Abstract
We describe a facile method to activate or repress transcription of endogenous genes in a quantitative and specific manner by treatment with designed regulatory proteins (DRPs), in which artificial transcription factors (ATFs) are fused to cell-penetrating peptides (CPPs). Penetration of DRPs into cells is mediated by an N-terminal CPP fused to a nuclear localization signal; a DNA-binding domain and a transactivation domain follow. The DNA-binding domain was targeted to the vascular endothelial growth factor (VEGF)-A gene. An agonist DRP was rapidly taken up by cells and transported to the nucleus; soon after, the cells began transcribing the gene and secreting VEGF-A protein in a dose-dependent manner. Multiple copies of a short oligopeptide derived from a minimal transactivation domain of human β-catenin was stronger than VP-16. The SRDX domain from the plant transcription factor, SUPERMAN, changed the DRP to a hypoxia-induced antagonist of VEGF-A. DRPs combine many of the potential benefits of transgenes with those of recombinant proteins.
With the advent of gene therapy, the disciplines of genetics and pharmacology have begun to merge. The genetic basis of disease is becoming clear and, consequently, gene-based tools have gained therapeutic potential. However, the use of nucleic acid technologies in humans remains largely experimental. In contrast, protein therapy has been used for decades. We describe a class of proteins that can activate or repress any desired gene when added to the medium outside human cells. We call these agents designed regulatory proteins (DRPs).
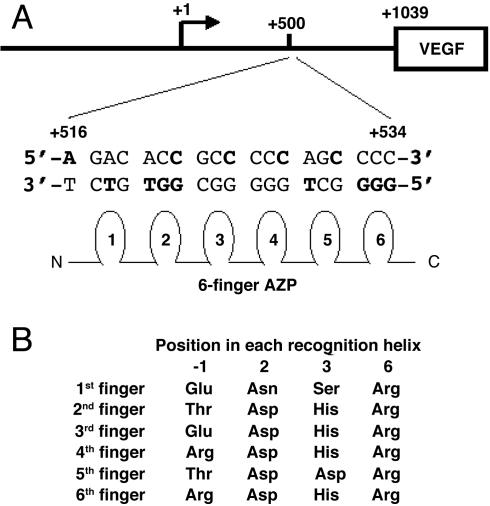
Artificial zinc finger protein (AZP) technology allows DNA sequences to be selectively targeted by the design or selection of a DNA-binding domain that is typically composed of three or six fingers. We have previously developed a rational method for designing AZPs using a nondegenerate recognition code table (1). We use our design method (1) and six fingers to bind 19-bp target sequences. By sandwiching the AZP domain between a nuclear localization signal (NLS) sequence and a transeffect domain, such as a transactivation or a transrepression domain, and expressing this artificial transcription factor (ATF) gene in a cell, any gene in the cell can be transcriptionally activated or repressed (2, 3). The effectiveness of ATFs has been demonstrated in plants (1, 4, 5) and animals (6).
Complementary technology arose from the discovery that many proteins have an intrinsic ability to cross cell membranes. Biological membranes are generally impermeable to large hydrophilic molecules such as proteins, and this limits the development of peptide drugs and therapeutic proteins. However, a series of small protein domains, termed cell-penetrating peptides (CPPs) or protein transduction domains (PTDs), has been shown to cross biological membranes (7-9). Since the TAT protein, a transactivation factor from HIV type 1 (HIV-1), was first reported to cross the plasma membrane when exogenously added to culture medium (10-12), a variety of CPPs have been investigated for delivering molecules into mammalian cells (7, 8, 13-16). When CPPs are fused to full-length proteins, they can deliver biologically active protein into cells (17, 18).
Vascular endothelial growth factor A (VEGF-A) levels are dramatically increased by hypoxia (19), triggering angiogenesis and microvascular permeability (20, 21). Treatments that reduce VEGF-A levels may prevent angiogenesis associated with tumor growth, rheumatoid arthritis, or diabetic retinopathy (22). In contrast, compounds that increase VEGF-A levels may stimulate neovascularization to treat ischemia, arteriosclerosis obliterans, or wound healing.
AZPs have been used to create ATFs that, when introduced as transgenes in mammalian cells, either repressed or activated the VEGF-A gene, depending on the type of transeffect domain that was fused to the AZP. When a repression domain was used, VEGF-A was repressed 20-fold, to normal levels, in a tumorigenic glioblastoma cell line (23). With an activation domain, VEGF-A was increased 4- to 5-fold, to levels greater than that induced by hypoxia, in human embryonic kidney 293 (HEK293) cells (24). The natural spectrum of splice variants was produced by an ATF, in contrast to cDNA expression. This is an important point, because the VEGF-A splice variants have different biological activities. Studies in mice showed that both an ATF and the cDNA for VEGF-A stimulated angiogenesis, but the cDNA also caused hyperpermeability of the vessels (6). Specificity and affinity of ATFs are high; GeneChip analysis indicated that only the target gene was repressed (25) and subnanomolar dissociation constants are typical.
We created DRPs by fusing CPPs to ATFs. The VEGF-A gene was the target in this study. We report that, when DRPs are added to the culture medium, they enter the nucleus in HEK293 cells, where they can both up- and down-regulate the endogenous VEGF-A gene.
Materials and Methods
Design and Construction of DRP-Encoding DNA. We designed and constructed an AZP gene by using our recognition code table as described (1). The target sequence was 5′-GGG GCT GGG GGC GGT GTC T-3′ (+516 to +534 in the human VEGF-A gene, where +1 is the transcription start site). The amino acids chosen to contact the 19-bp target (Fig. 1A) are listed in Fig. 1B. The DNA encoding the AZP was cloned into pTriEX-3 (Novagen) containing the CPP (residues 47-57) of HIV-1 TAT (9), PTD4 (16), or a 9-mer of arginine (9R; ref. 14); a nuclear localization signal (NLS) from the simian virus 40 large T antigen; a transeffect domain; and a FLAG epitope tag. As an activation domain, herpes simplex virus VP-16 activation domain (residues 415-490; ref. 26), a C-terminal transcription activation domain of β-catenin (residues 695-781; ref. 27), or one or more copies of the last five amino acids derived from the C-terminal transcription activation domain of β-catenin (FDTDL) was used. For repression domains, a Krüppel-associated box (KRAB) domain of KOX1 (residues 1-75; ref. 28), a minimal Sin3 interaction domain of human Mad1 protein (residues 1-35; ref. 29), or an SRDX domain from Arabidopsis thaliana SUPERMAN protein (30) was introduced into the DRP construct. Schematic representation and the complete amino acid sequence of TAT-ATF are shown in Fig. 2A and Fig. 8, which is published as supporting information on the PNAS web site.
Fig. 1.
Design of the AZP protein. (A) DNA sequence of the AZP target for VEGF-A gene activation. The 19-bp target is located from +516 to +534 (relative to the transcriptional start site, +1) in the 5′ UTR. DNA bases used for the design of the AZP are indicated in bold. (B) Amino acids in the six-finger domain used for the 19-bp recognition. The amino acid sequence of entire TAT-ATF construct including the AZP domain is shown in Fig. 8.
DRP Purification. The pTriEX-3 constructs were introduced into Escherichia coli Rosetta(DE3)pLac I (Novagen). A 600-ml culture was grown in LB medium supplemented with 30 mg of ampicillin and 18 mg of chloramphenicol at 37°C until OD600 reached 0.6-0.75. The culture was induced with 1 mM isopropyl β-d-thiogalactoside for 3 h, and the cells were lysed in 10 ml of cold buffer [50 mM phosphate buffer, pH 7.2/1 M NaCl/5 mM DTT/0.1 mM ZnCl2/complete EDTA-free medium (Roche Molecular Biochemicals)] by using an ultrasonicator. After centrifugation (4°C, 15,000 × g, 10 min), the supernatant was applied to a DEAE Sepharose Fast Flow (Amersham Pharmacia) column, and the flow-through fractions were then purified by chromatography on a Bio-Rex 70 column. The column was washed and eluted with 300 and 600 mM NaCl in buffer (50 mM phosphate buffer, pH 7.2/0.2 mM DTT/0.1 mM ZnCl2), respectively. The protein was purified to >95% homogeneity as judged by SDS/PAGE. The protein concentration was determined by using the Protein Assay ESL kit (Roche Molecular Biochemicals).
Electrophoretic Mobility-Shift Assay (EMSA). See Table 2, which is published as supporting information on the PNAS web site, for details.
Analysis of Cellular Uptake of DRP by Immunofluorescent Staining. A total of 2 × 104 HEK293 cells (American Type Culture Collection) was plated onto an eight-well culture slide coated with poly(d-lysine) (Becton Dickinson). The cells were incubated at 37°C for 36 h in 200 μl of DMEM (GIBCO/BRL) supplemented with 0.1 mM nonessential amino acids and 10% FBS (GIBCO/BRL). The culture medium was then changed to 200 μl of fresh DMEM containing 2 μM DRP. After incubation at 37°C for 3 h, the cells were fixed with 4% paraformaldehyde in Tris-buffered saline (TBS) (50 mM Tris/150 mM NaCl, pH 7.4) and permeabilized with 0.2% Triton X-100 in TBS for 5 min. After rinsing three times with TBS, the cells were incubated in 10% goat serum (Sigma) in TBS containing 100 mM NH4Cl for 15 min. This was followed by incubation with 50 μg/ml of anti-FLAG M2 monoclonal antibody-FITC conjugate (Sigma) in TBS containing 0.1% Tween 20 for 1 h. The resulting cells were then stained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) solution (100 ng/ml in TBS), and finally mounted in 70% glycerol in TBS containing 2.5% 1,4-diazabicyclo[2.2.2]octane (DABCO) (Sigma). Fluorescence was analyzed under an Olympus IX70 fluorescence microscope.
Assay for Endogenous VEGF-A Production by Transient Transfection of DRP-Encoding Plasmid into HEK293 Cells. HEK293 cells were plated and grown as described above. DRP expression vectors were cotransfected with pCMV-β-galalactosidase plasmid (Clontech) and Lipofectamine 2000 (Invitrogen) according to the protocol accompanying the reagent. Cell numbers and the amounts of the transfected plasmids in the experiments are indicated in the figure legends. The culture medium was collected, and the VEGF-A protein concentration was quantified as described above. The transfected cells were harvested, and the β-galactosidase activities were measured by using the Luminescent β-Galactosidase Detection kit (BD Biosciences). The experiments were carried out in duplicate and repeated independently at least three times.
Quantitative RT-PCR Analysis of Activated Endogenous VEGF-A Gene by DRP. Quantitative RT-PCR analysis of the VEGF-A mRNA expression level was carried out by real-time RT-PCR by using TaqMan chemistry on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). A total of 5 × 105 HEK293 cells per well was plated onto a 24-well tissue culture plate and incubated at 37°C for 36 h in 300 μl of DMEM supplemented with 0.1 mM nonessential amino acids and 10% FBS. Subsequently, 30 μl of 1 μM DRP solution in Opti-MEM I Reduced Serum Medium (GIBCO/BRL) was added to each well and incubated at 37°C for 5 h. The total RNA was isolated by using TRIzol (Invitrogen) according to the manufacturer's instructions. The reverse transcription of the total RNA (20 ng) was performed with 0.2 μM each primer, 0.1 μM probe at 48°C for 30 min with MultiScribe reverse transcriptase (Applied Biosystems). After denaturing at 95°C for 10 min, a PCR amplification reaction was conducted for 30 cycles at 95°C for 15 s and at 60°C for 1 min by using the One-Step RT-PCR Master Mix Reagent kit (Applied Biosystems). The primer set and a probe used were as follows: forward primer, 5′-TGGTGAAGTTCATGGATGTCTATCA-3′; reverse primer, 5′-TCAGGGTACTCCTGGAAGATGTC-3′; TaqMan probe, 5′-FAM-CAGCTACTGCCATCCAATCGAGACCCT-TAMR-3′. PCR amplification of the housekeeping gene, GAPDH, was performed to allow normalization between samples by using human GAPDH Control Reagents (Applied Biosystems). The primers and a probe used here recognize five known splice variants.
Endogenous VEGF-A Protein Production. A total of 5 × 104 HEK293 cells per well was plated onto a poly(d-lysine)-coated 96-well plate (BD Biosciences) with 100 μl of DMEM supplemented with 0.1 mM nonessential amino acids and 10% FBS. One day later, the medium was replaced with 100 μl of fresh medium and the cells were treated with various concentrations of DRPs. The culture medium was collected and frozen at -20°C until assayed. VEGF-A protein content in the culture medium was assayed by using a human VEGF-A ELISA kit (R & D Systems) according to the manufacturer's protocol.
Results
DNA Binding to the Target Sequence by TAT-ATF. EMSA experiments were performed to measure in vitro affinity. The control protein, TAT-ATFΔAla, substitutes alanine at the four positions (-1, 2, 3, and 6) in each finger that contact the DNA. TAT-ATFΔAla was unable to shift the target DNA even at 1 μM, whereas TAT-ATF caused a single shifted species (data not shown) with an apparent dissociation constant (Kd) of 870 pM (Table 1 and discussed below), showing that specific amino acids are required to activate the VEGF-A gene.
Table 1. Dissociation constants (Kd) for various DRPs binding to the wild-type target DNA probe.
| Protein | Domains | Kd, nM |
|---|---|---|
| PTD4-ATF | PTD4-NLS-AZP-VP16-FLAG | 1.08 |
| PTD4-ATFΔAla | PTD4-NLS-AZPΔAla-VP16-FLAG | >1,000 |
| PTD4-AZP | PTD4-NLS-AZP-FLAG | 0.08 |
| TAT-ATF | TAT-NLS-AZP-VP16-FLAG | 0.87 |
| R9-ATF | R9-NLS-AZP-VP16-FLAG | 0.33 |
The sequence of the wild-type oligonucleotides used is given in Table 2. Affinities for the binding sites were determined by EMSA. Values are the average of at least two independent determinations.
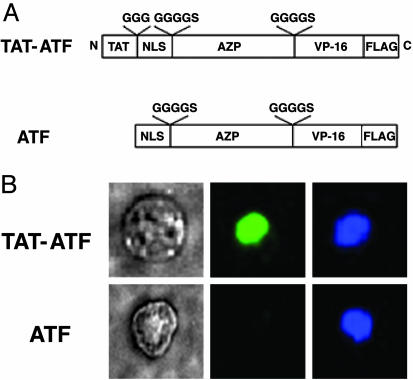
Cellular Uptake of TAT-ATF. TAT-ATF protein in the tissue culture medium was rapidly concentrated in the nuclei (compare Fig. 2B Upper Center with Upper Right), whereas ATF alone did not enter the cells (compare Fig. 2B Lower Center with Lower Right) underscoring the importance of a CPP for cell penetration. More than 80% and <5% of the cells were anti-FLAG-FITC-positive after a 2-h incubation with TAT-ATF and ATF, respectively.
Fig. 2.
Immunofluorescent staining of TAT-ATF and ATF in HEK293 cells. (A) Schematic representation of DRPs used in this study. A cell-penetrating peptide, TAT, was fused to an artificial transcription factor (ATF) comprising a NLS, a six-fingered AZP, a VP-16 activation domain, and a FLAG epitope tag to yield a TAT-ATF. The amino acids shown above the open boxes are linker peptides. (B) Visualization of DRPs transduced into HEK293 cells by immunofluorescent assay. HEK293 cells (2 × 104) were treated with 2 μM TAT-ATF (Upper) or ATF (Lower) for 2 h at 37°C, and the transduced proteins were identified by immunofluorescent staining as described in Materials and Methods. Phase-contrast image of a 293-H cell (Left) and a fluorescence image of the same cell stained with an anti-FLAG antibody-FITC conjugate (Center) are shown. The nucleus of the cell was also labeled with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Right).
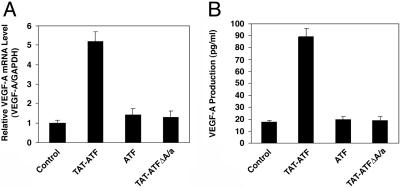
Reporter Gene Activation and mRNA Induction by TAT-ATF. TAT-ATF protein added exogenously to the tissue culture medium caused an 18-fold activation of the luciferase reporter gene under the control of the VEGF-A promoter/5′ UTR (-2279 to +1038 relative to the transcriptional start site; ref. 24). To investigate whether the TAT-ATF protein could also activate transcription of the VEGF-A gene from the endogenous chromosomal locus, real-time RT-PCR was performed. We observed an increase of mRNA for VEGF-A in a dose-dependent fashion (data not shown) with the maximum of 5-fold at 500 nM TAT-ATF (Fig. 3A). Two negative controls, ATF and TAT-ATFΔAla, failed to activate transcription.
Fig. 3.
Activation of the endogenous human VEGF-A gene by TAT-ATF. (A) Quantitative RT-PCR analysis of VEGF-A mRNA expression. Twelve hours after transduction with or without 500 nM protein, the total RNA was isolated from the cells, and the VEGF-A mRNA level was measured by TaqMan. The level of VEGF-A mRNA was normalized against GAPDH. (B) Activation of endogenous VEGF-A protein production by the DRP protein. HEK293 cells (2 × 104) were treated with or without 500 nM of the protein and then incubated for 24 h. Quantification of the VEGF-A protein in the culture medium was quantified by ELISA. Each point represents the mean ± SD obtained from three independent experiments.
Effect of TAT-ATF on the Secretion of VEGF-A Protein by HEK293 Cells. Incubation of cells in the presence of 500 nM TAT-ATF caused the accumulation of ≈5-fold elevated levels of VEGF-A protein, whereas ATF and TAT-ATFΔAla did not cause a significant change (Fig. 3B). None of the treatments affected the viability of the cells (data not shown).
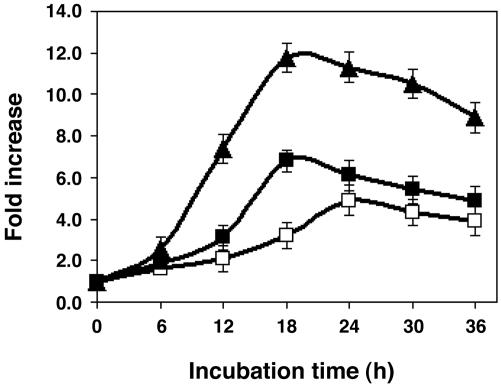
Effect of Different CPPs on the Production of VEGF-A Protein. To assess the effectiveness of different CPPs, we replaced the TAT peptide with PTD4 or a 9-mer of arginine (R9). These CPPs can be more effective than TAT (14, 16). As shown in Fig. 4, PTD4-ATF caused VEGF-A protein to accumulate faster and to higher levels than did TAT-ATF. The maximum accumulation caused by 500 nM PTD4-ATF and TAT-ATF were observed at 18 h (7-fold) and 24 h (5-fold), respectively. Treatment of cells with R9-ATF resulted in the highest level of VEGF-A protein accumulation; the maximum induction was observed at 18-h incubation, resulting in an ≈12-fold increase in VEGF-A protein. VEGF-A protein levels in the supernatant began to decrease after 18 h, reflecting their turnover and a reduction in biosynthesis. This reduction may be due to instability of the DRPs inside the cells, resulting in only one or a few rounds of transcription. Many transactivation domains, including VP-16, are substrates for ubiquitin-mediated proteolysis (27).
Fig. 4.
The potency of DRPs with different CPPs. Time course of the induction of VEGF-A protein production by TAT-ATF (open square), PTD4-ATF (filled square), and R9-ATF (filled triangle) in HEK293 cells. The cells (2 × 104) were incubated with DRP protein (500 nM) for indicated time intervals. Quantification of the VEGF-A protein in the culture medium was measured as described in Materials and Methods. Fold increase relative to the control was calculated by dividing the quantified concentrations of the VEGF protein from medium cultured in the presence of the protein by those in the absence of the protein. Each point represents the mean ± SD obtained from three independent experiments.
Dose Effects. Cells treated with R9- or PTD4-ATF accumulated VEGF-A protein in a dose-dependent manner (Fig. 9, which is published as supporting information on the PNAS web site). Control cells that had been treated with a “knockout” mutant containing R9 (R9-ATFΔAla) or PTD4 (PTD4-ATFΔAla) did not show significant changes, even with up to 5 μM DRP (data not shown).
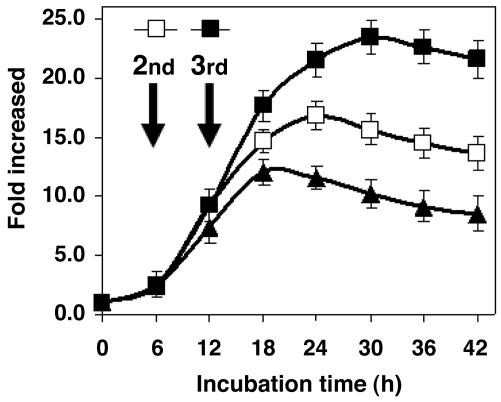
We repeated the addition of R9-ATF to the cells twice at 6-h intervals. As shown in Fig. 5, when the cells experienced the second administration of R9-ATF, maximum accumulation reached >17-fold; a third treatment caused a 23-fold accumulation.
Fig. 5.
Effect of multiple administration of R9-ATF on VEGF-A production in HEK293 cells. The cells (2 × 104) were incubated with 1.65 μg (500 nM final concentration) of R9-ATF protein for 6 h and then treated with an additional 1.65 μg of the protein (filled and open square) or buffer (filled triangle). After 6 h of incubation, a third aliquot of 1.65 μg of the protein (filled square) or buffer (open square and filled triangle) was added to the cells. The culture medium was harvested at the indicated time intervals. The concentration of VEGF-A in the medium was quantified by ELISA as described in Materials and Methods. Fold increase relative to the control was calculated as described in Fig. 4. The data represent an average of three independent experiments, and the SD is shown.
Sequence Specificity of the DRP and Effect of CPP and Transeffect Domains on DNA Binding. We studied the affinity and specificity of the AZP domain of PTD4-ATF by using EMSA. As shown in Table 2, the position of the mutations in the target DNA sequence had a significant effect on in vitro binding. Relatively weak effects were observed when the position of the mutation(s) was located within 4 nt from the end of the target sequence (Mut-4 to -6 in Table 1). Mutation(s) in the middle portion of the sequence, such as that seen in Mut-2, had a larger effect on affinity.
To test whether CPP or transeffect domains of the DRP affect binding affinity, apparent dissociation constants were determined for a series of DRPs with the wild-type target sequence (Table 1). The CPP domains used in this study are rich in basic amino acid residues, raising the theoretical pI of the PTD4, TAT, and R9 domains to 11.71, 12.31, and 12.9, respectively. The measured affinity for the target sequence increased in this order. Therefore, basic amino acids in the CPP domain may contribute to the overall affinity through an ionic interaction between the phosphate backbone and the basic amino acid residues. PTD4-AZP (PTD4-ATF without any transeffect domains such as VP-16) resulted in a 13.5-fold increased affinity compared to PTD4-ATF. These data demonstrate that both the CPP and transeffect domains contribute to the overall affinity of the DRP for its target DNA.
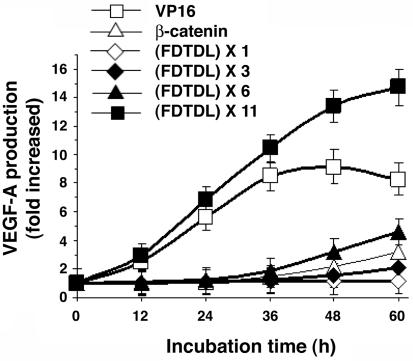
Activation of the VEGF-A Gene by DRPs Fused with Different Activator Domains. We created DRPs with domains derived from human β-catenin. Use of the reported minimal transactivation domain (MTAD; residues 695-781; ref. 27) resulted in 3-fold induction of the VEGF-A protein (Fig. 6). As shown in Fig. 6, one copy of the putative MTAD motif, FDTDL, of β-catenin did not activate transcription of VEGF-A protein production, whereas three and six tandem repeats of the motif induced 2- and 4-fold increases. Eleven copies of the MTAD motif caused the DRP to induce VEGF-A protein 15-fold; the activation potency was nearly twice that of the VP-16 transactivation domain. The increased potency was largely caused by sustaining the period of protein accumulation to >60 h, compared to a peak of accumulation at 48 h for VP-16; the rate of accumulation was also slightly greater.
Fig. 6.
Activation of VEGF-A protein production by tandem repeats of the putative minimal activation domain of β-catenin. The plasmid encoding DRP with various activation domains (20 ng) and pCMV-β-galactosidase plasmid (2 ng) were cotransfected into 2 × 104 HEK293 cells. The cells were incubated for the indicated time period, and then the culture medium and the transfected cells were collected separately. The VEGF concentration and the β-galactosidase activities were quantified as described in Materials and Methods. Results were normalized for transfection efficiency with β-galactosidase activities. Each point represents the mean ± SD obtained from three independent experiments.
Conversion of DRP Activators to Repressors. To convert the DRP to a transcriptional repressor, the VP-16 activation domain of PTD4-ATF was replaced with a KRAB repressor domain to yield PTD4-NLS-AZP-KRAB. As shown in Fig. 10, which is published as supporting information on the PNAS web site, significant repression of baseline VEGF-A protein production was observed with 500 nM PTD4-NLS-AZP-KRAB, producing ≈80% reduction in VEGF-A protein accumulation. PTD4-NLS-AZP alone caused a 10% repression in the VEGF-A protein level. Neither of the proteins was toxic to the cells (data not shown).
Repression of the VEGF-A Gene by DRPs Fused with Different Repression Domains. Cobalt treatment mimics hypoxia by stabilizing hypoxia-inducible factor α (HIF-α) (31-33). Cells cultured under normoxic conditions expressed low levels of VEGF-A protein (≈50 pg/ml) that were increased >4-fold by incubation in the presence of 150 μM CoCl2 (≈230 pg/ml; data not shown).
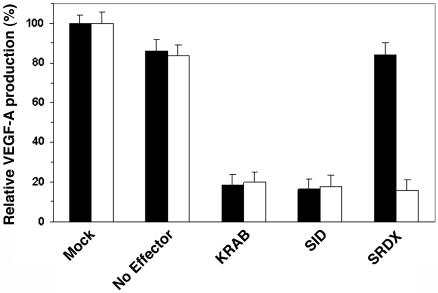
DRPs containing transcriptional repressor domains derived from human KRAB and Sin3 interaction domain repressed VEGF-A protein production by 80% under both normal and hypoxic conditions (Fig. 7). DRPs with a transcriptional repressor domain, SRDX, derived from the plant transcription factor SUPERMAN (30), selectively abolished hypoxia-induced VEGF-A production.
Fig. 7.
Repression of VEGF-A protein production by various transcription repression domains in normoxia and hypoxia. The plasmid encoding DRP with various repression domains (0.1 μg) and pCMV-β-galactosidase plasmid (0.01 μg) were cotransfected into 1 × 105 HEK293 cells. Cells were allowed to grow for 12 h and then treated with (hypoxia, open bar) or without (normoxia, filled bar) 150 μM CoCl2. After 24 h, the culture medium and the transfected cells were collected separately. The VEGF concentration and the β-galactosidase activities were quantified as described in Materials and Methods. Results were normalized for transfection efficiency with β-galactosidase activities. Each point represents the mean ± SD obtained from three independent experiments.
Discussion
DRPs provide a facile means to activate or repress endogenous gene expression by using proteins rather than nucleic acids. DRPs are specific for a given 19-bp target, which will generally enable any single gene to be controlled. We demonstrated that DRPs can successfully enter human cells, whereas the corresponding ATF fails to penetrate (Fig. 2B). Entry into the cells caused induction of VEGF-A mRNA synthesis and protein secretion (Fig. 3). One advantage to DRPs is that nearly all (>80%) treated cells are similarly affected.
A full understanding of specificity is not provided by our experiments, but it is clear that even single point mutations in the 19-bp target site significantly reduce its affinity for the DRP (Table 2). Changes at the 5′ end of the target site had a greater effect than changes at the 3′ end, and affinity decreased with increasing number of mutations.
We previously reported that the AZP alone has a very high binding affinity (Kd < 3 pM) (1). The affinities of DRPs were 1,000-fold lower. We found that the modules around the AZP affect target site affinity (Table 1). Removing the VP-16 domain increased binding 13.5-fold, possibly because the acidic residues of the DNA and VP-16 repel each other. DRPs made with the R9 CPP were more potent than DRPs made with PTD4 or TAT, in part because R9-CPP contributes to an increased affinity of the DRP for DNA (Table 1). R9-ATF may also be more efficiently taken up by HEK293 cells (Fig. 4).
The effect of DRPs on HEK293 cells follows a normal dose-response curve with saturation at ≈500 nM. The cells recovered sensitivity to DRPs soon after a saturating treatment. In fact, subsequent treatments raised secreted VEGF levels to more than double the concentration achieved with an initial saturating dose (Fig. 5). We speculate that DRPs are initially saturating the VEGF-A chromosomal binding sites and triggering one or a few rounds of transcription before being degraded. Subsequent doses apparently repeat the process. The VP-16 domain accelerates the degradation of transcription factors (34).
The FXXΦΦ motif (X is any amino acid and Φ is a hydrophobic amino acid) in the β-catenin transactivation domain is conserved in several transcriptional activation domains, including VP-16, p53, NF-κB, p65, and yeast transcriptional activator Adr1p, and it may be a general recognition element of acidic activation domains for TAFII31 (35). We hypothesized that the FDTDL motif in β-catenin may also recruit TAFII31 and act as an MTAD. The potency of the VN8 MTAD (DFDLDMLG) derived from VP-16 is increased in proportion to the copy number of the module (34, 36), leading us to predict that tandem repeats of the β-catenin MTAD may be effective.
Indeed, multiple copies of FDTDL in DRPs induced VEGF-A; the effect increased with copy number (Fig. 6). Eleven copies of the MTAD caused not only a stronger but also a longer effect, double that caused by VP-16; perhaps the MTAD increases the half-life of the DRP.
We demonstrated that the DRP can be used as either a transcriptional activator or a repressor depending on its transeffect domain (Fig. 10). In the absence of a transeffect domain, the DRP acted as a weak repressor (Figs. 7 and 10), which may be due to a cryptic repressor domain or steric interference.
Transactivation and repression domains from human transcription factors function in plant systems (37), so we tested the plant-derived SRDX domain in DRPs. SRDX not only repressed VEGF-A expression in human cells but, remarkably, repression was hypoxia-specific (Fig. 7).
Recently, it was demonstrated that a small interfering RNA (siRNA) directed against human VEGF-A reduced hypoxia-induced VEGF protein levels by approximately one-half in HEK293 cells (38). ATFs were more potent (23) and, as shown in Fig. 7, the DRP repressors were also more potent than siRNA. Similarly, reports of 60-75% repression of CHK2 by siRNA (39) indicate less potency than the full repression seen with ATFs (25). It will be interesting to learn whether ATFs and DRPs are consistently more potent than siRNA.
We have shown that a DNA binding protein with high affinity and specificity can be readily designed and fused to a CPP, NLS, and transeffect domain. The resulting protein, or DRP, becomes a potent exogenous regulator of endogenous genes. DRPs provide a convenient way to modulate the direction, time, location, magnitude, and duration of the desired change in gene expression. These properties should prove useful in functional genomics studies. The ability to up-regulate as well as down-regulate any gene by protein treatment should expand the application of functional genomics to organisms that are refractory to nucleic acid technologies. This may enable the creation of new animal models of disease. Because many human conditions could be treated by increasing or decreasing the expression of endogenous genes, DRPs may have therapeutic applications. For example, R9-NLS-AZP-SRDX, which selectively represses VEGF-A secretion by hypoxic cells, may be a selective inhibitor of tumor vascularization, permitting neovascularization to continue in healthy tissues. These properties could be useful in the treatment of cancer.
Supplementary Material
Acknowledgments
We thank Kim Vo for technical assistance; and the members in the sequencing department at the Torrey Mesa Research Institute and Diversa Corporation for their sequence analysis. The Torrey Mesa Research Institute and Diversa Corporation provided all financial support.
Abbreviations: DRP, designed regulatory protein; AZP, artificial zinc-finger protein; ATF, artificial transcription factor; CPP, cell-penetrating peptide; PTD, protein transduction domain; VEGF-A, vascular endothelial growth factor A; NLS, nuclear localization signal; KRAB, Krüppel-associated box; EMSA, electrophoretic mobility-shift assay; MTAD, minimal transactivation domain.
References
- 1.Sera, T. & Uranga, C. (2002) Biochemistry 41, 7074-7081. [DOI] [PubMed] [Google Scholar]
- 2.Beerli, R. R., Segal, D. J., Dreier, B. & Barbas, C. F., III (1998) Proc. Natl. Acad. Sci. USA 95, 14628-14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, L., Spratt, S. K., Liu, Q., Johnstone, B., Qi, H., Raschke, E. E., Jamieson, A. C., Rebar, E. J., Wolffe, A. P. & Case, C. C. (2000) J. Biol. Chem. 275, 33850-33860. [DOI] [PubMed] [Google Scholar]
- 4.Guan, X., Stege, J., Kim, M., Dahman, Z., Fan, N., Heifetz, P. Barbas, C. F., III, & Briggs, S. P. (2002) Proc. Natl. Acad. Sci. USA 99, 13296-13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ordiz, M. I., Barbas, C. F., III, & Beachy, R. N. (2002) Proc. Natl. Acad. Sci. USA 99, 13290-13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebar, E. J., Huang, Y., Hickey, R., Nath, A. K., Meoli, D., Nath, S., Chen, B., Xu, L., Liang, Y., Jamieson, A. C., et al. (2002) Nat. Med. 8, 1427-1432. [DOI] [PubMed] [Google Scholar]
- 7.Derossi, D., Joliot, A. H., Chassaing, G. & Prochiantz, A. (1994) J. Biol. Chem. 269, 10444-10450. [PubMed] [Google Scholar]
- 8.Elliott, G. & O'Hare, P. (1997) Cell 88, 223-233. [DOI] [PubMed] [Google Scholar]
- 9.Vives, E., Brodin, P. & Lebleu, B. (1997) J. Biol. Chem. 272, 16010-16017. [DOI] [PubMed] [Google Scholar]
- 10.Green, M. & Loewenstein, P. M. (1988) Cell 55, 1179-1188. [DOI] [PubMed] [Google Scholar]
- 11.Frankel, A. D. & Pabo, C. O. (1988) Cell 55, 1189-1193. [DOI] [PubMed] [Google Scholar]
- 12.Mann, D. A. & Frankel, A. D. (1991) EMBO J. 10, 1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschle, M., Schmidt, W., Zauner, W., Mechtler, K., Trska, B., Kirlappos, H. & Birnstiel, M. L. (1997) Proc. Natl. Acad. Sci. USA 94, 3256-3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wender, P. A., Mitchell, D. J., Pattabiraman, K., Pelkey, E. T., Steinman, L. & Rothbard, J. B. (2000) Proc. Natl. Acad. Sci. USA 97, 13003-13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futaki, S., Suzuki, T., Ohashi, W., Yagami, T., Tanaka, S., Ueda, K. & Sugiura, Y. (2001) J. Biol. Chem. 276, 5836-5840. [DOI] [PubMed] [Google Scholar]
- 16.Ho, A., Schwarze, S. R., Mermelstein, S. J., Waksman, G. & Dowdy, S. F. (2001) Cancer Res. 61, 474-477. [PubMed] [Google Scholar]
- 17.Schwarze, S. R., Ho, A., Vocero-Akbani, A. & Dowdy, S. F. (1999) Science 285, 1569-1572. [DOI] [PubMed] [Google Scholar]
- 18.Takenobu, T., Tomizawa, K., Matsushita, M., Li, S. T., Moriwaki, A., Lu, Y. F. & Matsui, H. (2002) Mol. Cancer Ther. 1, 1043-1049. [PubMed] [Google Scholar]
- 19.Dvorak, H. F. (1986) N. Engl. J. Med. 315, 1650-1659. [DOI] [PubMed] [Google Scholar]
- 20.Connolly, D. T., Heuvelman, D. M., Nelson, R., Olander, J. V., Eppley, B. L., Delfino, J. J., Siegel, N. R., Leimgruber, R. M. & Feder, J. (1989) J. Clin. Invest. 84, 1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung, D. W., Cachianes, G., Kuang, W. J., Goeddel, D. V. & Ferrara, N. (1989) Science 246, 1306-1309. [DOI] [PubMed] [Google Scholar]
- 22.Ferrara, N. (1999) J. Mol. Med. 77, 527-543. [DOI] [PubMed] [Google Scholar]
- 23.Snowden, A. W., Zhang, L., Urnov, F., Dent, C., Jouvenot, Y., Zhong, X., Rebar, E. J., Jamieson, A. C., Zhang, H. S., Tan, S., et al. (2003) Cancer Res. 63, 8968-8976. [PubMed] [Google Scholar]
- 24.Liu, P. Q., Rebar, E. J., Zhang, L., Liu, Q., Jamieson, A. C., Liang, Y., Qi, H., Li, P. X., Chen, B., Mendel, M. C., et al. (2001) J. Biol. Chem. 276, 11323-11334. [DOI] [PubMed] [Google Scholar]
- 25.Tan. S., Guschin, D., Davalos, A., Lee, Y. L., Snowden, A. W., Jouvenot, Y., Zhang, H. S., Howes, K., McNamara, A. R., Lai, A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 11997-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Triezenberg, S. J., Kingsbury, R. C. & McKnight, S. L. (1988) Genes Dev. 2, 718-729. [DOI] [PubMed] [Google Scholar]
- 27.Hecht, A., Litterst, C. M., Huber, O. & Kemler, R. (1999) J. Biol. Chem. 274, 18017-18025. [DOI] [PubMed] [Google Scholar]
- 28.Margolin, J. F., Friedman, J. R., Meyer, W. K-H., Vissing, H., Thiesen, H-J. & Rauscher, F. J., III (1994) Proc. Natl. Acad. Sci. USA 91, 4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eilers, A. L., Billin, A. N., Liu, J. & Ayer, D. E. (1999) J. Biol. Chem. 274, 32750-32756. [DOI] [PubMed] [Google Scholar]
- 30.Hiratsu, K., Matsui, K., Koyama, T. & Ohme-Takagi, M. (2003) Plant J. 34, 733-739. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, B. H., Zheng, J. Z., Leung, S. W., Roe, R. & Semenza, G. L. (1997) J. Biol. Chem. 272, 19253-19260. [DOI] [PubMed] [Google Scholar]
- 32.Pugh, C. W., O'Rourke, J. F., Nagao, M., Gleadle, J. M. & Ratcliffe, P. J. (1997) J. Biol. Chem. 272, 11205-11214. [DOI] [PubMed] [Google Scholar]
- 33.Semenza, G. L. (1999) Annu. Rev. Cell Dev. Biol. 15, 551-578. [DOI] [PubMed] [Google Scholar]
- 34.Salghetti, S. E., Muratani, M., Wijnen, H., Futcher, B. & Tansey, W. P. (2000) Proc. Natl. Acad. Sci. USA 97, 3118-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uesugi, M. & Verdine, G. L. (1999) Proc. Natl. Acad. Sci. USA 96, 14801-14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka, M. (1996) Proc. Natl. Acad. Sci. USA 93, 4311-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, I., Galweiler, L., Grosskopf, D., Schell, J. & Palme, K. (1998) Proc. Natl. Acad. Sci. USA 95, 376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reich, S. J., Fosnot, J., Kuroki, A., Tang, W., Yang, X., Maguire, A. M., Bennett, J. & Tolentino, M. J. (2003) Mol. Vis. 9, 210-216. [PubMed] [Google Scholar]
- 39.Ahn, J., Urist, M. & Prives, C. (2003) J. Biol. Chem. 278, 20480-20489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.