ≈ 0 or PSI > 0), several DEGs with highest PSI encoded cytokines, e.g., IL17A (PSI = −0.46), IL19 (PSI = −0.59), and IL36A (PSI = −0.73) (Figure 2C).
Figure 2. Psoriasis specificity index (PSI) discriminates among cutaneous DEGs based upon disease-specificity.
(A) PSI discriminates among DEGs with psoriasis-like gene expression shifts in other skin diseases (PSI < 0), DEGs unaltered in other skin diseases (PSI ≈ 0), and DEGs with psoriasis-opposite gene expression shifts in other diseases (PSI > 0). (B) Example PP-increased DEGs with low (top) and high (bottom) PSI and their expression shifts in other skin diseases. PSI for each gene is listed in parentheses (left margin). (C) 200 PP-increased DEGs with largest FC (PP/PN). DEGs were ranked from most (ARG1) to least (S100A9) psoriasis-specific. The mean PSI of genes listed in each row is shown (left). (D) 200 PP-decreased DEGs with lowest FC (PP/PN). DEGs were ranked from most (CPNE6) to least (BTC) psoriasis-specific. The mean PSI of genes listed in each row is shown (left).
The 100 most non-specific PP-increased DEGs (PSI ≤ −2.40) were enriched with respect to immune- and interferon-associated Gene Ontology (GO) biological process (BP) terms (Figure S8B). Genes driving this association included MX1, IFI27, IRF7 and OASL, all of which are commonly elevated in lesions from psoriasis and other diseases (Figure S8B). Psoriasis-specific DEGs, in contrast, were frequently linked to membrane-associated processes, including transmembrane transport of calcium and potassium (Figures S8A and S8C). Enrichment of immune and inflammation-associated GO BP terms among psoriasis DEGs thus appeared to be driven by non-specific, rather than psoriasis-specific, DEGs.
Increased gene expression in psoriasis lesions: Non-specific cutaneous inflammation and a psoriasis-specific KC response
Gene expression in psoriasis partly reflects histological differences between lesional and normal skin (e.g., due to immune cell infiltration or epidermal expansion) (Swindell et al., 2013a). These differences have been analyzed using in silico methods (Swindell et al., 2013a), but such approaches have not distinguished among DEGs based on psoriasis-specificity.
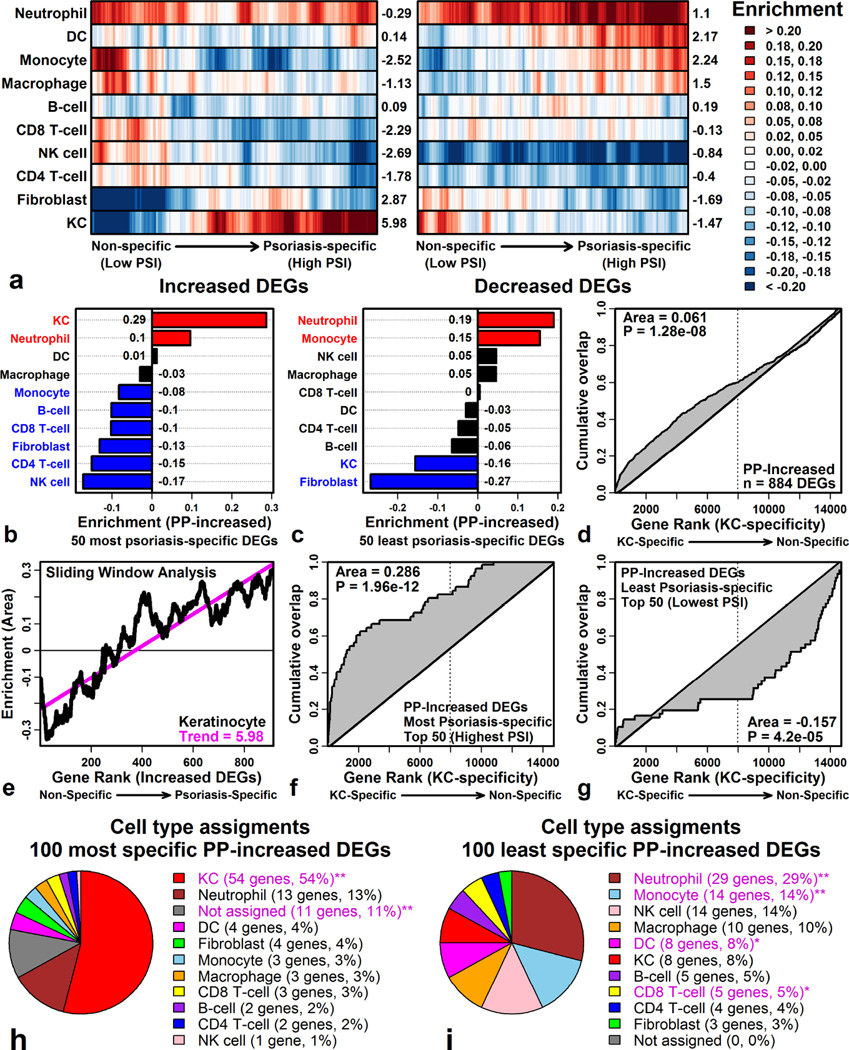
We thus used PSI to order psoriasis DEGs and then used a “sliding window” approach to evaluate associations with 10 cell types. This showed that only non-specific PP-increased DEGs are specifically expressed by inflammatory cells (e.g., monocytes and macrophages; Figure 3A). In contrast, psoriasis-specific PP-increased DEGs were KC-specific (Figure 3A). Of the 100 most psoriasis-specific PP-increased DEGs, more than half were expressed most highly in KCs (Figure 3H). In contrast, many non-specific PP-increased DEGs were expressed most strongly by inflammatory cells (e.g., neutrophils, monocytes, DC and CD8+ T-cells; Figure 3I). Distinctive features of the psoriasis transcriptome can thus be traced to KC-expressed genes, whereas immune cell infiltration appears to drive transcriptome convergence among skin diseases.
Figure 3. Psoriasis-specific DEGs elevated in psoriasis lesions are KC-associated.
(A) Sliding window analysis. DEGs were ranked by PSI and windows were evaluated for enrichment with respect to each cell type (Enrichment > 0: DEGs specifically expressed by cell type; 40 DEGs/window; Right margin: trend score). (B & C) Cell type enrichment for (B) psoriasis-specific and (C) non-specific DEGs. (D) Cumulative overlap between PP-increased DEGs and genes ranked according to their level of KC-specific expression. (E) Sliding window analysis (PP-increased DEGs and KCs; see part A). (F & G) Cumulative overlap for (F) psoriasis-specific and (G) non-specific DEGs. (H & I) Cell type assignments for (H) psoriasis-specific and (I) non-specific DEGs (**FDR < 0.05; Fisher’s Exact Test).
The unique psoriasis KC signature is associated with IL-17A but the non-specific inflammatory signature is associated with IFN-γ and TNF
Psoriasis-specific DEGs elevated in psoriasis lesions included cytokine-encoding genes (e.g., IL17A, IL19 and IL36A; Figure 2A). We thus screened 59 microarray experiments that measured KC expression responses following cytokine stimulation (Supplemental Data File 2), with the aim of identifying experiments in which psoriasis-specific and non-specific DEGs responded differently to cytokine stimulation.
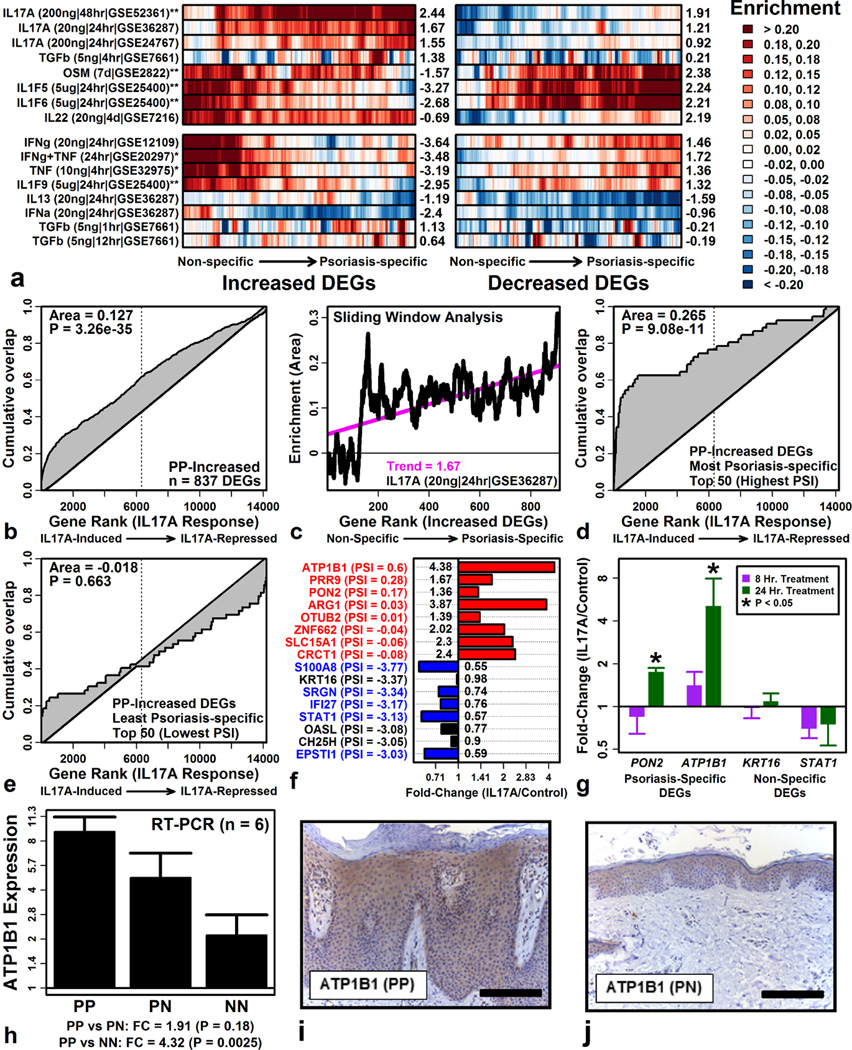
Psoriasis-specific and non-specific DEGs elevated in psoriasis lesions differed most strongly in their IL-17A response (Figure 4A). Psoriasis-specific PP-increased DEGs were disproportionately induced by IL-17A (e.g., ATP1B1, PRR9, PON2), whereas non-specific DEGs were unaltered or in some cases repressed (e.g., S100A8, IFI27, STAT1) (Figures 4B – 4F). Using RT-PCR, we confirmed that psoriasis-specific DEGs (ATP1B1, PON2) are IL-17A-induced, whereas non-specific DEGs (KRT16, STAT1) show no induction or slight repression (Figure 4G). RT-PCR also confirmed trends toward elevated ATP1B1 expression in psoriasis lesions (Figure 4H). Immunohistochemical staining localized ATP1B1 to lesional epidermis (Figures 4I and 4J), consistent with KC-specific expression (Figure 3).
Figure 4. Psoriasis-specific DEGs elevated in lesions are IL17A-induced.
(A) Sliding window analysis. DEGs were ranked by PSI and windows were evaluated for enrichment with respect to each cytokine (Enrichment > 0: DEGs cytokine-induced; 40 DEGs/window; Right margin: trend score). (B) Cumulative overlap between PP-increased DEGs and genes ranked by IL-17A response. (C) Sliding window analysis (PP-increased DEGs and IL-17A; see part A). (D & E) Cumulative overlap for (D) psoriasis-specific and (E) non-specific DEGs. (F) Effects of IL-17A on PP-increased DEGs (GSE36287; top: psoriasis-specific; bottom: non-specific; parentheses: PSI). (G) Effects of IL-17A stimulation in cultured KCs (RT-PCR; n = 3). (H) ATP1B1 expression in PP, PN and normal skin from control subjects (NN). (I and J) Immunohistochemical staining with ATP1B1-specific antibody (bar: 100 µM).
Non-specific DEGs elevated in psoriasis lesions were, in contrast, disproportionately induced by IFN-γ, TNF, and the combination IFN-γ + TNF, whereas this wasn’t the case among psoriasis-specific DEGs (Figure 4A). Psoriasis-specific and non-specific DEGs therefore exhibit differing cytokine responses, with enrichment for IL-17A targets observed only among psoriasis-specific DEGs.
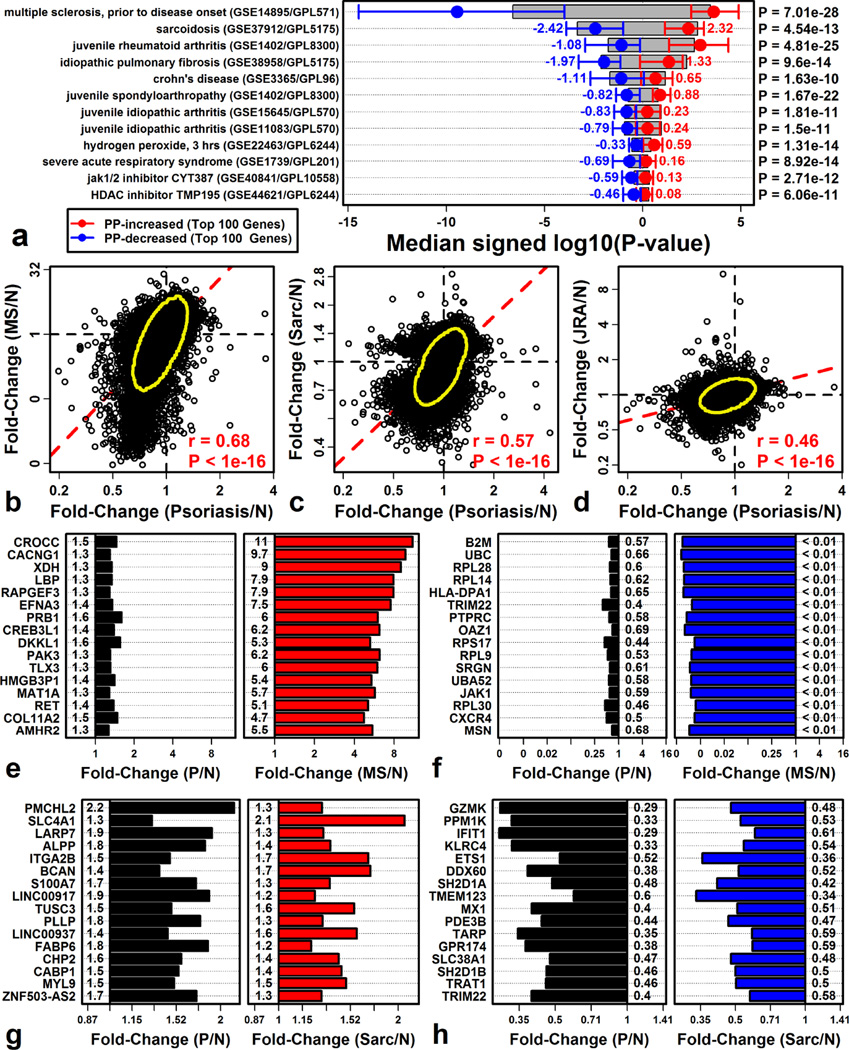
A PBMC expression signature shared between psoriasis and other autoimmune diseases (multiple sclerosis, sarcoidosis, rheumatoid arthritis)
We next considered whether DEGs identified from psoriasis patient PBMC show greater psoriasis-specificity than cutaneous DEGs. We therefore analyzed PBMC from psoriasis patients (n = 5) and healthy controls (n = 5) (GSE40263), which led to the identification of 1930 DEGs, including 693 psoriasis-increased DEGs and 1237 psoriasis-decreased DEGs (P < 0.05). We examined how these DEGs were altered in 421 PBMC microarray comparisons (Supplemental Data File 2).
PBMC-derived DEGs were more psoriasis-specific than cutaneous DEGs, but still overlapped significantly with PBMC differential expression patterns in other diseases (Figure 5). We identified autoimmune diseases for which PBMC expression shifts mirrored those in psoriasis, including multiple sclerosis (MS) (P = 7.01 × 10−28), sarcoidosis (P = 4.54 × 10−13), juvenile rheumatoid arthritis (P = 4.81 × 10−25), and Crohn’s disease (P = 1.63 × 10−10) (Figure 5A). For each of these diseases, FC estimates (disease/control) were correlated with those calculated for psoriasis (psoriasis/control) (rs ≥ 0.46; Figure 5B – 5D).
Figure 5. The psoriasis PBMC differential expression signature overlaps significantly with other autoimmune diseases.
(A) PBMC treatments with psoriasis-like gene expression changes. Shifts in gene expression were quantified using signed log10-transformed p-values (Log10P). Log10P was compared between psoriasis-increased and psoriasis-decreased DEGs (right margin, Wilcoxon rank sum test). Red and blue bars span the middle 50% of Log10P values among psoriasis-increased and psoriasis-decreased DEGs, respectively (grey boxes: non-DEGs). Only the 100 psoriasis top-ranked DEGs were considered for each DEG group (selected based upon FC). (B – D) FC comparison between psoriasis and (B) MS (C) sarcoidosis and (D) rheumatoid arthritis. (E – H) Genes similarly altered in psoriasis and (E & F) MS or (G & H) sarcoidosis (red/blue bars: FDR < 0.05).
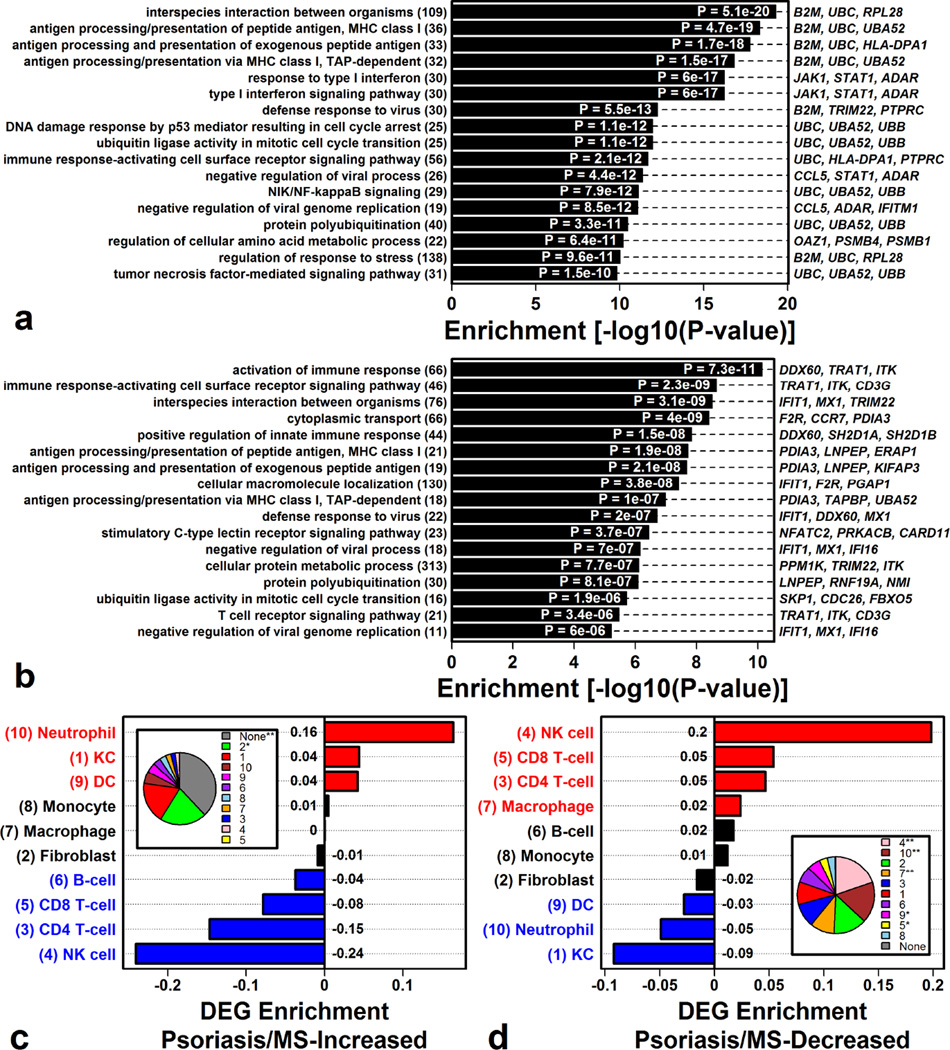
Genes decreased in both psoriasis and MS (Figure 5F), or both psoriasis and sarcoidosis (Figure 5H), were associated with antigen processing and presentation (via MHC Class I) and interferon signaling (Figures 6A and 6B). DEGs decreased in psoriasis and MS patients were specifically expressed by NK cells (Figure 6D), and such DEGs overlapped significantly with genes near (1–200 kb) psoriasis-associated SNPs from genome-wide association studies (Figure S8E).
Figure 6. The PBMC signature common to psoriasis and other autoimmune diseases is enriched with MHC- and NK-cell-associated genes.
(A) Gene ontology (GO) biological process (BP) terms enriched among genes with decreased expression in PBMC from psoriasis and MS patients. (B) GO BP terms enriched among genes with decreased expression in PBMC from psoriasis and sarcoidosis patients. (C & D) Cell type associated for genes (C) increased in psoriasis and MS patient PBMC or (D) decreased in psoriasis and MS patient PBMC (enrichment > 0: DEGs specifically expressed by cell type; enrichment < 0: DEGs not specifically expressed by cell type; red/blue: FDR < 0.05). Pie charts indicate the percentage of genes assigned to each cell type (*P< 0.05; **FDR < 0.05; Fisher’s Exact Test).
Discussion
Skin disease transcriptome studies have often followed a one-disease-at-a-time strategy without emphasizing comparisons to other disease states. This has been effective, but data sharing in the modern era now enables “cross-disease transcriptomics”, which is broader in scope, integrative, and capable of distinguishing between disease-specific and non-specific mechanisms (Haider et al., 2006; Inkeles et al., 2015; Swindell et al., 2015a; Wong et al., 2012). We here used RNA-seq meta-analysis to identify DEGs in lesional psoriasis skin, but found that most DEGs are non-specific and show similar differential expression in other skin conditions (Figure 1). Discrimination among DEGs using a psoriasis-specificity index (PSI), however, identified IL-17A pathway activation as a unique feature of psoriasis lesions. This activation is accompanied by circulating immune cell (PBMC) responses paralleling those observed in other autoimmune diseases, which involve down-regulation of MHC- and NK cell-associated genes (Figures 5 and 6). These findings highlight shared and distinctive features of psoriasis that can only be discerned from concurrent analysis of multiple diseases simultaneously. In future work, the integrative approach applied here can be extended to identify disease-specific drug targets, or potentially, to guide re-purposing of existing drugs based upon knowledge of shared disease mechanisms.
Psoriasis lesion development is closely tied to an underlying cytokine network, which directs and responds to signals from cutaneous cells, including KCs and infiltrating immunocytes (Perera et al., 2014). Previous psoriasis transcriptome studies have highlighted IL-1a, IL-1b, IL-17A and IL-20R1/R2 cytokines (i.e., IL-19, IL-20, IL-24) as in vivo drivers of gene expression in psoriasis lesions (Swindell et al., 2013a; Swindell et al., 2013b; Swindell et al., 2014a; Yao et al., 2008). Quantitative distinctions among these cytokines have been slight, however, based upon enrichment of cytokine-induced gene sets among PP-increased DEGs. Our current analysis provides sharper distinction along these lines and identifies IL-17A as an inducer of DEGs most uniquely elevated in psoriasis lesions (Figure 4). Consistent with this, elevated IL17A mRNA was one of the most unique characteristics of psoriasis lesions (Figure 2). These findings agree with our previous analysis (focused on a subset of psoriasis DEGs) (Swindell et al., 2015a), and also resonate with clinical data demonstrating efficacy of anti-IL-17A therapy for moderate-to-severe psoriasis (e.g., secukinumab, ixekizumab, brodalumab) (Griffiths et al., 2015; Langley et al., 2014; Lebwohl et al., 2015). This also provides proof-of-concept for the idea that cross-disease transcriptomics will point towards promising drug targets, suggesting that similar approaches might be applied to other skin diseases for which effective anti-cytokine treatments remain unknown.
Transcriptome studies of psoriasis have increasingly utilized RNA-seq technology (Jabbari et al., 2012; Keermann et al., 2015; Li et al., 2014; Swindell et al., 2014b; Tsoi et al., 2015), but thus far meta-analysis has not been used to compare and integrate RNA-seq data generated from these studies. To our knowledge, this study provides the first meta-analysis of RNA-seq data from lesional psoriatic skin. Our results suggest good agreement between RNA-seq datasets and moderate-to-good correspondence to microarray findings (0.678 ≤ rs ≤ 0.798), with improved microarray correspondence obtained by pooling RNA-seq data across studies (rs = 0.837). This suggests that, as with microarrays, meta-analysis of RNA-seq data will be instrumental for establishing robust inferences based upon the largest possible sample size (Swindell et al., 2015b; Tian et al., 2012). Notably, however, low-expressed DEGs identified by RNA-seq were biased towards decreased expression. Given this trend, future RNA-seq studies of psoriasis may benefit from stringent detection thresholds to filter out low-expressed transcripts (Swindell et al., 2014b). This may be especially important for studies focused on non-coding RNAs expressed at low levels, since a recent study noted greater differential expression among lncRNAs (as compared to protein-coding mRNAs) (Tsoi et al., 2015). Our current findings suggest that this may reflect a systemic trend among all low-expressed features (protein coding and non-coding).
Transcriptome analysis of PBMC and circulating leukocytes remains a relatively unexplored frontier in psoriasis. Most studies have focused on expression differences between lesional and normal skin (Li et al., 2014; Swindell et al., 2013a; Swindell et al., 2013b; Swindell et al., 2015b; Swindell et al., 2014a; Tian et al., 2012), but as we have shown, many such differences are non-specific, inflammation-associated, and generic. PBMC-derived DEGs, in contrast, overlap less prominently with other diseases, although we observed correspondence to autoimmune disease signatures (e.g., MS, sarcoidosis and juvenile rheumatoid arthritis). Genes decreased in PBMC from both psoriasis and MS patients, in particular, are MHC class I-associated, overlap significantly with genes near psoriasis-associated SNPs, and are NK cell-specific (Figure 6). Loss of NK-cell-associated expression in PBMC from psoriasis and MS patients appears consistent with NK-cell degeneration, as previously described for psoriasis and some other autoimmune diseases (Cameron et al., 2003; Shi and Zhou, 2011). This degeneration may be driven by cytokines released from autoreactive T-cells (e.g., IL-21) (Liu et al., 2006; Shi and Van Kaer, 2006). Our findings here demonstrate that this phenomenon can be analyzed based upon the peripheral blood transcriptome of autoimmune disease patients. Future large-cohort studies of the psoriasis PBMC transcriptome (and component PBMC cell types) may be beneficial for understanding mechanisms contributing to the shared autoimmune signature identified in this study.
Most mRNA profiling studies of psoriasis have, thus far, focused on cutaneous lesions rather than the peripheral blood transcriptome (Li et al., 2014; Swindell et al., 2013a; Swindell et al., 2013b; Swindell et al., 2015b; Swindell et al., 2014a; Tian et al., 2012). Only a minority of cutaneous DEGs discovered by this approach, however, will approach a disease-specific expression pattern, and unique properties of such DEGs can be overlooked when all DEGs are analyzed in aggregate (Swindell et al., 2015a). This is true for psoriasis lesions and, most likely, lesions from other skin diseases as well. The bioinformatic approach developed here provides one possible solution to this problem, in which a specificity index is first used to discriminate among DEGs, which then permits analysis of the most disease-specific DEGs apart from those that are non-specific. We expect that similar disease-specificity indices can be applied in future studies of other skin disorders to provide insights not obtained from aggregate DEG analysis. This may highlight cytokines or pathways representing effective drug targets, as demonstrated here in the context of psoriasis and IL-17A (Figure 4). Furthermore, as new expression profiling datasets are generated by the skin research community, this approach should grow more powerful, ultimately serving to facilitate data integration, enhance bioinformatic method development, and define shared mechanisms among skin diseases with otherwise dissimilar appearance and etiology.
Methods
RNA-seq meta-analysis was performed using reads from prior studies of PP and PN skin (GSE41745, GSE54456/GSE63979 and GSE67785; n = 44 patients) (Jabbari et al., 2012; Li et al., 2014; Swindell et al., 2015a; Tsoi et al., 2015). Statistical methods were similar to a pipeline described previously (Swindell et al., 2014b). Tophat was used to map reads to the UCSC hg19 genome (Kim et al., 2013). Read counts for protein-coding genes were then normalized using the voom algorithm (Law et al., 2014), and differential expression was assessed using Bayesian linear models with moderated t-statistics (Smyth, 2004). DEGs identified by RNA-seq were cross-referenced with microarray-based gene signatures associated with human skin diseases (98 signatures), KC cytokine responses (59), and PBMC tissue (421) (Supplemental Data File 2).
PSI was calculated using Equation (1) and p-values generated from n = 98 microarray comparisons involving human skin disease (diseased versus normal skin).
| [1] |
The p-value from the jth experiment (Pj) was log10-transformed and multiplied by a directionality indicator (dj), defined as −1 (psoriasis-consistent expression shift) or 1 (psoriasis-inconsistent). Missing p-values were imputed using a nearest neighbor (NN) strategy. A complete description of RNA-seq read processing, differential expression analysis, and PSI calculation is provided as supplementary material.
Supplementary Material
Acknowledgments
This work was supported by NIH K08 grant AR060802 (JEG), NIH RO1 grant AR069071 (JEG), Doris Duke Charitable Foundation grant 2013106 (JEG), the Babcock Endowment Fund (JEG), the A. Alfred Taubman Medical Research Institute (JEG), the Kenneth and Frances Eisenberg Emerging Scholar Award (JEG), and the American Skin Association Carson Family Research Scholar Award in Psoriasis (WRS).
Abbreviations
- DEG
Differentially expressed gene
- FC
fold-change
- FPKM
Fragments per kilobase of transcript per million mapped reads
- KC
keratinocyte
- MS
Multiple sclerosis
- PBMC
Peripheral blood mononuclear cell
- PN
Uninvolved skin from psoriasis patient
- PP
Lesional skin from psoriasis patient
- PSI
psoriasis specificity index
- SCC
Squamous cell carcinoma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors state no conflict of interest.
References
- Arlette JP, Trotter MJ. Squamous cell carcinoma in situ of the skin: history, presentation, biology and treatment. Australas J Dermatol. 2004;45:1–9. doi: 10.1111/j.1440-0960.2004.00025.x. [DOI] [PubMed] [Google Scholar]
- Cameron AL, Kirby B, Griffiths CE. Circulating natural killer cells in psoriasis. Br J Dermatol. 2003;149:160–164. doi: 10.1046/j.1365-2133.2003.05319.x. [DOI] [PubMed] [Google Scholar]
- Choy DF, Hsu DK, Seshasayee D, Fung MA, Modrusan Z, Martin F, et al. Comparative transcriptomic analyses of atopic dermatitis and psoriasis reveal shared neutrophilic inflammation. J Allergy Clin Immunol. 2012;130:1335.e5–1343.e5. doi: 10.1016/j.jaci.2012.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Erme AM, Wilsmann-Theis D, Wagenpfeil J, Hölzel M, Ferring-Schmitt S, Sternberg S, et al. IL-36gamma (IL-1F9) Is a Biomarker for Psoriasis Skin Lesions. J Invest Dermatol. 2015;135:1025–1032. doi: 10.1038/jid.2014.532. [DOI] [PubMed] [Google Scholar]
- Dhingra N, Suarez-Farinas M, Fuentes-Duculan J, Gittler JK, Shemer A, Raz A, et al. Attenuated neutrophil axis in atopic dermatitis compared to psoriasis reflects TH17 pathway differences between these diseases. J Allergy Clin Immunol. 2013;132:498.e3–501.e3. doi: 10.1016/j.jaci.2013.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths CE, Reich K, Lebwohl M, van de Kerkhof P, Paul C, Menter A, et al. Comparison of ixekizumab with etanercept or placebo in moderate-to-severe psoriasis (UNCOVER-2 and UNCOVER-3): results from two phase 3 randomised trials. Lancet. 2015;386:541–551. doi: 10.1016/S0140-6736(15)60125-8. [DOI] [PubMed] [Google Scholar]
- Haider AS, Peters SB, Kaporis H, Cardinale I, Fei J, Ott J, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126:869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- Inkeles MS, Scumpia PO, Swindell WR, Lopez D, Teles RM, Graeber TG, et al. Comparison of molecular signatures from multiple skin diseases identifies mechanisms of immunopathogenesis. J Invest Dermatol. 2015;135:151–159. doi: 10.1038/jid.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari A, Suarez-Farinas M, Dewell S, Krueger JG. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol. 2012;132:246–249. doi: 10.1038/jid.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keermann M, Koks S, Reimann E, Prans E, Abram K, Kingo K. Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC genomics. 2015;16:322. doi: 10.1186/s12864-015-1508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med. 2014;371:326–338. doi: 10.1056/NEJMoa1314258. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebwohl M, Strober B, Menter A, Gordon K, Weglowska J, Puig L, et al. Phase 3 Studies Comparing Brodalumab with Ustekinumab in Psoriasis. N Engl J Med. 2015;373:1318–1328. doi: 10.1056/NEJMoa1503824. [DOI] [PubMed] [Google Scholar]
- Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Van Kaer L, La Cava A, Price M, Campagnolo DI, Collins M, et al. Autoreactive T cells mediate NK cell degeneration in autoimmune disease. J Immunol. 2006;176:5247–5254. doi: 10.4049/jimmunol.176.9.5247. [DOI] [PubMed] [Google Scholar]
- Nomura I, Gao B, Boguniewicz M, Darst MA, Travers JB, Leung DY. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J Allergy Clin Immunol. 2003;112:1195–1202. doi: 10.1016/j.jaci.2003.08.049. [DOI] [PubMed] [Google Scholar]
- Perera GK, Ainali C, Semenova E, Hundhausen C, Barinaga G, Kassen D, et al. Integrative biology approach identifies cytokine targeting strategies for psoriasis. Sci Transl Med. 2014;6:223ra222. doi: 10.1126/scitranslmed.3007217. [DOI] [PubMed] [Google Scholar]
- Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- Shi FD, Van Kaer L. Reciprocal regulation between natural killer cells and autoreactive T cells. Nat Rev Immunol. 2006;6:751–760. doi: 10.1038/nri1935. [DOI] [PubMed] [Google Scholar]
- Shi FD, Zhou Q. Natural killer cells as indispensable players and therapeutic targets in autoimmunity. Autoimmunity. 2011;44:3–10. doi: 10.3109/08916931003782122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC genomics. 2013a;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Johnston A, Xing X, Voorhees JJ, Elder JT, Gudjonsson JE. Modulation of epidermal transcription circuits in psoriasis: new links between inflammation and hyperproliferation. PloS ONE. 2013b;8:e79253. doi: 10.1371/journal.pone.0079253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Remmer HA, Sarkar MK, Xing X, Barnes DH, Wolterink L, et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015a;7:86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Sarkar MK, Stuart PE, Voorhees JJ, Elder JT, Johnston A, et al. Psoriasis drug development and GWAS interpretation through in silico analysis of transcription factor binding sites. Clin Transl Med. 2015b;4:13. doi: 10.1186/s40169-015-0054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Stuart PE, Sarkar MK, Voorhees JJ, Elder JT, Johnston A, et al. Cellular dissection of psoriasis for transcriptome analyses and the post-GWAS era. BMC Med Genomics. 2014a;7:27. doi: 10.1186/1755-8794-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Xing X, Voorhees JJ, Elder JT, Johnston A, Gudjonsson JE. Integrative RNA-seq and microarray data analysis reveals GC content and gene length biases in the psoriasis transcriptome. Physiol Genomics. 2014b;46:533–546. doi: 10.1152/physiolgenomics.00022.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Krueger JG, Li K, Jabbari A, Brodmerkel C, Lowes MA, et al. Meta-analysis derived (MAD) transcriptome of psoriasis defines the "core" pathogenesis of disease. PloS ONE. 2012;7:e44274. doi: 10.1371/journal.pone.0044274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D, Kea B, Pesich R, Higgs BW, Zhu W, Brown P, et al. Interferon and biologic signatures in dermatomyositis skin: specificity and heterogeneity across diseases. PloS ONE. 2012;7:e29161. doi: 10.1371/journal.pone.0029161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Richman L, Morehouse C, de los Reyes M, Higgs BW, Boutrin A, et al. Type I interferon: potential therapeutic target for psoriasis? PloS ONE. 2008;3:e2737. doi: 10.1371/journal.pone.0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.