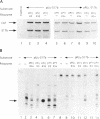
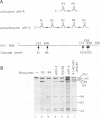
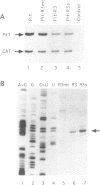
Abstract
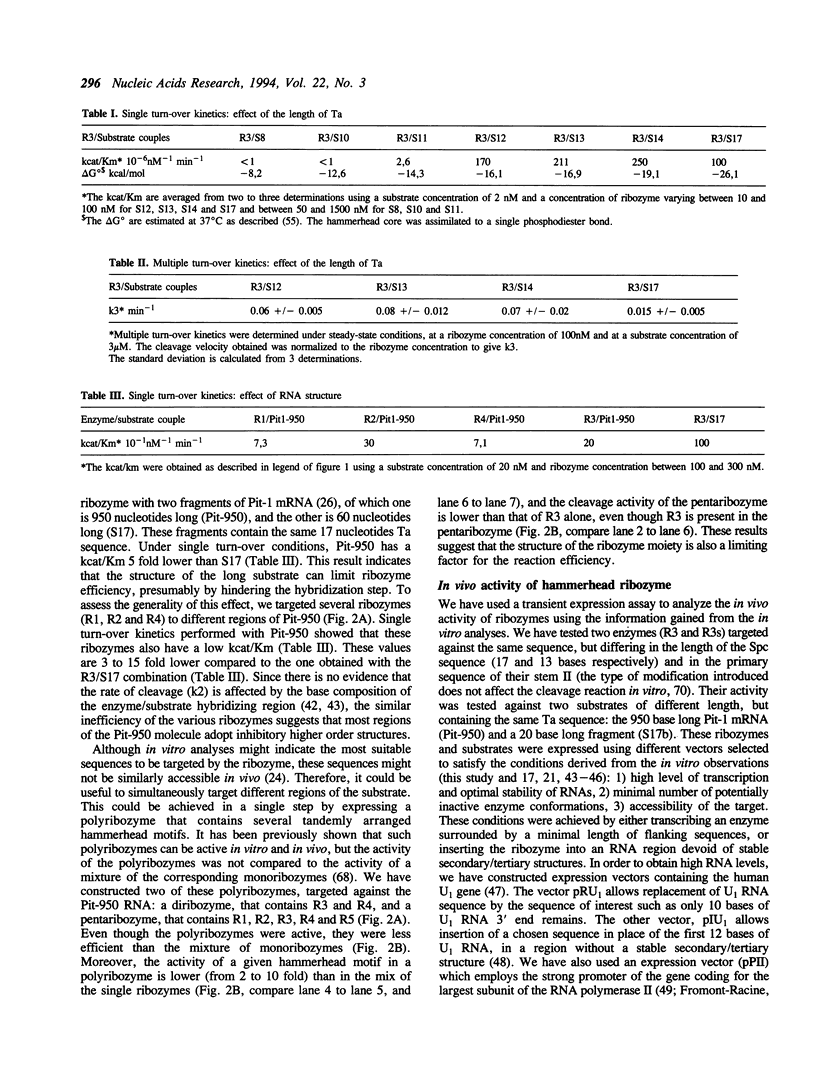
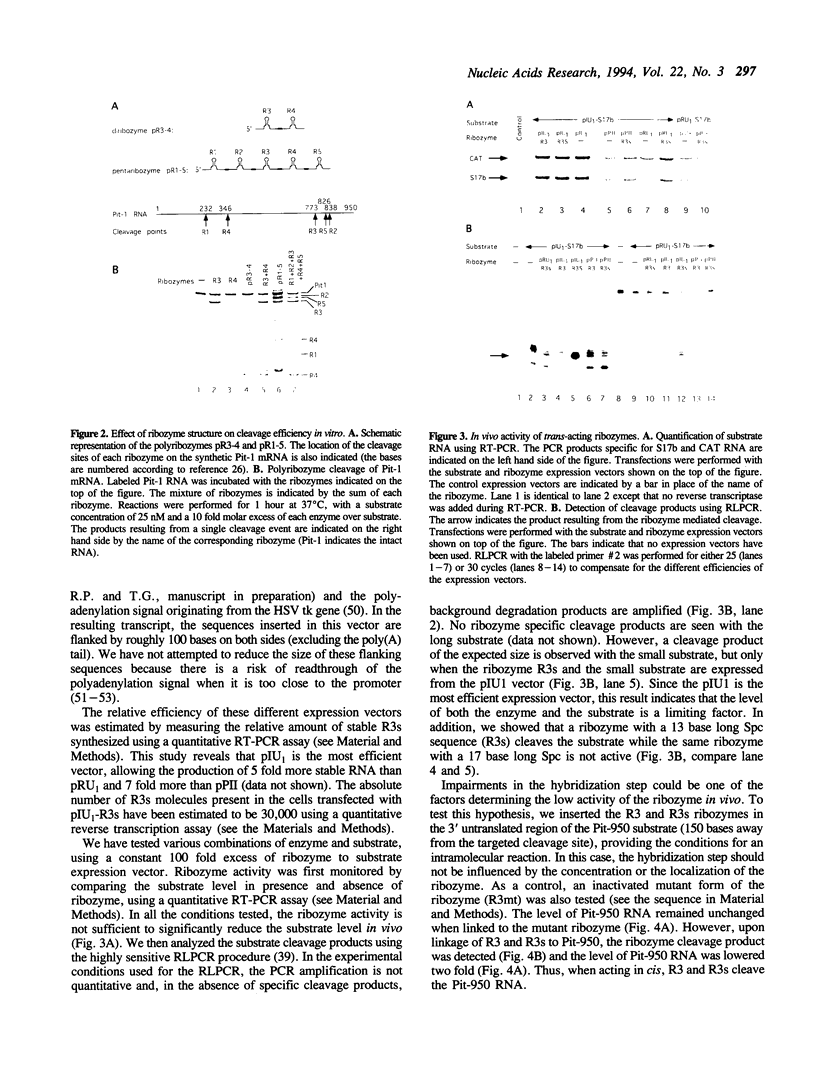
In order to improve hammerhead ribozyme efficiency and specificity, we have analyzed, both in vitro and in vivo, the activity of a series of ribozyme/substrate combinations that have the same target sequence but differ in the length of the ribozyme/substrate duplex or in their structure, i.e., the total length of the RNA. In vitro, we have found that optimal kcat/Km (at 37 degrees C) is obtained when the ribozyme/substrate duplex has a length of 12 bases, which according to the base composition represents a calculated free energy of binding of -16 kcal/mol. We discuss the importance of this value for ribozyme specificity and present strategies that may improve it. Increasing the length of the duplex from 14 to 17 bases (from -19 to -26 kcal/mol) produces a reduced ribozyme activity which is probably due to a slower rate of product dissociation. In addition, inclusion of either the substrate or the ribozyme in a long transcript produces a reduction (10 fold) of the kcat/Km, probably because of a different accessibility of the target sequence. In vivo, the activity of the trans-acting ribozyme was extremely low and detected in only one case: with a ribozyme/substrate duplex length of 13 bases and with both ribozyme and substrate embedded in short RNAs expressed at a very high level. The similarity of the results obtained in vitro and in vivo indicates that it is possible to use an in vitro system to optimize ribozymes which are to be used in vivo. Satisfactory results were obtained in vivo only with cisacting ribozymes. Altogether these results suggest that the ribozyme/substrate hybridization step is the limiting step in vivo and therefore it is not clear if ribozymes represent an improvement over antisense RNAs.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Aldovini A., Young R. A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990 May;64(5):1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Fromont-Racine M., Pictet R., Grange T. Visualization of the interaction of a regulatory protein with RNA in vivo. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3496–3500. doi: 10.1073/pnas.90.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan J. M., Feldstein P. A., Segrelles C., Bruening G. Autolytic processing of a phosphorothioate diester bond. Nucleic Acids Res. 1988 May 11;16(9):4009–4023. doi: 10.1093/nar/16.9.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan J. M., van Tol H., Feldstein P. A., Bruening G. Identification of a non-junction phosphodiester that influences an autolytic processing reaction of RNA. Nucleic Acids Res. 1990 Aug 11;18(15):4447–4451. doi: 10.1093/nar/18.15.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet J. P., Myers J. A. In-vivo secondary structure analysis of the small nuclear RNA U1 using psoralen cross-linking. J Mol Biol. 1987 Oct 5;197(3):543–553. doi: 10.1016/0022-2836(87)90563-8. [DOI] [PubMed] [Google Scholar]
- Cameron F. H., Jennings P. A. Specific gene suppression by engineered ribozymes in monkey cells. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9139–9143. doi: 10.1073/pnas.86.23.9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., Rossi J. J., Deshler J. O. Biological and functional aspects of catalytic RNAs. Crit Rev Eukaryot Gene Expr. 1992;2(4):331–357. [PubMed] [Google Scholar]
- Chen C. J., Banerjea A. C., Harmison G. G., Haglund K., Schubert M. Multitarget-ribozyme directed to cleave at up to nine highly conserved HIV-1 env RNA regions inhibits HIV-1 replication--potential effectiveness against most presently sequenced HIV-1 isolates. Nucleic Acids Res. 1992 Sep 11;20(17):4581–4589. doi: 10.1093/nar/20.17.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Birnstiel M. L. Ribozyme mediated destruction of RNA in vivo. EMBO J. 1989 Dec 1;8(12):3861–3866. doi: 10.1002/j.1460-2075.1989.tb08564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotten M., Schaffner G., Birnstiel M. L. Ribozyme, antisense RNA, and antisense DNA inhibition of U7 small nuclear ribonucleoprotein-mediated histone pre-mRNA processing in vitro. Mol Cell Biol. 1989 Oct;9(10):4479–4487. doi: 10.1128/mcb.9.10.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J. L., Gabus C., Nugeyre M. T., Clavel F., Barré-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990 Dec 5;216(3):689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- Dropulić B., Lin N. H., Martin M. A., Jeang K. T. Functional characterization of a U5 ribozyme: intracellular suppression of human immunodeficiency virus type 1 expression. J Virol. 1992 Mar;66(3):1432–1441. doi: 10.1128/jvi.66.3.1432-1441.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Kinetics of intermolecular cleavage by hammerhead ribozymes. Biochemistry. 1992 Dec 8;31(48):12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnor C., Bertrand J. R., Thenet S., Lemaître M., Morvan F., Rayner B., Malvy C., Lebleu B., Imbach J. L., Paoletti C. alpha-DNA. VI: Comparative study of alpha- and beta-anomeric oligodeoxyribonucleotides in hybridization to mRNA and in cell free translation inhibition. Nucleic Acids Res. 1987 Dec 23;15(24):10419–10436. doi: 10.1093/nar/15.24.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goblet C., Prost E., Whalen R. G. One-step amplification of transcripts in total RNA using the polymerase chain reaction. Nucleic Acids Res. 1989 Mar 11;17(5):2144–2144. doi: 10.1093/nar/17.5.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick R. J., Nigida S. M., Jr, Bess J. W., Jr, Arthur L. O., Henderson L. E., Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990 Jul;64(7):3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Heidenreich O., Eckstein F. Hammerhead ribozyme-mediated cleavage of the long terminal repeat RNA of human immunodeficiency virus type 1. J Biol Chem. 1992 Jan 25;267(3):1904–1909. [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3' end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985 Jul;4(7):1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: more isn't always better. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6921–6925. doi: 10.1073/pnas.88.16.6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M., Tzortzakaki S., Rittner K., Sczakiel G., Tabler M. Incorporation of the catalytic domain of a hammerhead ribozyme into antisense RNA enhances its inhibitory effect on the replication of human immunodeficiency virus type 1. Nucleic Acids Res. 1993 Jun 25;21(12):2809–2814. doi: 10.1093/nar/21.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hélène C., Montenay-Garestier T., Saison T., Takasugi M., Toulmé J. J., Asseline U., Lancelot G., Maurizot J. C., Toulmé F., Thuong N. T. Oligodeoxynucleotides covalently linked to intercalating agents: a new class of gene regulatory substances. Biochimie. 1985 Jul-Aug;67(7-8):777–783. doi: 10.1016/s0300-9084(85)80167-x. [DOI] [PubMed] [Google Scholar]
- Ingraham H. A., Chen R. P., Mangalam H. J., Elsholtz H. P., Flynn S. E., Lin C. R., Simmons D. M., Swanson L., Rosenfeld M. G. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988 Nov 4;55(3):519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- Iwasaki K., Temin H. M. The efficiency of RNA 3'-end formation is determined by the distance between the cap site and the poly(A) site in spleen necrosis virus. Genes Dev. 1990 Dec;4(12B):2299–2307. doi: 10.1101/gad.4.12b.2299. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984 Apr;36(4):1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- Jeffries A. C., Symons R. H. A catalytic 13-mer ribozyme. Nucleic Acids Res. 1989 Feb 25;17(4):1371–1377. doi: 10.1093/nar/17.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Van Brunschot A., Asad S., van der Elst I., Read S. E., Bernstein A. Inhibition of human immunodeficiency virus type 1 multiplication by antisense and sense RNA expression. J Virol. 1991 Oct;65(10):5524–5530. doi: 10.1128/jvi.65.10.5524-5530.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R., Giedroc D. P. Recombinant human immunodeficiency virus type 1 nucleocapsid (NCp7) protein unwinds tRNA. J Biol Chem. 1992 Apr 5;267(10):6689–6695. [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980 Jul 25;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- L'Huillier P. J., Davis S. R., Bellamy A. R. Cytoplasmic delivery of ribozymes leads to efficient reduction in alpha-lactalbumin mRNA levels in C127I mouse cells. EMBO J. 1992 Dec;11(12):4411–4418. doi: 10.1002/j.1460-2075.1992.tb05541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Thomas K. R., Capecchi M. R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988 Nov 24;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleij C. W. Pseudoknots: a new motif in the RNA game. Trends Biochem Sci. 1990 Apr;15(4):143–147. doi: 10.1016/0968-0004(90)90214-v. [DOI] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost E., Moore D. D. CAT vectors for analysis of eukaryotic promoters and enhancers. Gene. 1986;45(1):107–111. doi: 10.1016/0378-1119(86)90138-1. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Rungger D., Rungger-Brändle E., Chaponnier C., Gabbiani G. Intranuclear injection of anti-actin antibodies into Xenopus oocytes blocks chromosome condensation. Nature. 1979 Nov 15;282(5736):320–321. doi: 10.1038/282320a0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanfaçon H., Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990 Jul 5;346(6279):81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- Sarver N., Cantin E. M., Chang P. S., Zaia J. A., Ladne P. A., Stephens D. A., Rossi J. J. Ribozymes as potential anti-HIV-1 therapeutic agents. Science. 1990 Mar 9;247(4947):1222–1225. doi: 10.1126/science.2107573. [DOI] [PubMed] [Google Scholar]
- Saxena S. K., Ackerman E. J. Ribozymes correctly cleave a model substrate and endogenous RNA in vivo. J Biol Chem. 1990 Oct 5;265(28):17106–17109. [PubMed] [Google Scholar]
- Scanlon K. J., Jiao L., Funato T., Wang W., Tone T., Rossi J. J., Kashani-Sabet M. Ribozyme-mediated cleavage of c-fos mRNA reduces gene expression of DNA synthesis enzymes and metallothionein. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10591–10595. doi: 10.1073/pnas.88.23.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M., Drlica K. Prevention of human immunodeficiency virus type 1 integrase expression in Escherichia coli by a ribozyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7303–7307. doi: 10.1073/pnas.88.16.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M., Natvig J. B., Førre O. Preformed ribozyme destroys tumour necrosis factor mRNA in human cells. J Mol Biol. 1992 Feb 20;223(4):831–835. doi: 10.1016/0022-2836(92)90244-e. [DOI] [PubMed] [Google Scholar]
- Steinecke P., Herget T., Schreier P. H. Expression of a chimeric ribozyme gene results in endonucleolytic cleavage of target mRNA and a concomitant reduction of gene expression in vivo. EMBO J. 1992 Apr;11(4):1525–1530. doi: 10.1002/j.1460-2075.1992.tb05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson M. L., Zamecnik P. C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc Natl Acad Sci U S A. 1978 Jan;75(1):285–288. doi: 10.1073/pnas.75.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger B. A., Cech T. R. Tethering ribozymes to a retroviral packaging signal for destruction of viral RNA. Science. 1993 Dec 3;262(5139):1566–1569. doi: 10.1126/science.8248806. [DOI] [PubMed] [Google Scholar]
- Taira K., Nakagawa K., Nishikawa S., Furukawa K. Construction of a novel RNA-transcript-trimming plasmid which can be used both in vitro in place of run-off and (G)-free transcriptions and in vivo as multi-sequences transcription vectors. Nucleic Acids Res. 1991 Oct 11;19(19):5125–5130. doi: 10.1093/nar/19.19.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z., Khosla M., Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993 Oct 1;262(5130):99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Eckstein F. Hammerhead ribozymes: importance of stem-loop II for activity. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):6991–6994. doi: 10.1073/pnas.90.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Weerasinghe M., Liem S. E., Asad S., Read S. E., Joshi S. Resistance to human immunodeficiency virus type 1 (HIV-1) infection in human CD4+ lymphocyte-derived cell lines conferred by using retroviral vectors expressing an HIV-1 RNA-specific ribozyme. J Virol. 1991 Oct;65(10):5531–5534. doi: 10.1128/jvi.65.10.5531-5534.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann L. M., Perry R. P. Characterization of the expressed gene and several processed pseudogenes for the mouse ribosomal protein L30 gene family. Mol Cell Biol. 1984 Nov;4(11):2518–2528. doi: 10.1128/mcb.4.11.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. V., Cho K. W., Hardwicke J., Collins R. H., De Robertis E. M. Interference with function of a homeobox gene in Xenopus embryos produces malformations of the anterior spinal cord. Cell. 1989 Oct 6;59(1):81–93. doi: 10.1016/0092-8674(89)90871-4. [DOI] [PubMed] [Google Scholar]
- Xing Z., Whitton J. L. Ribozymes which cleave arenavirus RNAs: identification of susceptible target sites and inhibition by target site secondary structure. J Virol. 1992 Mar;66(3):1361–1369. doi: 10.1128/jvi.66.3.1361-1369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik P. C., Stephenson M. L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A. 1978 Jan;75(1):280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A., Gruss P. Production of chimaeric mice containing embryonic stem (ES) cells carrying a homoeobox Hox 1.1 allele mutated by homologous recombination. Nature. 1989 Mar 9;338(6211):150–153. doi: 10.1038/338150a0. [DOI] [PubMed] [Google Scholar]