Abstract
Trimeric class I virus fusion proteins undergo a series of conformational rearrangements that leads to the association of C- and N-terminal heptad repeat domains in a “trimer-of-hairpins” structure, facilitating the apposition of viral and cellular membranes during fusion. This final fusion hairpin structure is sustained by protein–protein interactions, associations thought initially to be refractory to small-molecule inhibition because of the large surface area involved. By using a photoaffinity analog of a potent respiratory syncytial virus fusion inhibitor, we directly probed the interaction of the inhibitor with its fusion protein target. Studies have shown that these inhibitors bind within a hydrophobic cavity formed on the surface of the N-terminal heptad-repeat trimer. In the fusogenic state, this pocket is occupied by key amino acid residues from the C-terminal heptad repeat that stabilize the trimer-of-hairpins structure. The results indicate that a low-molecular-weight fusion inhibitor can interfere with the formation or consolidation of key structures within the hairpin moiety that are essential for membrane fusion. Because analogous cavities are present in many class I viruses, including HIV, these results demonstrate the feasibility of this approach as a strategy for drug discovery.
Fusion with the host-cell plasma membrane is a crucial stage in the life cycle of all enveloped viruses, because it is necessary to facilitate the intracellular deposition of the viral genome before replication (1). Two chief classes of fusion proteins that mediate this process have been identified. Class I fusion proteins include HIV gp120, influenza hemagglutinin, and the F proteins from paramyxoviruses. The E proteins of Dengue virus, tick-borne encephalitis virus, and Semliki Forest virus, represent class II fusion proteins (2, 3). These two virus groups have evolved a structurally divergent, yet mechanistically analogous, fusion apparatus (4).
The respiratory syncytial virus (RSV) class I fusion machinery is located within the F1 subunit, which possesses a hydrophobic fusogenic N terminus that is proposed to insert into the cellular target membrane during the fusion process (5). Immediately adjacent to the fusion peptide is the N-terminal heptad repeat (HR-N), whereas an analogous C-terminal heptad repeat (HR-C) region is located proximal to the transmembrane-spanning domain of the F1 protein (6–9). Heptad repeats assume an extended α-helical conformation that can readily oligomerize into coiled-coils composed of two or more α-helices that adopt an overall helical shape (10). During the fusion process, the HR-N and HR-C of the RSV F1 protein associate in a hairpin-like configuration that promotes the juxtaposition of the viral and cellular envelopes (8). Within the fusion-protein hairpin structure, the HR-N domain forms a central trimeric coiled-coil. This HR-N arrangement produces binding grooves that accommodate the three HR-C segments. Together, they associate in a tight complex in which the N- and C-peptides are aligned in an antiparallel fashion (6, 8). The resultant structure is a stable six-helix bundle, or “trimer-of-hairpins.” With structural data gathered from several viral class I fusion proteins, a burgeoning concordance has arisen, indicating that the formation of a fusion-protein trimer-of-hairpins motif is a critical prelude to fusion between viral and cellular membranes (11–13).
Synthetic HR-C and HR-N peptides have been shown to inhibit the fusion of cognate viruses, presumably by binding to the prehairpin intermediate and interfering with the formation of the six-helix bundle structure (9, 14–16). However, orally bioavailable, low-molecular-weight inhibitors of this process would be preferable and provide a potential platform for drug development against many different viruses with class I fusion proteins. We have recently described BMS-433771 (see Fig. 1 A) as an orally active, small-molecule inhibitor of RSV fusion (17, 18). Photoaffinity-labeling results presented here provide a deeper understanding of the mechanism of action and mode of binding of this inhibitor specifically and the inhibition of viral fusion by low-molecular-weight compounds in general.
Fig. 1.
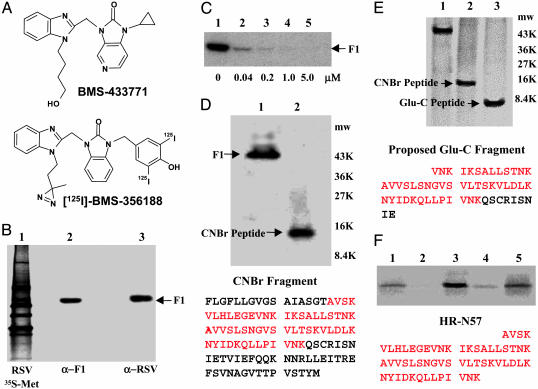
Specific labeling of F1 by [125I]BMS-356188. (A) The structures of BMS-433771 and [125I]BMS-356188. (B) Immunoprecipitation of affinity-labeled F1 polypeptide from intact virus by using either α-RSV polyclonal IgG to all RSV antigens (lane 3) or a mAb to F1 (lane 2). The F1 Mr (48,051) is indicated. Lane 1 is an α-RSV immunoprecipitation of [35S]Met-labeled RSV. (C) BMS-433771 inhibits [125I]BMS-356188 affinity labeling of the F1. The α-F1 protein immunoprecipitation analysis was performed as described for B. Increasing concentrations of BMS-433771 were used, as indicated below each lane. The [125I]BMS-356188 concentration is 4 nM. (D) CNBr cleavage of the F1 polypeptide after affinity labeling and immunoprecipitation. Lane 1, uncleaved F1; lane 2, CNBr-cleaved F1. Mr markers are indicated. Sequence of the CNBr peptide is illustrated with the HR-N region in red. (E) Endoprotease Glu-C digest of affinity-labeled F1 compared with CNBr fragment of the same preparation. The proposed sequence of the Glu-C peptide is shown with the HR-N portion in red. (F) Affinity labeling of the HR-N57 peptide. HR-N57 (8 μM) was labeled with 10 nM [125I]BMS-356188 (lane 1), in the presence of 1 μM of BMS-433771 (lane 2), with 50 μM RFI-461 (lane 3), with 50 μM RFI-461 in presence of 1 μM BMS-433771 competitor (lane 4), and in 20% trifluoroethanol (lane 5). The HR-N57 sequence is shown in red.
Materials and Methods
Compounds and RSV Assays. BMS-433771 was synthesized by the Medicinal Chemistry group at Bristol-Myers Squibb. We prepared [125I]BMS-356188 (19) as described in ref. 20. RFI-461 was synthesized as described in ref. 21. The growth of virus, plaque titration, and measurement of inhibition through a [35S]methionine incorporation assay were performed as described in ref. 17.
HR-N27, HR-N42, and HR-N57 Synthesis and Purification. HR-N27 and HR-N42 peptides were synthesized on a 9050 MilliGen peptide synthesizer by using Fmoc solid-phase chemistry on Peg resin (Applied Biosystems) and N-[dimethylamino)-1 H-1,2,3,-triazolo[4,5-b] pyridin-1-ylmethylene]-N-methylmethanaminium hexaflurophosphate N-oxide as the activator. Extended coupling for up to 60 min and increased deblocking up to 50% were used. The peptides were cleaved from the resin by 90% trifluoroacetic acid/5% thioanisole/3% ethanedithiol/2% anisole for 2 h, precipitated in diethyl ether, and washed extensively with diethyl ether before drying. HR-N57 was synthesized as described in ref. 8.
Photoaffinity Labeling of F1 Protein from Virus and Synthetic F1 HR-N Peptides. For RSV F1 protein labeling, 100 μl of RSV (≈1 × 107 plaque-forming units per ml) was incubated with 4 nM [125I]BMS-356188 (1.5 μM, 70 Ci/mmol in ethanol; 1 Ci = 37 GBq) for 30 min at 37°C. Samples were irradiated for 4–7 min from a 5-cm height (22). An equivalent volume of RIPA buffer (150 mM NaCl/50 mM Tris·HCl, pH 7.8/1% NP-40/0.5% sodium deoxycholate/0.1% SDS) was added, and immunoprecipitation analysis was performed (17). For large-scale F protein labeling, 0.5–1.0 ml of RSV and 20–40 nM [125I]BMS-356188 was used. After incubation, irradiation was carried out in 35-mm2 tissue-culture plates. After the addition of an equal volume of RIPA buffer, a 50- to 100-μl slurry of anti-RSV fusion protein-immunoaffinity resin was added. The immunoaffinity resin was made by covalently coupling the F protein mAb, Synagis (MedImmune, Gaithersburg, MD), to protein-G Sepharose 4 Fast Flow (Amersham Biosciences) by using standard protocols. Samples were incubated with gentle rocking for 0.5–1.0 h and then washed four times in RIPA buffer. Affinity-labeled RSV F1 was batch-eluted with 0.2 M glycine (pH 2.6) or 1% SDS, followed by heating at 95°C for 2 min. We affinity-labeled 8-μm concentrations of N27, N42, and N57 peptides in 25 μl of PBS by using 40 nM [125I]BMS-356188. PAGE analysis was performed as described in ref. 22.
Cyanogen Bromide (CNBr) and Endoproteinase Glu-C Fragment Generation and Analysis. After the elution of affinity-labeled F1 from the immunoaffinity resin, it was precipitated with eight volumes of acetone or methanol and pelleted by centrifugation at 16 × g for 5 min. CNBr treatment was performed as described in ref. 22. For Glu-C digestion, air-dried affinity-labeled F1 pellets were dissolved in a minimal volume of 1% SDS at 50°C, cooled to 25°C, and diluted 10-fold in 0.1 M ammonium bicarbonate (pH 8.1). We added 5 μm of endoproteinase Glu-C (Sigma), and incubation was carried out for 16–24 h at 37°C. SDS/PAGE sample buffer was added directly to the digest. Glu-C and CNBr-cleaved samples were electrophoresed on 15% Tris-Glycine Ready gels (Bio-Rad) and air-dried for analysis of molecular weight or electrophoretically blotted to poly(vinylidene difluoride) (Bio-Rad) membrane after SDS/PAGE and stained according to the manufacturer's instructions. Dried membranes were autoradiographed, and the stained bands corresponding to radiolabeled peptides were excised. Sequence analysis was performed on a 491 Procise protein sequencer (Applied Biosystems) by using the standard anilinothiazolinone method. For quantitation of coupled affinity label, each cycle was counted in 10 ml of Econofluor (NEN) scintillation fluid in a Beckman counter (Beckman Coulter).
CD. HR-N27, HR-N42, and HR-N57 peptides were dissolved in PBS (pH 7.4) at a dilution at which the maximum optical density in the 200- to 260-nm range was <1.2 absorbance units. CD spectra were obtained by using a Jasco 720 spectropolarimeter (Easton, MD).
Computer-Assisted Modeling. Within the crystal structure of the RSV trimer-of-hairpins, a binding pocket with the potential to accommodate small-molecule inhibitors was identified inside the deep groove of the HR-N57 heptad-repeat trimer (8). Several potential binding models were generated by docking BMS-433771 into the hydrophobic pocket by using dock 4.0 (23). During molecular dynamics (MD) simulations, the backbone heavy atoms of the protein were restrained. The average rms deviation for the protein (in Å; see Fig. 3B) is a reflection of protein side-chain motion during the MD. Average binding energy (kcal/mol) = (Σ protein–ligand complex total energy – Σprotein total energy – Σligand total energy)/1,000 frames. The rms deviation and binding energy were averaged over 1,000 frames taken every 0.5 ps from a 500-ps MD simulation.
Fig. 3.
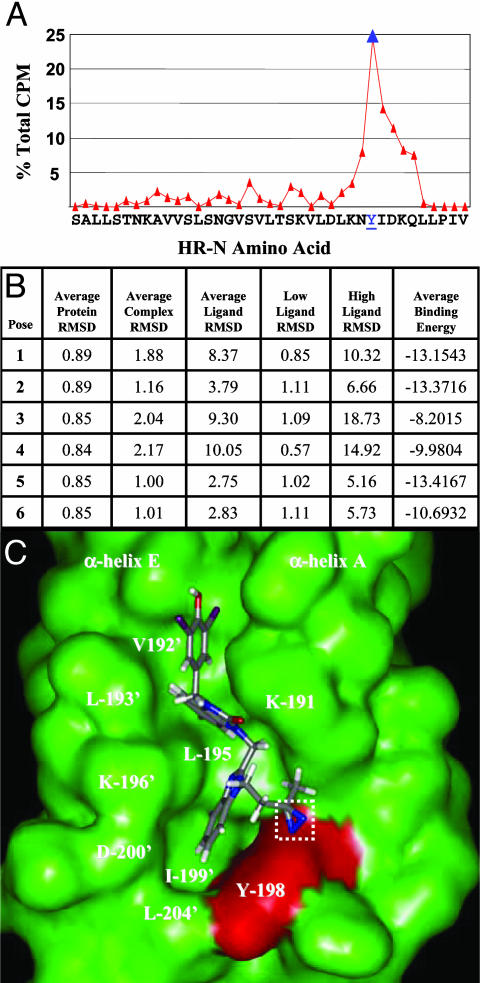
Covalent attachment site of [125I]BMS-356188 within the HR-N hydrophobic pocket. (A) Sequence analysis of affinity-labeled HR-N42. Percent of total [125I] counts recovered for each amino acid sequencing cycle is indicated. Tyr-198 is the most prevalently labeled amino acid (blue). (B) Molecular dynamic simulation scores for the docking of BMS-433771 into the hydrophobic pocket with dock 4.0.(C) Model (pose 6) of [125I]BMS-356188 within the HR-N hydrophobic cavity (green). The photoreactive diazerine (white box) is oriented toward Tyr-198 (red surface). HR-N α-helices A and E and selected amino acids are indicated.
Results
A radiolabeled analog of the RSV fusion inhibitor BMS-433771, [125I]BMS-356188, contains a photoreactive diazirine group (Fig. 1 A) (19, 20). Irradiation with UV light produces a highly reactive carbene species that can insert into amino acid residues proximal to the binding site. This photoaffinity probe exhibited potent inhibitory activity (EC50 = 38 nM) against the long strain of RSV (19). When the affinity probe was photoactivated in the presence of virus, the RSV F1 subunit of the fusion protein was the only covalently labeled viral protein (Fig. 1B). Anti-RSV antibody, immunoreactive to all RSV antigens (Fig. 1B, lane 1), immunoprecipitated a single radiolabeled protein that migrates at the molecular weight of the F1 polypeptide (Fig. 1B, lane 3). Immunoprecipitation with a mAb specific for the F1 also identified the same species (Fig. 1B, lane 2), indicating that the photoreactive compound modifies only the F1 polypeptide. BMS-433771 can inhibit the covalent cross-linking between [125I]BMS-356188 and the F1 polypeptide in a dose-dependent fashion, establishing that the binding of [125I]BMS-356188 to the F1 is specific and that it binds to the same site as BMS-433771 (Fig. 1C). To further map the photoaffinity-labeling site within F1, the polypeptide was treated with CNBr, producing a new, single radiolabeled peptide of ≈11,000–13,000 Da (Fig. 1D, lane 2). The peptide was sequenced and identified as the N terminus of the F1 polypeptide, comprising amino acids 137–251 of the fusion protein (Fig. 1D). This 115-aa fragment includes both the fusion peptide and the HR-N, which are two important domains that have been shown (24) to be critically involved in viral fusion (Fig. 1D).
Additional analysis of the binding-site elements covalently labeled by [125I]BMS-356188 was explored by treatment with endoprotease Glu-C, which generated a single predominant radiolabeled band with a molecular mass ranging between ≈5,000 and 7,000 Da (Fig. 1E, lane 3). This band is proposed to be the 5,998-Da F1 peptide fragment comprising amino acids 164–218 of the F protein, which encompasses most the HR-N peptide (Fig. 1E).
Relatively short peptides corresponding to the RSV HR-N have been shown to spontaneously associate into trimeric coiled-coils, mirroring the conformation adopted by the intact fusion protein at the time of fusion (8). Consequently, we tested the ability of the photoaffinity probe to label the 57-residue peptide of the RSV HR-N (HR-N57) used to solve the crystal structure of the RSV fusion core (8). Initial experiments indicated that the labeling of HR-N57 by [125I]BMS-356188 was relatively weak, but binding could readily be inhibited by BMS-433771 (Fig. 1F, lanes 1 and 2). During competition studies, it was observed that a significant increase in labeling could be achieved by adding the RSV fusion inhibitor RFI-641 (Fig. 1F, lane 3, and ref. 25). RFI-641 has been shown to bind to the F1 N-terminal heptad repeat (21). However, we have observed that this compound does not bind to the same site as BMS-433771 (unpublished data). Whereas it was anticipated that RFI-641 would not interfere with affinity labeling, the inclusion of RFI-641 significantly enhanced the radiolabeling of F1 by [125I]BMS-356188. Interestingly, RFI-641 has been reported to bind to purified RSV F-protein and prevent aggregation, possibly by stabilizing the trimers (25). The RFI-641-enhanced labeling was inhibited by the addition of BMS-433771, indicating that the photoaffinity probe was binding in the same mode under both conditions (Fig. 1F, lane 4). A similar, but weaker stimulatory effect on affinity labeling was seen when 20% trifluoroethanol was used in lieu of RFI-641 (Fig. 1F, lane 5). Trifluoroethanol is used as a cosolvent to mimic a membrane environment (26). Trifluoroethanol has also been shown to stabilize another RSV F1 peptide (27), as well as many other α-helices (28). Therefore, RFI-461 and trifluoroethanol may be functioning in a similar fashion with respect to the stimulation of photoaffinity labeling.
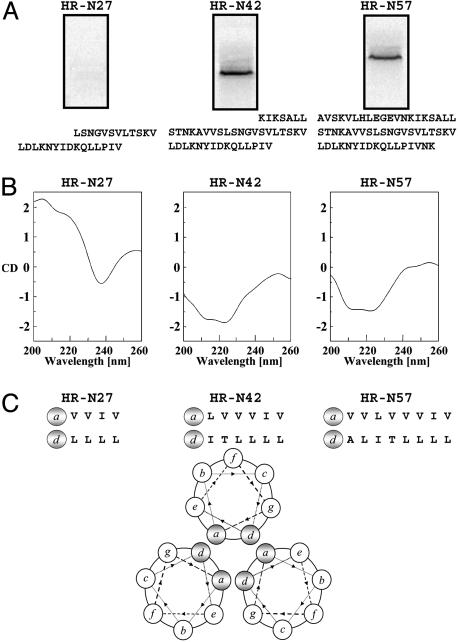
Labeling experiments were also performed with shorter HR-N peptides containing 42 (HR-N42) and 27 (HR-N27) amino acids. The HR-N42 peptide was radiolabeled by the photoaffinity reagent, but the shorter HRN-27 peptide was not (Fig. 2A). These results correlate with the ability of HR-N57 and HR-N42 to fold into α-helical structures in PBS, as revealed by CD measurements and the failure of HR-N27 to adopt a similar conformation (Fig. 2B). The HR-N42 and HR-N57 peptides exhibit typical α-helix CD spectra, with ≈222-nm and ≈209-nm minima. This signature α-helix CD pattern is absent from the HR-N27 tracing, most likely due to the reduced number of hydrophobic residues available to participate in intramolecular associations that stabilize α-helices and intermolecular interactions that favor trimer formation, as illustrated in the heptad repeat helical wheel projections (Fig. 2C). The HR-N27 peptide has only 8 hydrophobic residues per α-helix available for interaction, whereas the HR-N42 and HR-N57 peptides possess 12 and 16 hydrophobic amino acids, respectively.
Fig. 2.
HR-N27, HR-N42, and HR-N57 peptide affinity labeling, CD spectra, and hydrophobic heptad repeat pattern. (A) HR-N peptides (8 μM) were labeled with 40 nM [125I]BMS-356188 in the presence of 50 μM RFI-461 and analyzed by 15% SDS/PAGE. HR-N27, HR-N42, and HR-N57 labeling results and sequences are indicated. Efficient affinity labeling was seen for HR-N42 and HR-N57 only. (B) CD spectra for HR-N peptides in PBS alone. RFI-461 is omitted because compound autofluorescence interferes with CD measurements. The α-helical signatures are exhibited by HR-N42 and HR-N57 only. (C) Heptad repeat amino acids indicated for the three HR-N peptides. The a and d positions of hydrophobic amino acids shown to participate in timer formation are predicted from the helical wheel projection.
With the ability to achieve specific covalent binding using an HR-N peptide, direct peptide sequencing was used to locate the affinity probe attachment site within this structure. The shorter HR-N42-labeled peptide was covalently labeled and sequenced. [125I]BMS-356188 was found to be attached primarily to Tyr-198, which participates in the formation of a hydrophobic cavity within the HR-N trimer (Fig. 3A). From an examination of the RSV fusion core crystal structure, this pocket was proposed as a potential target for small-molecule inhibitors (8). Also revealed in the x-ray structure were two key aromatic residues from the HR-C occupying this cavity, Phe-483 and Phe-488 (8). Molecular Dynamics simulations were used to model the compound into this pocket. The top 100 scoring poses were clustered into six binding modes (Fig. 3B). The best scoring pose from each cluster was used as the starting point for restrained MD scoring using charmm (29), the CFF98 force field (30), and a generalized born solvation model (31). Each pose was assessed based on the MD ensemble averaged binding energy and MD stability. The small binding energy range between the top poses made it difficult to select a single binding mode using energy alone. For poses 1–4, the ligand dissociates from the hydrophobic pocket and slides along either the groove (poses 1 and 2) or surface (poses 3 and 4) of the protein during the MD simulation (Fig. 3B), whereas in poses 5 and 6, the inhibitor binds solely within the hydrophobic pocket. The prominent labeling of Tyr-198 is most consistent with pose 6 (Fig. 3C), where this small molecule is in the hydrophobic cavity with the photoreactive diazirine oriented toward the Tyr-198 residue. In this model, the benzimidazolone ring system occupies the C-terminal Phe-488 binding site, with the pyridine moiety buried deep in the groove and sandwiched between HR-N α-helices A and E, near Leu-195 of α-helix A and Leu-193′ and Val-192′ of α-helix E. The diiodo-hydroxybenzyl ring is proximal to the aliphatic chain of Lys-191, mimicking the C-terminal Ile-492. The benzimidazole ring system occupies the C-terminal Phe-483 binding position and is sandwiched between Tyr-198, Lys-196′, Ile-199′, Asp-200′, and Leu-204′ of the N-terminal α-helices A and E, respectively. The leading and trailing labeling that is observed around Tyr-198 could essentially be explained by a secondary binding mode (pose 5) and the rocking motion observed for the benzimidazole ring that would allow for an alternate labeling opportunity.
Although this study provides insight into the mechanism of action for this series of small-molecule inhibitors, it does not provide a complete understanding of where these molecules act in the activation pathway. We speculate that these inhibitors interfere with the interaction of key amino acids (residues Phe-483 and Phe-488) from the HR-C peptide with the prominent cavity in the HR-N trimer and that this interference alone is sufficient to abrogate fusion (Fig. 4). By analogy with the recently solved crystal structure of the Newcastle disease virus F protein (32, 33), this cavity may exist in the native fusion protein conformation of the RSV fusion protein and is preserved in the fusogenic trimer-of-hairpins. Thus, it is possible that both the native fusion protein from intact virus and a trimeric HR-N peptide were specifically labeled by [125I] BMS-356188. However, we cannot rule out the possibility that photoaffinity labeling of the intact protein occurs during a conformational transition of the F1 protein.
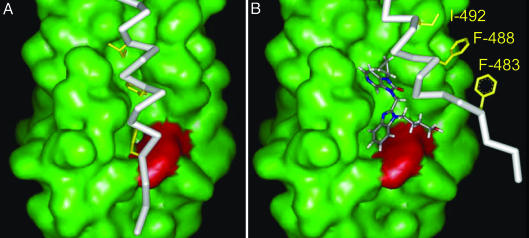
Fig. 4.
Proposed model for the mechanism of action of this inhibitor series. (A) The HR-C (white stick display; Phe-483, Phe-488, and Ile-492 aa side chains highlighted in yellow) association with the HR-N hydrophobic pocket (green surface display; Tyr-198 is shown in red). The HR-C amino acids Phe-483 and Phe-488 bind in the HR-N hydrophobic pocket. (B) The fusion inhibitor, BMS-433771, residing in the HR-N pocket, blocks the HR-C Phe-483, Phe-488, and Ile-492 associations.
Discussion
The fusion of viral and cellular membranes is an essential early stage of virus infection that is amenable to intervention as an antiviral strategy. Despite extensive sequence disparities, the class I fusion proteins of certain enveloped animal viruses share many common structural and functional properties. Evidence from the study of several viruses suggests that class I fusion proteins can assume three distinct conformational states, which are (i) the nonfusogenic native structure, in which the fusion peptide is buried within the trimeric protein; (ii) the transient prehairpin intermediate, in which the N-terminal fusion peptide is extended to penetrate the host-cell target membrane; and (iii) the fusogenic trimer-of-hairpins structure, in which the HR-C and HR-N peptides are associated in a six-helix-bundle conformation that is a critical element in the union of viral and cellular envelopes (11, 12, 34). The transitions from the native state to the trimer-of-hairpin conformations via the prehairpin intermediate offer opportunities for interference and inhibition. Several influenza fusion inhibitors have been reported to prevent the conformational change of hemagglutinin from the native, metastable form to the prehairpin intermediate (22, 35, 36). Another type of influenza fusion inhibitor has been shown to prematurely trigger the conformational change of hemagglutinin (37). Inhibition through targeting of the transient prehairpin intermediate can be accomplished in many viruses with synthetic HR-C peptide derivatives, as described for RSV (9, 15, 16) and HIV (38). For HIV, T-20 (Fuzeon, Enfuvirtide) was the first synthetic HR-C peptide derivative to be approved by the Food and Drug Administration for the treatment of HIV (38). HR-N peptides can also inhibit virus fusion, but they are generally less potent and not universally effective (15).
The fusion trimer-of-hairpins structure is maintained through protein–protein interactions, which are associations that have been believed traditionally to be refractory to small-molecule inhibition because of the vast number of contacts involved between the various peptides. However, at the C-terminal of the HR-N trimer surface, a deep groove exists that opens into a cavity, forming a hydrophobic pocket recognized as a potential binding site for small-molecule inhibitors in HIV, simian virus 5, RSV, and Visna virus (8, 39, 40). In HIV, because only three highly conserved amino acids from the C-terminal (Trp-628, Trp-631, and Ile-635) occupy the HR-N pocket, it was theorized that small organic molecules could mimic the interactions of this triplet and interfere with stability of the hairpin structure (41). Virtual screens and ELISA-based assays employing HIV HR-C/HR-N peptides have identified small-molecule compounds that may bind inside the hydrophobic N-terminal cavity (42–44). However, to our knowledge, the direct demonstration of binding to the cavity by cocrystallization or affinity-labeling experiments has yet to be accomplished.
This article demonstrates directly the binding of a virus fusion inhibitor within the hydrophobic pocket formed by the HR-N coiled-coil by using photoaffinity-labeling technology. This series of small molecule fusion inhibitors is exemplified by BMS-433771, which is orally active in rodent models of RSV infection (17, 18) and a candidate for clinical evaluation for a disease for which current therapy is inadequate. The high potency of this class of inhibitor suggests that the interruption of the association of Phe-483 and Phe-488 of the C-terminal with the N-terminal pocket is sufficient to disrupt the precise conformation within the fusion hairpin configuration that is critical for the fusion of viral and cellular envelopes. Studies with the related paramyxovirus simian virus 5 support this model. Within the simian virus 5 six-helix bundle, Leu-447 and Ile-449 of the HR-C domains bind to the hydrophobic pockets within the HR-N timers. A single I449G substitution in the C-terminal domain destabilizes a six-helix bundle structure composed of HR-N and HR-C peptides, as demonstrated by CD measurements and thermal melting analysis (45).
A common theme among class I enveloped viruses is that analogous HR-N and HR-C associations form within a trimer-of-hairpins structure as a prelude to virus-cell fusion. In a number of class I fusion proteins, key amino acid residues from the HR-C make important contacts with hydrophobic pockets of the HR-N. The identification of the binding site for BMS-433771 within the hydrophobic pocket in the HR-N of the RSV fusion protein establishes proof of concept that it is possible to identify an orally bioavailable, small-molecule fusion inhibitor that binds in this crucial pocket.
Author contributions: C.C., R.C., N.A.M., and M.K. designed the research; C.C., K.-L.Y., J.R., Z.L., and R.D. performed research; D.D.D., A.S., and R.D. contributed new reagents or analytical tools; C.C., D.R.L., Y.S., N.A.M., and M.K. analyzed the data; C.C., D.R.L., and M.K . wrote the paper; and N.A.M. edited the manuscript.
Abbreviations: RSV, respiratory syncytial virus; HR-N, N-terminal heptad repeat; HR-C, C-terminal heptad repeat; MD, molecular dynamics; CNBr, cyanogen bromide.
References
- 1.Lamb, R. A. (1993) Virology 197, 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. (2004) Nature 427, 313–319. [DOI] [PubMed] [Google Scholar]
- 3.Gibbons, D. L., Vaney, M. C., Roussel, A., Vigouroux, A., Reilly, B., Lepault, J., Kielian, M. & Rey, F. A. (2004) Nature 427, 320–325. [DOI] [PubMed] [Google Scholar]
- 4.Jardetzky, T. S. & Lamb, R. A. (2004) Nature 427, 307–308. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., Huang, Y. T. & Wertz, G. W. (1984) Proc. Natl. Acad. Sci. USA 81, 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawless-Delmedico, M. K., Sista, P., Sen, R., Moore, N. C., Antczak, J. B., White, J. M., Greene, R. J., Leanza, K. C., Matthews, T. J. & Lambert, D. M. (2000) Biochemistry 39, 11684–11695. [DOI] [PubMed] [Google Scholar]
- 7.Matthews, J. M., Young, T. F., Tucker, S. P. & Mackay, J. P. (2000) J. Virol. 74, 5911–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao, X., Singh, M., Malashkevich, V. N. & Kim, P. S. (2000) Proc. Natl. Acad. Sci. USA 97, 14172–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert, D. M., Barney, S., Lambert, A. L., Guthrie, K., Medinas, R., Davis, D. E., Bucy, T., Erickson, J., Merutka, G. & Petteway S. R., Jr. (1996) Proc. Natl. Acad. Sci. USA 93, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutch, R. E., Leser, G. P. & Lamb, R. A. (1999) Virology 254, 147–159. [DOI] [PubMed] [Google Scholar]
- 11.Dutch, R. E., Jardetzky, T. S. & Lamb, R. A. (2000) Biosci. Rep. 20, 597–612. [DOI] [PubMed] [Google Scholar]
- 12.Eckert, D. M. & Kim, P. S. (2001) Annu. Rev. Biochem. 70, 777–810. [DOI] [PubMed] [Google Scholar]
- 13.Colman, P. M. & Lawrence, M. C. (2003) Nat. Rev. Mol. Cell. Biol. 4, 309–319. [DOI] [PubMed] [Google Scholar]
- 14.Wild, T. F. & Buckland, R. (1997) J. Gen. Virol. 78, 107–111. [DOI] [PubMed] [Google Scholar]
- 15.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, E., Sun, X., Qian, Y., Zhao, L., Tien, P. & Gao, G. F. (2003) Biochem. Biophys. Res. Commun. 302, 469–475. [DOI] [PubMed] [Google Scholar]
- 17.Cianci, C., Yu, K. Y., Combrink, K., Sin, N., Pearce, B., Wang, A., Civiello, R., Voss, S., Luo, G., Kadow, K., et al. (2004) Antimicrob. Agents Chemother. 48, 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cianci, C., Genovesi, E. V., Lamb, L., Medina, I., Yang, Z., Zadjura, L., Yang, H., D'Arienzo, C., Sin, N., Yu, K. L., et al. (2004) Antimicrob. Agents Chemother. 48, 2448–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu, K. L., Zhang, Y., Civiello, R. L., Trehan, A. K., Pearce, B. C., Yin, Z., Combrink, K. D., Gulgeze, H. B., Wang, X. A., Kadow, K. F., et al. (2004) Bioorg. Med. Chem. Lett. 14, 1133–1137. [DOI] [PubMed] [Google Scholar]
- 20.Dischino, D. D., Cianci, C. W., Civiello, R., Krystal, M., Meanwell, N. A., Morimoto, H., Williams, P. G. & Yu, K.-L. (2003) J. Labelled Comp. Radiopharm. 46, 1105–1116. [Google Scholar]
- 21.Aulabaug, A., Ding, W., Ellestad, G. A., Gazumyan, A., Hess, C. D., Krishnamurthy, G., Mitsner, B. & Zaccardi, J. (2000) Drugs of the Future 25, 287–294. [Google Scholar]
- 22.Cianci, C., Yu, K. L., Dischino, D. D., Harte, W., Deshpande, M., Luo, G., Colonno, R. J., Meanwell, N. A. & Krystal, M. (1999) J. Virol. 73, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing, T. J. A. & Kuntz, I. D. (1997) J. Comput. Chem. 18, 1175–1189. [Google Scholar]
- 24.Chambers, P., Pringle, C. R. & Easton, A. J. (1992) J. Gen. Virol. 73, 1717–1724. [DOI] [PubMed] [Google Scholar]
- 25.Ding, W., Mitsner, B., Krishnamurthy, G., Aulabaugh, A., Hess, C. D., Zaccardi, J., Cutler, M., Feld, B., Gazumyan, A., Raifeld, Y., et al. (1998) J. Med. Chem. 41, 2671–2675. [DOI] [PubMed] [Google Scholar]
- 26.Vranken, W. F., Fant, F., Budesinsky, M. & Borremans, F. A. (2001) Eur. J. Biochem. 268, 2620–2628. [DOI] [PubMed] [Google Scholar]
- 27.Toiron, C., Lopez, J. A., Rivas, G., Andreu, D., Melero, J. A. & Bruix, M. (1996) Biopolymers 39, 537–548. [DOI] [PubMed] [Google Scholar]
- 28.Roccatano, D., Colombo, G., Fioroni, M. & Mark, A. E. (2002) Proc. Natl. Acad. Sci. USA 99, 12179–12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4, 187–217. [Google Scholar]
- 30.Maple, J. R., Hwang, M.-J., Jalkanen, K. J., Stockfisch, T. P. & Hagler, A. T. (1998) J. Comp. Chem. 19, 430–458. [Google Scholar]
- 31.Dominy, B. & Brooks, C. L. R. (1999) J. Phys. Chem. 103, 3765–3773. [Google Scholar]
- 32.Chen, L., Gorman, J. J., McKimm-Breschkin, J., Lawrence, L. J., Tulloch, P. A., Smith, B. J., Colman, P. M. & Lawrence, M. C. (2001) Structure (London) 9, 255–266. [DOI] [PubMed] [Google Scholar]
- 33.Smith, B. J., Lawrence, M. C. & Colman, P. M. (2002) Protein Eng. 15, 365–371. [DOI] [PubMed] [Google Scholar]
- 34.Hernandez, L. D., Hoffman, L. R., Wolfsberg, T. G. & White, J. M. (1996) Annu. Rev. Cell Dev. Biol. 12, 627–661. [DOI] [PubMed] [Google Scholar]
- 35.Bodian, D. L., Yamasaki, R. B., Buswell, R. L., Stearns, J. F., White, J. M. & Kuntz, I. D. (1993) Biochemistry 32, 2967–2978. [DOI] [PubMed] [Google Scholar]
- 36.Plotch, S. J., O'Hara, B., Morin, J., Palant, O., LaRocque, J., Bloom, J. D., Lang, S. A., Jr., DiGrandi, M. J., Bradley, M., Nilakantan, R. & Gluzman, Y. (1999) J. Virol. 73, 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman, L. R., Kuntz, I. D. & White, J. M. (1997) J. Virol. 71, 8808–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilby, J. M., Hopkins, S., Venetta, T. M., DiMassimo, B., Cloud, G. A., Lee, J. Y., Alldredge, L., Hunter, E., Lambert, D., Bolognesi, D., et al. (1998) Nat. Med. 4, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 39.Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. (1997) Cell 89, 263–273. [DOI] [PubMed] [Google Scholar]
- 40.Malashkevich, V. N., Singh, M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan, D. C., Chutkowski, C. T. & Kim, P. S. (1998) Proc. Natl. Acad. Sci. USA 95, 15613–15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrer, M., Kapoor, T. M., Strassmaier, T., Weissenhorn, W., Skehel, J. J., Oprian, D., Schreiber, S. L., Wiley, D. C. & Harrison, S. C. (1999) Nat. Struct. Biol. 6, 953–960. [DOI] [PubMed] [Google Scholar]
- 43.Jiang, S. & Debnath, A. K. (2000) Biochem. Biophys. Res. Commun. 270, 153–157. [DOI] [PubMed] [Google Scholar]
- 44.Ernst, J. T., Kutzki, O., Debnath, A. K., Jiang, S., Lu, H. & Hamilton, A. D. (2002) Angew. Chem. Int. Ed. Engl. 41, 278–281. [DOI] [PubMed] [Google Scholar]
- 45.Russell, C. J., Kantor, K. L., Jardetzky, T. S. & Lamb, R. A. (2003) J. Cell Biol. 163, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]