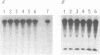
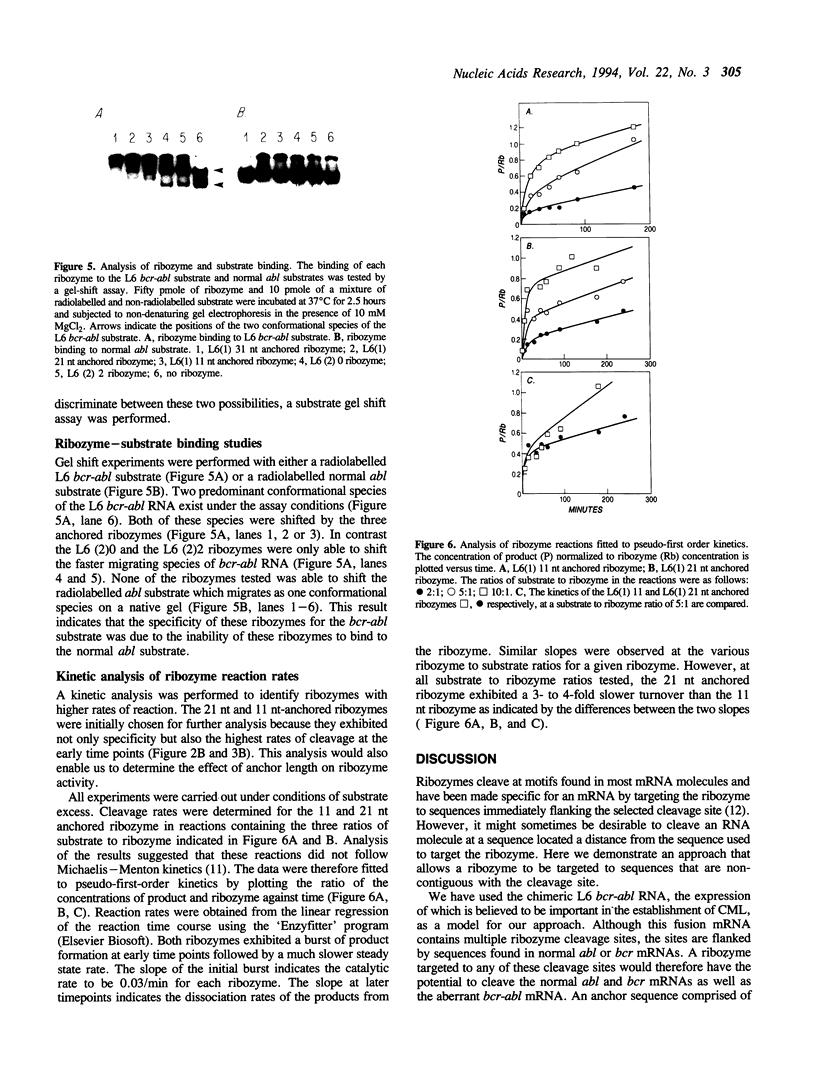
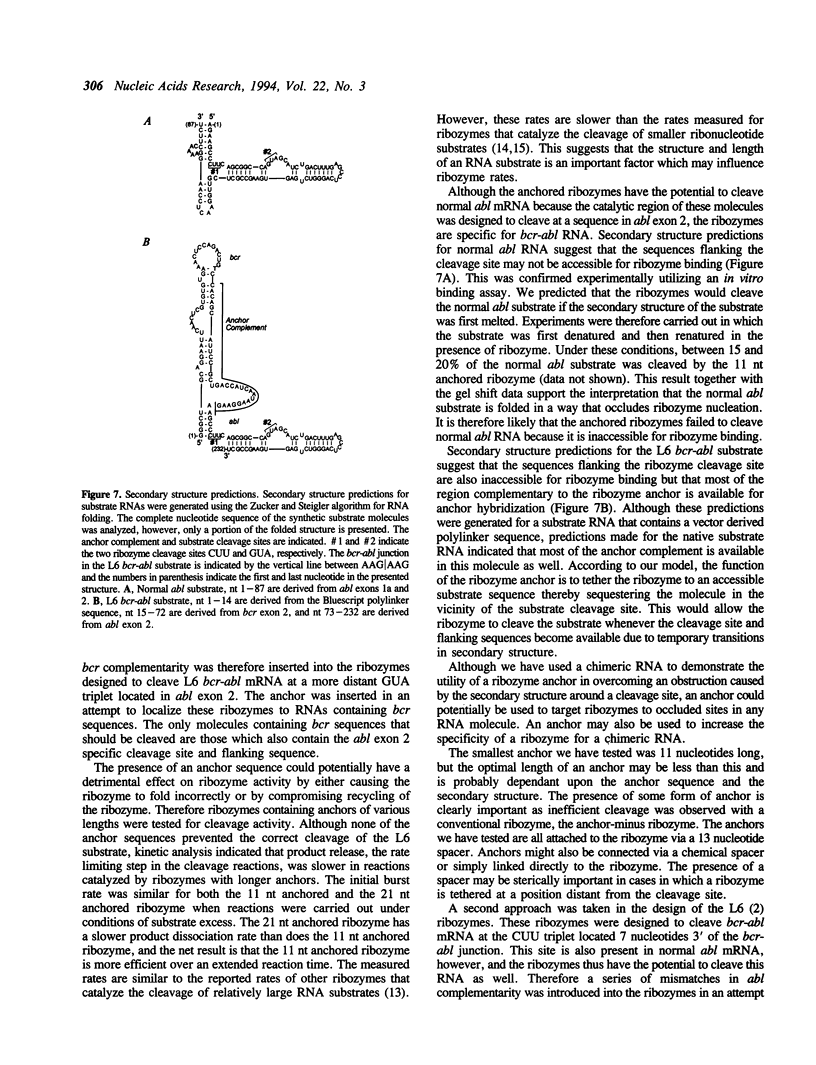
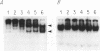Abstract
Conventionally designed ribozymes may be unable to cleave RNA at sites which are inaccessible due to secondary structure. In addition, it may also be difficult to specifically target a conventionally designed ribozyme to some chimeric RNA molecules. Novel approaches for ribozyme targeting were developed by using the L6 bcr-abl fusion RNA as a model. Using one approach, we successfully directed ribozyme nucleation to a site on the bcr-abl RNA that is distant from the GUA cleavage site. These ribozymes bound to the L6 substrate RNA via an anchor sequence that was complementary to bcr sequences. The anchor was necessary for efficient cleavage as the anchor minus ribozyme, a conventionally designed ribozyme, was inefficient at catalyzing cleavage at this same site. The effect of anchor sequences on catalytic rates was determined for two of these ribozymes. Ribozymes generated by a second approach were designed to cleave at a CUU site in proximity to the bcr-abl junction. Both approaches have led to the development of a series of ribozymes specific for both the L6 and K28 bcr-abl chimeric RNAs, but not normal abl or bcr RNAs. The specificity of the ribozyme correlated in part with the ability of the ribozyme to bind substrate as demonstrated by gel shift analyses. Secondary structure predictions for the RNA substrate support the experimental results and may prove useful as a theoretical basis for the design of ribozymes.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fedor M. J., Uhlenbeck O. C. Kinetics of intermolecular cleavage by hammerhead ribozymes. Biochemistry. 1992 Dec 8;31(48):12042–12054. doi: 10.1021/bi00163a012. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groffen J., Stephenson J. R., Heisterkamp N., de Klein A., Bartram C. R., Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984 Jan;36(1):93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N., Stam K., Groffen J., de Klein A., Grosveld G. Structural organization of the bcr gene and its role in the Ph' translocation. 1985 Jun 27-Jul 3Nature. 315(6022):758–761. doi: 10.1038/315758a0. [DOI] [PubMed] [Google Scholar]
- Heisterkamp N., Stephenson J. R., Groffen J., Hansen P. F., de Klein A., Bartram C. R., Grosveld G. Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature. 1983 Nov 17;306(5940):239–242. doi: 10.1038/306239a0. [DOI] [PubMed] [Google Scholar]
- Lozzio C. B., Lozzio B. B. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975 Mar;45(3):321–334. [PubMed] [Google Scholar]
- Rowley J. D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973 Jun 1;243(5405):290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Shtivelman E., Lifshitz B., Gale R. P., Roe B. A., Canaani E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 1986 Oct 24;47(2):277–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- Taylor N. R., Rossi J. J. Ribozyme-mediated cleavage of an HIV-1 gag RNA: the effects of nontargeted sequences and secondary structure on ribozyme cleavage activity. Antisense Res Dev. 1991 Summer;1(2):173–186. doi: 10.1089/ard.1991.1.173. [DOI] [PubMed] [Google Scholar]