Abstract
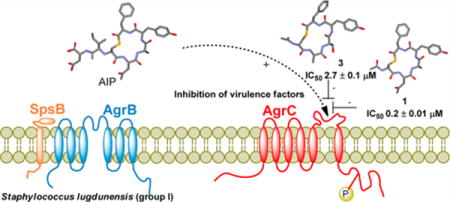
Competitive quorum sensing antagonisim (QS) offers a novel strategy for attenuating current multidrug resistant staphylococcal infections. To this end, a series of 10 truncated analogues based on the parent autoinducing peptides (AIPs) of Staphylococcus lugdunensis (groups I and II) and Staphylococcus epidermidis (groups I–III) were sequentially assessed against a newly developed Staphylococcus lugdunensis group I QS reporter strain. The truncated analogues based upon Staphylococcus lugdunensis AIP-1 (1) and AIP-2 (2) displayed respective IC50 values of 0.2 ± 0.01 μM and 0.3 ± 0.01 μM, while the truncated analogue of the Staphylococcus epidermidis AIP-1 (3) elicited an IC50 value of 2.7 ± 0.1 μM. These findings demonstrate the potential of cognate and “crosstalk” competitive quorum sensing inhibition using truncated AIPs as a means of attenuating staphylococcal infections in species beyond Staphylococcus aureus.
Graphical abstract
INTRODUCTION
The Staphylococcus genus comprises a number of extraordinary bacterial pathogens capable of causing a diverse array of acute and chronic infections leading to boils, pneumonia, toxic shock, endocarditis, and osteomyelitis.1–4 This pathogenesis is primarily mediated by virulence factors that include exotoxins, cell-wall binding proteins, proteases, lipase, and superantigens.2 These factors enable the organism to evade host defenses, to adhere to cells and the tissue matrix, to spread within the host, and to degrade cells and tissues. Hence the suppression of virulence and cell adhesion factors presents a novel approach to attenuate infection.5
Virulence factor expression is tightly regulated by intricate gene regulatory networks incorporating transcriptional regulators and two-component signal transduction system (TCSTS).6–8 At least 16 TCSTSs have been identified in Staphylococcus aureus (S. aureus), and blockade of these systems has been demonstrated to significantly attenuate infection; hence this provides a promising approach for the development of next generation antibiotic agents.9–13
Currently the most extensively characterized TCSTS in S. aureus is the QS system known as the accessory gene regulator (agr). Activation of this system up-regulates virtually all S. aureus toxins and exoenzymes (Figure 1). While the agr locus has primarily been studied in S. aureus, it appears to be conserved across the firmicutes14 and is known to contain two divergent transcripts named RNAII and RNAIII that are controlled by the P2 and P3 promoters, respectively (Figure 1).2,12,14,15
Figure 1.
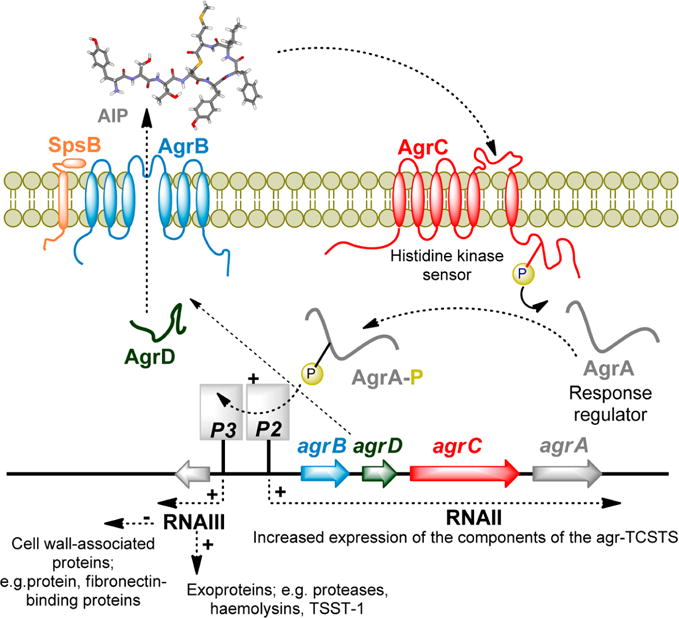
The agr locus is composed of two divergent transcripts called RNAII and RNAIII, driven by the P2 and P3 promoters, respectively. The RNAII transcript is an operon of four genes, agrBDCA, which encode the core machinery of the system. AgrD is the peptide precursor and is processed and exported through AgrB and SpsB at the cytoplasmic membrane. At the threshold concentration, autoinducing peptide (AIP) binds to the AgrC receptor, a membrane-bound histidine kinase. AIP binding activates the AgrC kinase, resulting in phosphorylation of the AgrA response regulator which in turn leads to activation of the P2 and P3 promoters.
The P3 transcript, a 517-nucleotide termed RNAIII, is the effector of the agr response, initiating the production of virulence factors. The RNAII transcript covers a four-gene operon containing agrBDCA, which encodes for the cytosolic, transmembrane, and extracellular component of the density-sensing TCSTC.
Among staphylococci, the AIPs are macrocyclic thiolactones or lactones consisting of 7–12 amino acids in which a central cysteine or serine residue is covalently linked to the C-terminal α-carboxylate to form a macrocycle comprising five amino acid residues.3,14–17 The sequence of the AIPs is highly variable; for example, on the basis of AIP primary amino acid sequence, S. aureus can be subdivided into four different agr groups (I–IV)14 while Staphylococcus epidermidis (S. epidermidis) can be subdivided into three different agr groups (I–III).18 Intriguingly, most cross-group AIP-AgrC interactions are inhibitory with AIPs activating their cognate receptor and competitively inhibiting noncognate receptors. Further cross-species inhibition of S. aureus AgrC-1, -2, and -3 by S. epidermidis AIP-1 was first observed by Otto and colleagues,19 and more recent quantification indicates S. epidermidis AIP-1 competitively inhibits S. aureus AgrC-2 and -3 with IC50 values of approximately 160 nM and 13 nM, respectively.20 Staphylococcus lugdunensis (S. lugdunensis) AIP-1 has also been shown to competitively inhibit S. aureus AgrC-1, -2, and -3 (IC50 ≈ 380 nM, IC50 ≈ 420 nM, and IC50 ≈ 40 nM, respectively).16,18,20
Although the precise evolutionary and physiological relevance of this crosstalk inhibition is yet to be elucidated, it presents significant therapeutic potential. Inhibition of the agr TCSTS by noncognate AIPs has been shown to abolish the production of the enterotoxin C3, lipase, and toxic shock syndrome toxin-1. Additionally, interference of AIP signaling through the use of competing AIPs or AIP-sequestering antibodies reduces abscess formation in S. aureus skin and soft tissue infections.21–23 These findings indicate that agr-targeted therapies, such as competitive AIP inhibition, constitute a unique therapeutic approach based on the attenuation of bacterial virulence.
The AIP macrocyclic thiolactone scaffold has been subjected to a handful of structure–activity relationship (SAR) studies that have focused on the S. aureus AIP-I,4,16,24 AIP-II,25 and AIP-III20,26,27 scaffolds. A consistent observation within these studies was the macrocycle is critical for AIP function as linear peptides do not activate agr, and further hydrolysis of the thioester blocks function. However, in relation to potent agr inhibition the most significant compound emerged from site-directed mutagenesis with alanine-scanning on S. aureus AIP-I affording the D5A variant which is a potent inhibitor of AgrC-1 (IC50 ≈ 20 nM) and later shown to be a universal inhibitor across the S. aureus species.4,16 Further, more recently it has been reported that mutation of aspartic acid to alanine at position 4 within the S. aureus AIP-III macrocyclic also affords a pan-group inhibitor with subnanomolar potency.27 Subsequent investigations focused on truncated (tr) analogues that comprised only the macrocyclic portion of the scaffold. These investigations culminated in the development of N-acetylated tr-(Ala5)-AIP-I which elicits IC50 values of approximately 0.1–5 nM across all four S. aureus agr systems.16 Further highlighting the potential of utilizing truncated analogues is the recent report of tr-(Ala2/Trp3)-AIP-III and tr-(Ala2/Tyr5)-AIP-III which both display pan-group inhibitory activity at subnanomolar concentrations.27
Despite these advances, the potential of utilizing truncated AIPs as AgrC inhibitors in staphylococcal species beyond S. aureus has received little attention as has the potential of developing a pan-staphylococcal AgrC inhibitor. Hence our attention turned to S. lugdunensis, one of the most virulent pathogens of the coagulase-negative staphylococci. S. lugdunensis is capable of the spectrum of infections usually associated with S. aureus, such as skin and soft tissue, bone, joint, cardiovascular, and CNS infections.28 At present two agr types (I and II) have been identified for S. lugdunensis,29 and molecular typing of all S. lugdunensis isolates, both commensal and infectious from a Twaiwanese tertiary hospital over a 10-year window from 2003 to 2013, found that the agr types occurred with roughly similar frequencies and did not show any statistically significant correlation with infection type.30
Although many of the toxins typically associated with S. aureus have not been identified in S. lugdunensis, various virulence factors are shared and may explain the similar pathogenic potential. Further S. lugdunensis has been associated with severe endophthalmitis, particularly postcataract surgery endophthalmitis, resulting in visual loss and dense infiltration of the vitreous.31,32 Presently the incidence rates of S. lugdunensis infection remain relatively low;33 however increasing numbers of patients with S. lugdunensis infective endocarditis have recently been reported,34,35 and these are associated with a high mortality rate compared with other coagulase-negative staphylococci.36 Further while the bacteria are sensitive to a number of antibiotics, isolated case reports of resistance to erythromycin,37 streptomycin,37 tetracycline,33 penicillin,38,39 gentamicin,40 ceftazidime,40 aminoglycosides,41 and macrolides42 have been reported. Hence S. lugdunensis has been often referred to “as a wolf in sheep’s clothing”43 and is most certainly a coagulase-negative staphylococcus that cannot be ignored.28
Hence we were of the opinion that the development of truncated derivatives based on the native macrocyclic portions of S. lugdunensis AIP-I and -II may provide an effective approach to reduce virulence in both species. This inference was supported by the recent discovery that a truncated analogue based on S. epidermidis AIP-I can function as a cognate AgrC-1 inhibitor.44 Hence in a bid to develop S. lugdunensis AgrC- I receptor inhibitors, we proposed examining the N-acetylated truncated analogues 1 and 2, which comprised the native macrocyclic portion of S. lugdunensis AIP-I and AIP-II, respectively.14 With regard to further examining cross-species interactions we also aimed to prepare the N-acetylated truncated analogues 3–5 (Figure 2), which comprised the native macrocyclic portions of S. epidermidis AIP-I, AIP-II, and AIP-III, respectively.18 From here the AgrC inhibitory activity of each of these five analogues would be sequentially assessed against a newly developed S. lugdunensis group I QS reporter strain.
Figure 2.
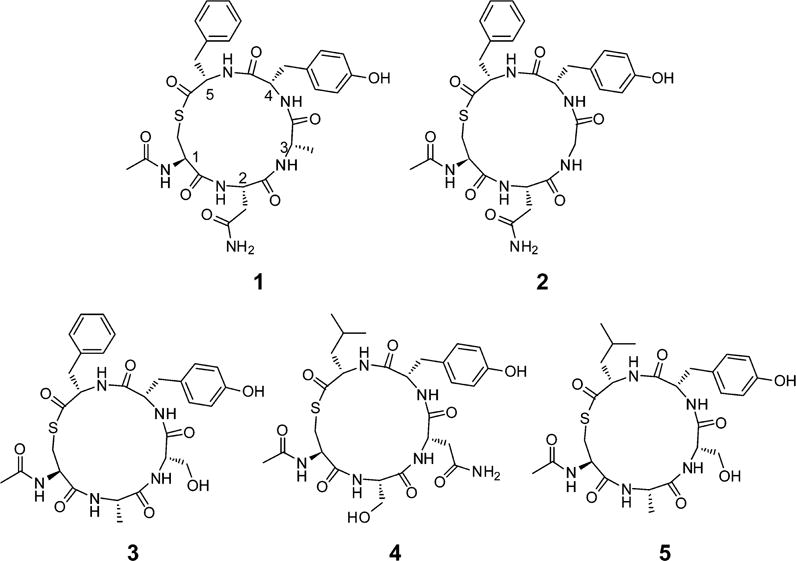
Structure N-acetylated truncated analogues 1 and 2 which comprised the native macrocyclic sequence of S. lugdunensis AIP-I and AIP-II, respectively,14 along with the N-acetylated truncated analogues 3–5, which comprised the native macrocyclic sequence of S. epidermidis AIP-I, AIP-II, and AIP-III, respectively.18 Each of these analogues was to be sequentially assessed against a newly developed S. lugdunensis group I QS reporter strain.
CHEMISTRY
As outlined in Scheme 1, the linear peptide sequences required to access the macrocycles were prepared via a standard Fmoc solid phase synthetic approach utilizing 2-chlorotrityl chloride resin. Initially a solution of resin, C-terminal amino acid residue (4 equiv), and DIPEA (8 equiv) in DCM was agitated at rt for 2 h prior to the addition of MeOH to cap unreacted 2-chlorotrityl chloride moieties. Fmoc deprotection was effected with 20% piperidine in DMF with the remainder of the linear sequences constructed using a standard Fmoc approach. Final amide capping of the cysteine N-terminal amine was achieved with 10 equiv of acetic anhydride in 10 equiv of DIPEA. Finally, concurrent cleavage of the linear peptide from the resin and deprotection of the cysteine S-4-methoxytrityl protecting group were effected using a dilute TFA cocktail to afford the linear protected pentapeptide 6.
Scheme 1. General Synthetic Protocol Utilized To Access the Protected and Deprotected Truncated Autoinducing Peptide Analoguesa.
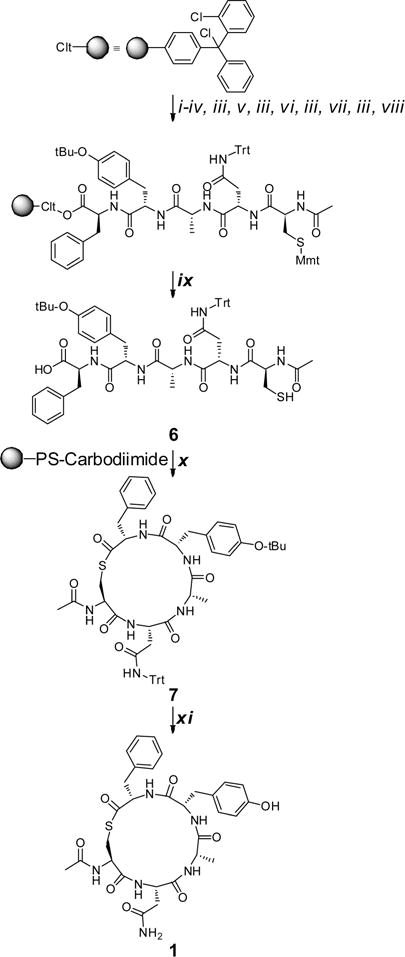
aReagents and conditions: (i) Fmoc-Phe-OH (4 equiv), DIPEA (8 equiv), DCM, rt, 2 h; (ii) MeOH, rt, 20 min; (iii) 20% piperidine/DMF; (iv) Fmoc-Tyr(O-t-Bu)-OH (4 equiv), HCTU (4 equiv), DIPEA (8 equiv), DMF, 40 °C, 1 h; (v) Fmoc-Ala-OH (4 equiv), HCTU (4 equiv), DIPEA (8 equiv), DMF, 40 °C, 1 h; (vi) Fmoc-Asn(Trt)-OH (4 equiv), HCTU (4 equiv), DIPEA (8 equiv), DMF, 40 °C, 1 h; (vii) Fmoc-Cys(Mmt)-OH (4 equiv), HCTU (4 equiv), DIPEA (8 equiv), DMF, 40 °C, 1 h; (viii) acetic anhydride/DIPEA (1:2), rt, 0.5 h; (ix) DCM/TFA/Et3SiH (96:2:2); (x) N′-polystyrene methyl-N′-cyclohexylcarbodiimide (3 equiv), CHCl3, rt; (xi) TFA/Et3SiH/H2O (95:2.5:2.5), rt, 6 h.
Turning to macrocyclization, essentially two previous approaches have been utilized, either carbodiimde based coupling4,24,45 or solution phase chemoselective thioligation.16,22,25,27 The latter approach entails cyclization of a deprotected linear sequence within a guanidinium chloride phosphate buffer solution, and while it requires two additional synthetic steps, compared to the carbodiimde approach, the thioligation tactic is amendable with all residues and is highly efficient typically effecting complete cyclization within 2h.16,22,25,27 In contrast the carbodiimde approach, which typically involves cyclization of a protected sequence in the presence of HOAt,24 is purported to be limited by low cyclization efficiency due to the steric bulk of the protecting groups.27 Nonetheless, despite this reported limitation, we were of the opinion that biological evaluation of the protected cyclized analogues (i.e., compound 7) may potentially provide additional SAR insights and consequently we were curious to assess the efficiency of this method.
To gauge the utility of the methodology and to explore possible potential for optimization, initially three polymer supported carbodiimides were assessed, the EDCI resin 8, the benzyl tethered DCC analogue resin 9, and the ether tethered DCC analogue resin 10 (Figure 3). Here the rationale for examining these resins was based investigating PS-DDC adducts with differing polarities. In separate experiments an amount of 3 mol equiv of the respective polymer supported carbodiimide was added to a 1 mM solution of the linear pentapeptide 6 in chloroform notably without the addition of HOAt. In order to ascertain a qualitative measure of cyclization efficacy, aliquots were extracted from each experiment at various time points across a 3 h period and analyzed via HPLC. As outlined in Figure 3, the EDCI resin (PS-EDCI) was ineffective affording ~10% conversion in 3 h and ~65% conversion after a 2-day period. The DCC resin (PS-DCC) proved slightly more efficient effecting ~50% conversion within 3 h while the ether tethered DCC (PS-OCD) resin promoted ~95% conversion in 3 h and complete macrocyclization within 5 h without the addition of HOAt. However, we must note that alone PS-OCD resin was ineffective for cyclization of polar nonprotected sequences. Nonetheless with the addition of 3 equiv of HOAt, completed cyclization of unprotected linear sequences was observed within 8 h (data contained in Supporting Information).
Figure 3.
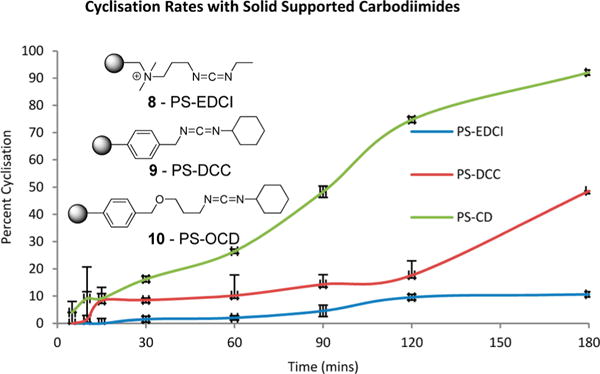
Plot of the percent conversion of the linear peptide sequence 6 to the thioester based macrocycle 7 using the solid supported carbodiimide reagent PS-EDCI, PS-DCC, and PS-OCD.
Thus, in our hands the carbodiimide approach appears considerably more effective than previously purported, and while slightly less efficient and robust than the reported chemoselective thioligation approaches,16,22,25,27 we were of the opinion that the rapid reaction workup (i.e., simple removal of the PS-OCD followed by concentration in vacuo) and access to additional protected analogues justified utilizing PS-OCD for the syntheses of the remaining analogues. Hence 11–14 (Figure 4) were synthesized utilizing this approach (e.g., Scheme 1) with the protected cyclized analogues (exemplified by compound 7, Scheme 1) purified via RP semipreparative HPLC. Subsequent global deprotection of 11–14 was effected with a TFA cocktail (TFA/H2O/Et3SiH, 95:2.5:2.5) with each final peptide subjected to RP semipreparative HPLC with purities of ≥95% as judged via analytical HPLC analysis.
Figure 4.
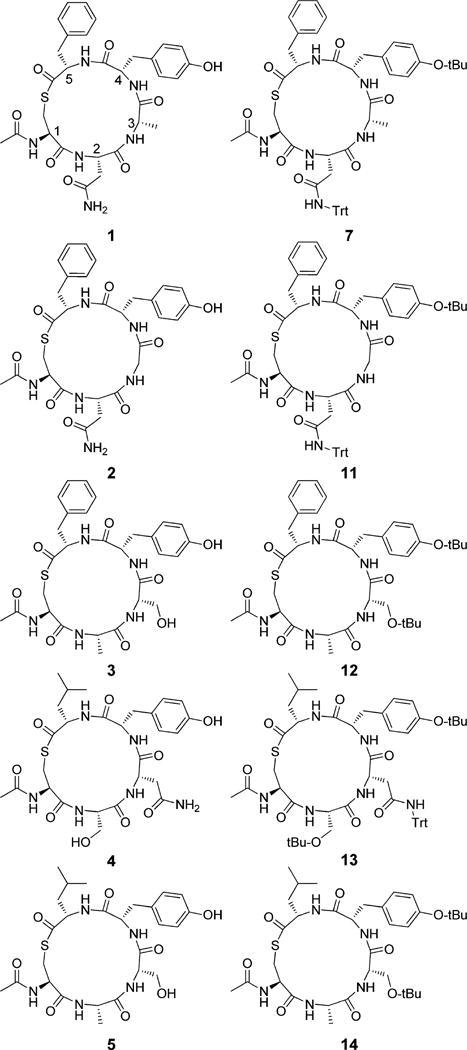
Structure of truncated deprotected analogues 1–5 and the protected analogues 7, 11–14 which were separately biologically evaluated against the S. lugdunensis group I QS reporter.
S. lugdunensis QS INHIBITORY ASSAY
In order to test inhibition of the S. lugdunensis group I QS system, we engineered a GFP reporter strain that responds to activation of the agr P3 promoter. For this purpose, the P3-GFP reporter plasmid pCM4018 was transformed into the sequenced N920143 strain,46 and a time course demonstrated GFP induction during exponential phase (see Supporting Information) as has been observed for other staphylococcal agr reporters.47 While none of the compounds impacted on bacterial growth, a number of derivatives inhibited QS as illustrated in Table 1.
Table 1.
IC50 Values Determined for the Deprotected Analogues 1–5 and the Protected Analogues 7, 11–14 against S. lugdunensis AgrC-1
| compd | IC50 (μM)a | species/agr type origin of truncated AIP | macrocycle sequenceb |
|---|---|---|---|
| S. lugdunensis I | CNAYF | ||
| 1 | 0.2 ± 0.01 | S. lugdunensis I | Ac-CNAYF |
| 2 | 0.3 ± 0.01 | S. lugdunensis II | Ac-CNGYF |
| 3 | 2.7 ± 0.1 | S. epidermidis I | Ac-CASYF |
| 4 | ≫100 | S. epidermidis II | Ac-CSNYL |
| 5 | ≫100 | S. epidermidis III | Ac-CASYL |
| 7 | 152 ± 65 | S. lugdunensis I | Ac-CN*AY*F |
| 11 | 0.9 ± 0.1 | S. lugdunensis II | Ac-CN*GY*F |
| 12 | ≫100 | S. epidermidis I | Ac-CAS*Y*F |
| 13 | ≫100 | S. epidermidis II | Ac-CS*N*Y*L |
| 14 | ≫100 | S. epidermidis III | Ac-CAS*Y*L |
Dose response curves and growth curves for the active compounds are contained in Supporting Inforamtion.
N* indicates trityl protected N, Y* indicates -O-t-Bu protected Y, and S* indicates -O-t-Bu protected S.
RESULTS AND DISCUSSION
As illustrated in Table 1, 1 inhibited its cognate receptor with an IC50 value of 0.2 μM while 2 was near equipotent eliciting an IC50 of 0.3 μM. Further the S. epidermidis AIP-I derivative 3 displayed an IC50 value of 2.7 μM further demonstrating the potential for cross-species inhibition. Hence this initial small library of compounds demonstrates that analogues comprising the macrocyclic component of native AIP scaffolds can function as inhibitors of S. lugdunensis AgrC-1, thus serving as value leads for further structural modification. In relation to SAR data for the significantly differing IC50 values of compounds 3 and 5 (e.g., 2.7 μM to ≫100 μM) indicate an essential requirement for an aromatic moiety at endocyclic position 5, which is further supported by the inhibitory activity of the protected analogues 7 and 11. Additionally based on the IC50 values of 1 versus 7, 2 versus 11, and 3 versus 12, protection of Asn-2 and Tyr-4 has moderate impact on binding. It is likely that the affinity of 2 is impacted less than that of 1 as Gly (vs Ala) at position 3 allows for much greater conformational space within the macrocycle that can accommodate/relieve any strain associated with fitting the protected Asn-2 and Tyr-4 side chains within the binding site on AgrC. When Ser-3 is protected (e.g., 3 vs 12), inhibition is completely lost, suggesting that there is insufficient room in the binding site to accommodate the t-Bu and/or the macrocycle can no longer relieve stress associated with fitting the t-Bu protected Tyr-4 within the binding site. It is proposed that this decreased affinity is due to insufficient room for the Ser-t-Bu, since the constraints placed on the conformational space available to the backbone by Ser and Ser-t-Bu are likely to be very similar. The addition of a β-carbon (moving from Gly to Ala) severely restricts the φ/ψ conformational space that is accessible to the main chain, but what gets attached to the β-carbon has very little impact in comparison.
CONCLUSIONS
This study demonstrates that truncated analogues based on the macrocyclic portion of S. lugdunensis AIP-I display inhibitory activity against cognate AgrC-1, while truncated analogues comprising the macrocyclic portions of the noncognate AIPs S. lugdunensis AIP-II and S. epidermidis AIP-I also display S. lugdunensis AgrC-1 inhibitory activity. Further in relation to structure–activity relationships and particularly the crucial requirement of a Phe residue at endocyclic position 5, we note that this is also a key structural requirement for inhibitory activity against S. aureus AgrC-1 and AgrC-2.25 Consequently this provides additional support for the possibility of generating a broad spectrum staphylococcus quorum sensing antibiotic.
Although the inhibitory activities of the analogues examined in this study are considerably lower than the subnanomolar activities reported for numerous truncated analogues against S. aureus AgrC1–4,4,16,19,25,26,48 the activities are in line with truncated sequences based on native S. aureus AIPs. For example, the macrocyclic portion of S. aureus AIP-I is reported to inhibit S. aureus AgrC1, AgrC2, and AgrC3 with respective IC50 values of 4.5, 0.05, and 0.2 μM.16 Yet a single alteration at endocyclic position 4 from Asp to Ala afforded the pan S. aureus AgrC inhibitor tr-AIP-I-D2A, which inhibits AgrC1–4 with near-subnanomolar activity. Hence current efforts within our research laboratories are focused on replicating such potency improvements against S. lugdunensis through systematic alternations of the tr-S.lug-AIP-I and tr-S.lug-AIP-II scaffolds.
Studies pertaining to the pharmaceutical potential of the truncated AIPs remain ongoing; however in relation to stability within physiological environments, initial results are promising. Recently the hydrolytic and proteolytic stability of a thioester truncated AIP was examined and it was reported that in PBS, 30% of the peptide had degraded in 72 h while in alkaline ammonium carbonate solution complete degradation was observed in 8 h.49 Similar results were observed upon peptide treatment with trypsin/chymotrypsin in ammonium carbonate solution. However, this study also demonstrated that conversion of the thioester linkage to an amide significantly enhanced stability within all of the aforementioned conditions (<40% degradation after 72 h) while imparting an insignificant decrease on IC50 value.49
These data, along with the findings of the present study, underlie the potential of developing broad-spectrum AgrC inhibitors to treat numerous staphylococcal infections and potentially other Gram-positive infections. For example, Clostridium difficile, Clostridium botulinum, and Listeria monocytogenes are all known to utilize similar agr TCSTSs.14 The potential of developing broad-spectrum AgrC highlighted in this study is further supported by the previous findings that full length S. epidermidis AIP-I and S. lugdunensis AIP-I display inhibitory activity against S. aureus AgrC-1 and AgrC-3 (IC50 values ranging from 13 to 419 nM)20 in addition to full length S. epidermidis AIP-I being reported to competitively interfere with S. epidermidis AgrC-II and AgrC-III.18 Therefore, in addition to improving potency against S. lugdunensis AgrC-1 a central focus of our current directions is to investigate the inhibitory activity of next generation tr-S.lug-AIP-I and tr-S.lug-AIP-II analogues against S. aureus and S. epidermidis in a bid to developed a broad spectrum AgrC inhibitor and to investigate the effectiveness of these analogues skin models. The results of these efforts will be reported in due course.
EXPERIMENTAL SECTION
Chemistry. General Methods
Chemicals and solvents were purchased from standard suppliers and used without further purification. Fmoc-protected amino acids were purchased for Auspep as was 2-chlorotrityl chloride resin (loading 1.5 mmol/g) and (2-(6-chloro-1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate) (HCTU). Polymer-bound 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide and N-benzyl-N′-cyclohexylcarbodiimide were purchased from Sigma-Aldrich with respective loadings of approximately 1–2 mmol/g). The PS-carbodiimde was obtained from Biotage (loading 1.27 mmol/g). All solvents were used as supplied (analytical, HPLC, or peptide grade) without prior purification. Milli-Q water was used for chemical reactions. Deuterated DMSO-d6 was purchased from Sigma-Aldrich. Reactions were monitored by using analytical RP-HPLC and MS.
Mass spectra (ES-TOF) were recorded on a Waters 2795 separation module/Micromass LCTTM platform. 1H and 13C spectra were recorded at 25 °C on a Varian mercury 300 operating at 299.8 and 75.4 MHz, respectively. Chemical shifts (δ) are reported in parts per million (ppm), referenced to TMS (1H, 0.0 ppm). Coupling constants (J) are recorded in Hz and significant multiplicities described by singlet (s), doublet (d), doublet of doublets (dd), doublet of triplets (dt), triplet (t), quadruplet (q), broad (br), multiplet (m). Spectra were assigned using appropriate gCOSY sequences.
Analytical RP-HPLC was performed using a Waters instrument comprising two 510 pumps, a 486 detector, and Millenium software. The systems outlined below were used for purification and to confirm purity. Analytical RP-HPLC was performed using Phenomenex Onyx Monolithic reversed-phase C18 column (4.6 mm × 100 mm). Solvent A: 0.06% TFA in water. Solvent B: 0.06% TFA in CH3CN/H2O (90:10). Flow rate of 1.0 mL/min, gradient 10–100 (% B), curve = 6, over 15.0 min, and detection at 216 and 254 nm (system 1).
Semipreparative RP-HPLC was performed using a Waters 2525 binary gradient pump equipped with a Waters 2487 dual λ absorbance detector and a ChromolithSemiPrep RP-18e 100−10 mm column. A flow rate of 10 mL/min was used with solvent A, 0.06% TFA in water, and solvent B, 0.06% TFA in CH3CN/H2O (90:10). Gradient 10–75 (% B) over 15 min, curve = 6, with UV detection at 216 and 254 nm. Final compound purity was judged via analytical HPLC analysis with all compound final compounds purities of ≥95%.
General Procedure 1: Resin Loading
Fmoc-Phe-2-ClTrt Resin
2-Chlorotrityl chloride resin (100–200 mesh), 1% DVB (5.00 g, loading = 1.5 mmol/g) (Merck Chemicals Ltd. no. 8.55017) was swelled in DCM (50.0 mL) for 0.5 h prior to the addition of 4.0 equiv of Fmoc-Phe-OH (11.62 g, 0.03 mol) and 8.0 equiv of DIPEA (10.45 mL, 0.06 mmol). The resulting suspension was gently stirred at rt for 2 h before MeOH (5.0 mL) was added. After an additional 20 min of gentle stirring the resin was filtered and sequentially washed with DMF (2 × 50 mL), acetonitrile (2 × 50 mL), hexanes (2 × 50 mL), and DCM (2 × 50 mL). The resin was dried overnight under high vacuum, and standard UV Fmoc quantification revealed a resin loading of 0.83 mmol/g (loading = 86%).
Fmoc-Leu-2-ClTrt Resin
Prepared utilizing general procedure 1 with 2-chlorotrityl chloride resin (100–200 mesh), 1% DVB (5.00 g, loading = 1.5 mmol/g), 4.0 equiv of Fmoc-Leu-OH (10.60 g, 0.03 mol), and 8.0 equiv of DIPEA (10.45 mL, 0.06 mmol). Fmoc quantification revealed a resin loading of 1.07 mmol/g (loading = 94%).
General Procedure 2: Synthesis of Linear Sequences (Compound 7)
Fmoc-Phe-2-chlorotrityl chloride resin (0.14 g, 0.12 mmol, loading = 0.83 mmol/g) was placed in an Omnifit BenchMark column assembly and swelled with DMF (2.5 mL) for 0.5 h. Utilizing a NovaSyn manual peptide synthesizer, Fmoc-depotection and washing were achieved using 20% piperidine in DMF and DMF (2.8 mL/min), respectively. The column was drained, and a solution of 4.0 equiv of Fmoc-(t-Bu)Try-OH (0.22 g, 0.48 mmol), 4.0 equiv of HCTU (0.19 g, 0.48 mmol), and 8.0 equiv of DIPEA (0.16 mL, 3.84 mmol) in DMF (1.5 mL) was added. The column was gently agitated at ~40 °C for 1 h after which the resin was washed (DMF 2.8 mL/min), Fmoc deprotected (20% piperidine in DMF 2.8 mL/min) and washed (DMF 2.8 mL/min). The remaining linear peptide sequence was prepared via subsequent rounds of acylation, washing (DMF 2.8 mL/min), Fmoc deprotection (20% piperidine in DMF, 2.8 mL/min), and washing (DMF 2.8 mL/min). Each acylation was achieved using a solution of 4.0 equiv of HCTU (0.19 g, 0.48 mmol), 8.0 equiv of DIPEA (0.16 mL, 3.84 mmol), DMF (1.5 mL) at ~40 °C over 1 h and 4.0 equiv of Fmoc-Ala-OH (0.14 g, 0.48 mmol), and 4.0 equiv of Fmoc-(Trt)Asn-OH (0.28 g, 0.48 mmol), and Fmoc-(Mmt)Cys-OH (0.29 g, 0.48), respectively. The N-terminal Cys-amine was acetylated via addition of an acetic anhydride (0.27 mL, 2.94 mmol), DIPEA (0.51 mL, 2.94 mmol) solution. After 0.5 h the resin was washed with MeOH (2 × 5.0 mL), acetonitrile (2 × 5.0 mL), hexanes (2 × 5.0 mL), and DCM (2 × 5.0 mL) and dried in vacuo. Concurrent cleavage of the linear peptide from the resin and deprotection of the Cys(Mmt) protecting group were achieved via flushing the column with a TFA cocktail (10 × 2 mL, DCM/TFA/TIPS 96:2:2). The resulting solution was concerted in vacuo and the residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. MS (ESI+) m/z 957 (M + 1, 100%). HRMS (ESI+) for C53H61N6O9S: calculated 957.4142, found, 957.41426. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 40–100% B in 15 min, tR = 9.5 min.
General Procedure 3: Cyclization of Linear Sequence AIPs (Compound 7)
A suspension of the crude linear peptide and 3.0 equiv of PS-carbodiimide (0.28g, 0.36 mmol) and CH3Cl (120 mL, 1.0 mM) was then stirred under an atmosphere of nitrogen for 5 h. After this period the solution was filtered, concentrated in vacuo, purified via semipreparative RP-HPLC, and lyophilized to afford Pro-N-Ac-Tr S.lug AIP-1 (12 mg, 10.6%). (Note: 3 mg of this sample was collect for biological analysis.) MS (ESI+) m/z 939 (M + 1, 100%). HRMS (ESI+) for C53H59N6O8S: calculated 939.4037, found, 939.4042. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 13.75 min.
General Procedure 4: Deprotection of Cyclized AIPs (Compound 1)
A solution of a TFA (10.0 mL), TIPS (0.25 mL), H2O (0.25 mL), and 12 (9 mg, 0.009 mmol) was stirred at rt for 6 h. The resulting mixture was concentrated in vacuo, and the residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. The crude material was purified by semipreparative RP-HPLC to afford compound 1 (5.2 mg, 85%) as a white solid. MS (ESI+) for C30H37N6O8S m/z 641.56 (M + 1, 100%). HRMS (ESI+): calculated 641.2315, found 641.2309; RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 6.08 min. 1H NMR (300 MHz, DMSO) δ 9.13 (s, 1H), 8.27 (d, J = 7.6 Hz, 1H), 8.10 (d, J = 7.7 Hz, 1H), 7.95 (d, J = 7.8 Hz, 1H), 7.93−7.88 (m, 2H), 7.43 (s, 1H), 7.30−7.14 (m, 5H), 6.98 (d, J = 8.4 Hz, 3H), 6.60 (d, J = 8.4 Hz, 2H), 4.49 (q, J = 6.7 Hz, 1H), 4.42−4.27 (m, 3H), 4.15−4.02 (m, 1H), 3.05 (d, J = 5.9 Hz, 1H), 3.00 (d, J = 5.8 Hz, 1H), 2.95−2.79 (m, 3H), 2.79−2.50 (m, 5H), 2.46−2.38 (m, 1H), 2.30 (t, J = 8.4 Hz, 1H), 1.84 (d, J = 3.4 Hz, 3H), 1.07 (d, J = 7.1 Hz, 3H). 13C NMR (75 MHz, DMSO) δ 172.98, 172.35, 172.23, 172.04, 171.38, 171.09, 170.95, 170.32, 170.12, 156.09, 137.77, 130.52, 129.55, 128.64, 128.35, 126.87, 115.27, 55.51, 54.63, 53.97, 50.26, 49.14, 37.29, 37.19, 36.72, 26.57, 22.92, 18.23.
Compounds 2 and 11
The linear sequence required for 11 was prepared using general procedure 2. The residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. MS (ESI+) m/z 943 (M + 1, 100%). HRMS (ESI+) for C52H59N6O9S: calculated 943.3986, found, 943.3981. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 40–100% B in 15 min, tR = 9.1 min.
Compound 11
Compound 11 was synthesized utilizing general produce 3. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 13 (15 mg, 13.5%). (Note: 3 mg of this sample was collect for biological analysis.) MS (ESI+) m/z 925 (M + 1, 100%). HRMS (ESI+) for C52H57N6O8S: calculated 925.3860, found, 925.3867. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 13.03 min.
Compound 2
Compound 2 was prepared utilizing general procedure 4. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 2 (7.2 mg, 88.5%). MS (ESI+) for C52H57N6O8S, m/z 627.2 (M + 1, 100%). HRMS (ESI+): calculated 627.2159, found, 627.2204. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 6.48 min. 1H NMR (300 MHz, DMSO) δ 9.12 (s, 1H), 8.28 (d, J = 7.5 Hz, 1H), 8.17 (d, J = 7.8 Hz, 1H), 8.09 (d, J = 7.8 Hz, 1H), 7.97 (t, J = 5.5 Hz, 1H), 7.89 (d, J = 8.6 Hz, 1H), 7.39 (s, 1H), 7.28−7.16 (m, 5H), 6.98 (d, J = 8.5 Hz, 2H), 6.59 (d, J = 8.5 Hz, 2H), 4.47 (dd, J = 13.6, 7.0 Hz, 1H), 4.42−4.30 (m, 3H), 3.58 (t, J = 5.9 Hz, 2H), 3.06 (d, J = 5.5 Hz, 1H), 3.01 (d, J = 5.4 Hz, 1H), 2.92 (d, J = 8.7 Hz, 1H), 2.89−2.52 (m, 7H), 2.43 (d, J = 7.0 Hz, 1H), 2.29 (t, J = 8.5 Hz, 1H), 1.85 (s, 3H). 13C NMR (75 MHz, DMSO) δ 173.08, 172.07, 171.57, 171.46, 170.44, 170.15, 168.63, 156.09, 137.84, 130.55, 129.54, 128.65, 128.35, 126.89, 115.28, 55.50, 54.54, 54.51, 54.00, 50.39, 42.41, 37.26, 37.14, 37.04, 26.49, 22.92.
Compounds 3 and 12
The linear sequence required for 12 was prepared using general procedure 2. The residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. MS (ESI+) m/z 744 (M + 1, 100%). HRMS (ESI+) for C37H54N5O9S: calculated 744.3564, found, 744.3559. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 10.28 min.
Compound 12
Compound 12 was synthesized utilizing general produce 3. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 12 (12 mg, 13.8%). (Note: 3 mg of this sample was collect for biological analysis.) MS (ESI+) m/z 726 (M + 1, 100%). HRMS (ESI+) for C37H52N5O8S: calculated 726.3458, found, 726.3451. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 11.47 min.
Compound 3
Compound 3 was prepared utilizing general procedure 4. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 3 (6.2 mg, 81.5%). MS (ESI+) for C29H36N5O8S m/z 614.40. HRMS (ESI+) calculated 614.2206, found, 614.2231; RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 6.82 min. 1H NMR (300 MHz, DMSO) δ 9.13 (s, 1H), 8.23 (d, J = 7.8 Hz, 1H), 8.15 (d, J = 7.1 Hz, 1H), 8.07 (d, J = 7.9 Hz, 1H), 7.82 (d, J = 7.7 Hz, 1H), 7.73 (d, J = 8.2 Hz, 1H), 7.29−7.17 (m, 5H), 6.95 (d, J = 8.5 Hz, 2H), 6.58 (d, J = 8.5 Hz, 2H), 4.45−4.30 (m, 3H), 4.30−4.16 (m, 2H), 3.05 (d, J = 5.5 Hz, 1H), 3.00 (d, J = 5.4 Hz, 1H), 2.93−2.81 (m, 3H), 2.80−2.53 (m, 4H), 2.32 (t, J = 8.4 Hz, 1H), 1.85 (s, 3H), 1.16 (d, J = 7.1 Hz, 3H). 13C NMR (75 MHz, DMSO) δ 173.05, 172.52, 171.26, 170.12, 170.00, 169.88, 156.14, 137.79, 130.57, 129.53, 128.66, 127.93, 126.90, 115.23, 62.12, 55.40, 54.31, 54.03, 48.77, 37.20, 36.89, 26.62, 22.92, 18.35.
Compounds 4 and 13
The linear sequence required for 13 was prepared using general procedure 2. The residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. MS (ESI+) m/z 995 (M + 1, 100%). HRMS (ESI+) for C54H71N6O10S: calculated 995.4874, found, 995.4879. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 13.91 min.
Compound 13
Compound 13 was synthesized utilizing general produce 3. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 15 (13 mg, 10.9%). (Note: 3 mg of this sample was collect for biological analysis.) MS (ESI+) m/z 977 (M + 1, 100%). HRMS (ESI+) for C54H69N6O9S: calculated 977.4768, found, 977.4752. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 15.38 min.
Compound 4
Compound 4 was prepared utilizing general procedure 4. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 4 (4.7 mg, 75.1%). MS (ESI+) for C27H39N6O9S, m/z 623.3 (M + 1, 100%). HRMS (ESI+) calculated 623.2421, found, 623.2425; RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 6.26 min. 1H NMR (300 MHz, DMSO) δ 9.16 (s, 1H), 8.10 (d, J = 8.0 Hz, 2H), 8.06 (d, J = 7.4 Hz, 1H), 8.01 (d, J = 7.8 Hz, 1H), 7.81 (d, J = 8.3 Hz, 1H), 7.33 (s, 1H), 6.98 (d, J = 8.5 Hz, 2H), 6.89 (s, 1H), 6.61 (d, J = 8.5 Hz, 2H), 4.51−4.30 (m, 3H), 4.26 (dd, J = 13.2, 6.0 Hz, 1H), 4.20−4.09 (m, 1H), 3.65−3.50 (m, 4H), 2.92 (dd, J = 14.2, 3.9 Hz, 1H), 2.79 (d, J = 5.3 Hz, 1H), 2.75 (d, J = 5.2 Hz, 1H), 2.69−2.56 (m, 2H), 2.44 (d, J = 5.4 Hz, 1H), 2.35 (d, J = 7.5 Hz, 1H), 2.30 (d, J = 7.3 Hz, 1H), 1.85 (d, J = 7.0 Hz, 3H), 1.67−1.40 (m, 3H), 0.87 (d, J = 6.3 Hz, 2H), 0.82 (d, J = 6.3 Hz, 3H). 13C NMR (75 MHz, DMSO) δ 174.29, 172.96, 172.88, 172.03, 171.29, 170.85, 170.56, 170.26, 170.09, 156.13, 130.51, 128.24, 128.20, 115.34, 115.34, 62.04, 55.49, 53.24, 53.02, 50.84, 50.34, 32.85, 26.62, 26.61, 24.64, 23.30, 22.92, 21.82.
Compounds 5 and 14
The linear sequence required for 14 was prepared using general procedure 2. The residual crude material was triturated with ice-cold ether (×3) to furnish an off-white solid. MS (ESI+) m/z 710 (M + 1, 100%). HRMS (ESI+) for C34H56N5O9S: calculated 710.3720, found, 710.3721. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 10.03 min.
Compound 14
Compound 14 was synthesized utilizing general produce 3. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 14 (16 mg, 18.8%). (Note: 3 mg of this sample was collect for biological analysis.) MS (ESI+) m/z 692 (M + 1, 100%). HRMS (ESI+) for C34H54N5O8S: calculated 692.3615, found, 692.3610. RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 11.43 min.
Compound 5
Compound 5 was prepared utilizing general procedure 4. The crude material was purified via semipreparative RP-HPLC and lyophilized to afford 5 (9.1 mg, 71.8%). MS (ESI+) for C27H38N5O8S, m/z 580.16. HRMS (ESI+) calculated 580.2363, found, 580.2367; RP-HPLC Onyx Monolithic C18 100 mm × 4.6 mm, 10–100% B in 15 min, tR = 6.55 min. 1H NMR (300 MHz, DMSO) δ 9.16 (s, 1H), 8.10 (d, J = 8.0 Hz, 2H), 8.06 (d, J = 7.4 Hz, 1H), 8.01 (d, J =7.8 Hz, 1H), 7.81 (d, J = 8.3 Hz, 1H), 7.33 (s, 1H), 6.98 (d, J = 8.5 Hz, 2H), 6.89 (s, 1H), 6.61 (d, J = 8.5 Hz, 2H), 4.51−4.30 (m, 3H), 4.26 (dd, J = 13.2, 6.0 Hz, 1H), 4.20−4.10 (m, 1H), 3.65−3.49 (m, 6H), 2.92 (dd, J = 14.2, 3.9 Hz, 2H), 2.79 (d, J = 5.3 Hz, 1H), 2.75 (d, J = 5.2 Hz, 1H), 2.70−2.56 (m, 2H), 2.44 (d, J = 5.4 Hz, 1H), 2.35 (d, J = 7.5 Hz, 1H), 2.30 (d, J = 7.3 Hz, 1H), 1.86 (s, 3H), 1.67−1.39 (m, 3H), 0.87 (d, J = 6.3 Hz, 3H), 0.82 (d, J = 6.3 Hz, 3H). 13C NMR (75 MHz, DMSO) δ 176.39, 174.74, 174.59, 173.46, 172.42, 172.30, 172.18, 172.11, 158.33, 132.76, 130.16, 117.41, 64.30, 57.70, 57.57, 56.42, 52.91, 50.95, 42.51, 38.98, 28.79, 26.80, 25.46, 25.10, 23.93, 20.65, 20.49.
General Biology Information
In order to assess the AgrC inhibitory activity of the AIP analogues, a S. lugdunensis I agr reporter strain, AH4031, was constructed by moving agrP3sGFP reporter plasmid pCM4018 into S. lugdunensis strain N92014346 using the electroporation protocol described by Heilbronner and colleagues.50 Prior to electroporation into S. lugdunensis N920143, pCM40 was passaged through E. coli strain SL01B,50 which is engineered to express the specificity (HsdS) and methylation (HsdM) subunits of the S. lugdunensis N920143 type I restriction system.
Overnight cultures of AH4031, which were grown in Tryptic Soy Broth (TSB) supplemented with Cam at a concentration of 10 μg/mL, were inoculated at a dilution of 1:500 into fresh TSB containing Cam. 100 μL aliquots were added to 96-well microtiter plates (Costar 3603) and combined with 100 μL aliquots of TSB containing Cam and 2-fold serial dilutions (either from 10 μM to 0.004 μM or from 40 μM to 0.2 μM) of the AIP analogs (dissolved in DMSO). After mixing, the effective inoculum dilution was 1:1000 and the final AIP concentrations ranged from 5 μM to 0.002 μM or from 20 μM to 0.1 μM, with a final DMSO concentration of 2% (v/v) in all wells. Four dilution series were prepared for each AIP, and four mock vehicle (DMSO) dilution series were also prepared. Microtiter plates were incubated at 37 °C with shaking (1000 rpm) in a Stuart SI505 incubator (Bibby Scientific, Burlington, NJ) with a humidified chamber. Fluorescence (top reading, 493 nm excitation, 535 nm emission, gain 60) and optical density (OD) readings at 600 nm were recorded at 30 min increments using a Tecan Systems (San Jose, CA) Infinite M200 plate reader. Data were transformed into units of percent of vehicle, and IC50 values were obtained by subjecting the transformed fluorescent data from 8 h of growth to four-parameter logistic fits (4PL) using GraphPad Prism, version 6.0g. In cases where the fluorescence at the highest AIP concentration tested was insufficient to fully inhibit quorum-sensing, the bottom baseline was fixed at 0%.
Supplementary Material
Acknowledgments
The authors thank Timothy Foster for supplying S. lugdunensis and E. coli strains. C.P.G. thanks the ARC for project support with the award of a DECRA (Grant DE130100513). A.R.H. is supported by Merit Award I01 BX002711 from the Department of Veteran Affairs
ABBREVIATIONS
- QS
quorum sensing
- AIP
autoinducing peptide
- agr
accessory gene regulator
- TCSTS
two-component signal transduction system
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmed-chem.6b00727.
Molecular formula strings and some data (CSV) Growth curves and dose–response curves; HLPC and MS data for all analogues; HPLC data for peptide cyclization trials and 1H NMR and 13C NMR spectra and HRMS data presented for final cyclized deprotected analogues; protocol for quorum sensing assay, reporter development, and IC50 calculations (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon CP, Williams P, Chan WC. Attenuating Staphylococcus aureus virulence gene regulation: a medicinal chemistry perspective. J Med Chem. 2013;56:1389–1404. doi: 10.1021/jm3014635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan WC, Coyle BJ, Williams P. Virulence Regulation and Quorum Sensing in Staphylococcal Infections: Competitive AgrC antagonists as quorum sensing inhibitors. J Med Chem. 2004;47:4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- 4.McDowell P, Affas Z, Reynolds C, Holden MTG, Wood SJ, Saint S, Cockayne A, Hill PJ, Dodd CER, Bycroft BW, Chan WC, Williams P. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 5.Cascioferro S, Cusimano MG, Schillaci D. Antiadhesion agents against Gram-positive pathogens. Future Microbiol. 2014;9:1209–1220. doi: 10.2217/fmb.14.56. [DOI] [PubMed] [Google Scholar]
- 6.Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111–2127. doi: 10.1128/JB.01524-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong D-W, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol. 2011;193:4672–4684. doi: 10.1128/JB.00353-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol. 2005;187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams P. Quorum sensing: An emerging target for antibacterial chemotherapy? Expert Opin Ther Targets. 2002;6:257–274. doi: 10.1517/14728222.6.3.257. [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006;296:149–161. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Costerton JW, Montanaro L, Arciola CR. Bacterial communications in implant infections: a target for an intelligence war. Int J Artif Organs. 2007;30:757–763. doi: 10.1177/039139880703000903. [DOI] [PubMed] [Google Scholar]
- 12.Pan J, Ren D. Quorum sensing inhibitors: a patent overview. Expert Opin Ther Pat. 2009;19:1581–1601. doi: 10.1517/13543770903222293. [DOI] [PubMed] [Google Scholar]
- 13.Yin S, Chang Y, Deng S, Wang Q, Yu W, Gong Q. Research progress of new antibacterial drugs that target bacterial quorum sensing systems. Yaoxue Xuebao. 2011;46:613–621. [PubMed] [Google Scholar]
- 14.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 16.Lyon GJ, Wright JS, Muir TW, Novick RP. Key Determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry. 2002;41:10095–10104. doi: 10.1021/bi026049u. [DOI] [PubMed] [Google Scholar]
- 17.Thoendel M, Horswill AR. Identification of Staphylococcus aureus AgrD residues required for autoinducing peptide biosynthesis. J Biol Chem. 2009;284:21828–21838. doi: 10.1074/jbc.M109.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson ME, Todd DA, Schaeffer CR, Paharik AE, Van Dyke MJ, Buettner H, Dunman PM, Rohde H, Cech NB, Fey PD, Horswill AR. Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J Bacteriol. 2014;196:3482–3493. doi: 10.1128/JB.01882-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto M, Echner H, Voelter W, Gotz F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:1957–1960. doi: 10.1128/IAI.69.3.1957-1960.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tal-Gan Y, Ivancic M, Cornilescu G, Blackwell HE. Characterization of structural elements in native autoinducing peptides and non-native analogs that permit the differential modulation of AgrC-type quorum sensing receptors in Staphylococcus aureus. Org Biomol Chem. 2016;14:113–121. doi: 10.1039/c5ob01735a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchdoerfer RN, Garner AL, Flack CE, Mee JM, Horswill AR, Janda KD, Kaufmann GF, Wilson IA. Structural basis for ligand recognition and discrimination of a quorum-quenching antibody. J Biol Chem. 2011;286:17351–17358. doi: 10.1074/jbc.M111.231258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick RP, Muir TW. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci U S A. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Jagasia R, Kaufmann GF, Mathison JC, Ruiz DI, Moss JA, Meijler MM, Ulevitch RJ, Janda KD. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem Biol. 2007;14:1119–1127. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott RJ, Lian L-Y, Muharram SH, Cockayne A, Wood SJ, Bycroft BW, Williams P, Chan WC. Side-chain-to-tail thiolactone peptide inhibitors of the staphylococcal quorum-sensing system. Bioorg Med Chem Lett. 2003;13:2449–2453. doi: 10.1016/s0960-894x(03)00497-9. [DOI] [PubMed] [Google Scholar]
- 25.George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- 26.Tal-Gan Y, Stacy DM, Blackwell HE. N-Methyl and peptoid scans of an autoinducing peptide reveal new structural features required for inhibition and activation of AgrC quorum sensing receptors in Staphylococcus aureus. Chem Commun. 2014;50:3000–3003. doi: 10.1039/c4cc00117f. [DOI] [PubMed] [Google Scholar]
- 27.Tal-Gan Y, Stacy DM, Foegen MK, Koenig DW, Blackwell HE. Highly potent inhibitors of quorum sensing in Staphylococcus aureus revealed through a systematic synthetic study of the group-III autoinducing peptide. J Am Chem Soc. 2013;135:7869–7882. doi: 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- 28.Babu E, Oropello J. Staphylococcus lugdunensis: the coagulase-negative staphylococcus you don’t want to ignore. Expert Rev Anti-Infect Ther. 2011;9:901–907. doi: 10.1586/eri.11.110. [DOI] [PubMed] [Google Scholar]
- 29.Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, Etienne J, Lina G. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002;184:1180–1186. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh C-F, Liu T-P, Cheng C-W, Chang S-C, Lee M-H, Lu J-J. Molecular characteristics of disease-causing and commensal Staphylococcus lugdunensis isolates from 2003 to 2013 at a tertiary hospital in Taiwan. PLoS One. 2015:10–e0134859. doi: 10.1371/journal.pone.0134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph CR, Lalitha P, Sivaraman KR, Ramasamy K, Behera UC. Real-time polymerase chain reaction in the diagnosis of acute postoperative endophthalmitis. Am J Ophthalmol. 2012;153:1031–1037. doi: 10.1016/j.ajo.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Murad-Kejbou S, Kashani AH, Capone A, Jr, Ruby A. Staphylococcus lugdunensis endophthalmitis after intravitreal injection: a case series. Retinal Cases Brief Rep. 2014;8:41–44. doi: 10.1097/ICB.0b013e3182a85a4f. [DOI] [PubMed] [Google Scholar]
- 33.Choi S-H, Chung J-W, Lee EJ, Kim TH, Lee MS, Kang JM, Song EH, Jun J-B, Kim M-N, Kim YS, Woo JH, Choi S-H. Incidence, characteristics, and outcomes of Staphylococcus lugdunensis bacteremia. J Clin Microbiol. 2010;48:3346–3349. doi: 10.1128/JCM.00609-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chopra A, Gulati D, Woldenberg N, Singh M. Intracardiac lead endocarditis due to Staphylococcus lugdunensis. Int J Infect Dis. 2010;14(Suppl 3):e291–e293. doi: 10.1016/j.ijid.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Shin JH, Jung HJ, Lee HR, Kim JH, Kim HR, Lee JN. Prevalence, identification, and antimicrobial susceptibility of Staphylococcus lugdunensis from various clinical specimens in Korea. Jpn J Infect Dis. 2007;60:312–313. [PubMed] [Google Scholar]
- 36.Liu P-Y, Huang Y-F, Tang C-W, Chen Y-Y, Hsieh K-S, Ger L-P, Chen Y-S, Liu Y-C. Staphylococcus lugdunensis infective endocarditis: a literature review and analysis of risk factors. J Microbiol Immunol Infect. 2010;43:478–484. doi: 10.1016/S1684-1182(10)60074-6. [DOI] [PubMed] [Google Scholar]
- 37.Vandenesch F, Etienne J, Reverdy ME, Eykyn SJ. Endocarditis due to Staphylococcus lugdunensis: report of 11 cases and review. Clin Infect Dis. 1993;17:871–876. doi: 10.1093/clinids/17.5.871. [DOI] [PubMed] [Google Scholar]
- 38.Higaki S, Kitagawa T, Morohashi M, Yamagishi T. Distribution and antimicrobial susceptibility of coagulase-negative staphylococci from skin lesions. J Int Med Res. 1999;27:191–195. doi: 10.1177/030006059902700406. [DOI] [PubMed] [Google Scholar]
- 39.Goldstein EJC, Citron DM, Merriam CV, Warren YA, Tyrrell KL, Fernandez HT. In vitro activity of ceftobiprole against aerobic and anaerobic strains isolated from diabetic foot infections. Antimicrob Agents Chemother. 2006;50:3959–3962. doi: 10.1128/AAC.00722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleurette J, Bes M, Brun Y, Freney J, Forey F, Coulet M, Reverdy ME, Etienne J. Clinical isolates of Staphylococcus lugdunensis and S. schleiferi: bacteriological characteristics and susceptibility to antimicrobial agents. Res Microbiol. 1989;140:107–118. doi: 10.1016/0923-2508(89)90044-2. [DOI] [PubMed] [Google Scholar]
- 41.Anguera I, Del Rio A, Miro JM, Matinez-Lacasa X, Marco F, Guma JR, Quaglio G, Claramonte X, Moreno A, Mestres CA, Mauri E, Azqueta M, Benito N, Garcia-de la Maria C, Almela M, Jimenez-Exposito MJ, Sued O, De Lazzari E, Gatell JM. Staphylococcus lugdunensis infective endocarditis: description of 10 cases and analysis of native valve, prosthetic valve, and pacemaker lead endocarditis clinical profiles. Heart. 2005;91:e10. doi: 10.1136/hrt.2004.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kragsbjerg P, Bomfim-Loogna J, Tornqvist E, Soderquist B. Development of antimicrobial resistance in Staphylococcus lugdunensis during treatment - report of a case of bacterial arthritis, vertebral osteomyelitis and infective endocarditis. Clin Microbiol Infect. 2000;6:496–499. doi: 10.1046/j.1469-0691.2000.00103.x. [DOI] [PubMed] [Google Scholar]
- 43.Frank KL, del Pozo JL, Patel R. From clinical microbiology to infection pathogenesis: how daring to be different works for Staphylococcus lugdunensis. Clin Microbiol Rev. 2008;21:111–133. doi: 10.1128/CMR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T, Tal-Gan Y, Paharik AE, Horswill AR, Blackwell HE. Structure-function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for agrc-type receptor modulation. ACS Chem Biol. 2016;11:1982–1991. doi: 10.1021/acschembio.6b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otto M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides. 2001;22:1603–1608. doi: 10.1016/s0196-9781(01)00495-8. [DOI] [PubMed] [Google Scholar]
- 46.Heilbronner S, Holden MTG, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, Bentley SD. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol Lett. 2011;322:60–67. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–309. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 48.Tal-Gan Y, Ivancic M, Cornilescu G, Cornilescu CC, Blackwell HE. Structural characterization of native autoinducing peptides and abiotic analogues reveals key features essential for activation and inhibition of an agrc quorum sensing receptor in Staphylococcus aureus. J Am Chem Soc. 2013;135:18436–18444. doi: 10.1021/ja407533e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the agrC quorum sensing receptor in Staphylococcus aureus. Angew Chem, Int Ed. 2016;55:8913–8917. doi: 10.1002/anie.201602974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heilbronner S, Hanses F, Monk IR, Speziale P, Foster TJ. Sortase A promotes virulence in experimental Staphylococcus lugdunensis endocarditis. Microbiology. 2013;159:2141–2152. doi: 10.1099/mic.0.070292-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


