Abstract
Adriamycin is known to induce the formation of adducts with DNA when reacted under in vitro transcription conditions. The factors affecting the extent of adduct formation were examined in order to establish the critical components and optimal conditions required for the reaction, and to gain insight into the nature of the DNA-adduct complex. There was a strong dependence on reaction temperature (with a 40-fold increase of adducts at 40-50 degrees C compared to 10 degrees C), pH (maximum adducts at pH 7), but little dependence on the oxygen level. There was an absolute requirement for a reducing agent, with adducts detected with DTT, beta-mercaptoethanol and glutathione, maximal adducts were formed at high levels of DTT (5-10 mM). Adducts were also formed with a xanthine oxidase/NADH reducing system, with increasing amounts of adducts detected with increasing NADH; no adducts were detected in the absence of either the enzyme or NADH. Of fourteen derivatives studied, only four yielded a similar extent of adduct formation as adriamycin; there was no absolute requirement for a carbonyl at C13 or hydroxyl at C14. Adducts were also observed with ssDNA but required a longer reaction time compared to dsDNA. The sequence specificity of adduct formation with ssDNA was examined using a primer-extension assay; almost all adducts were associated with a guanine residue. Overall, the results are consistent with a two-step reaction mechanism involving reductive activation of adriamycin, with the activated species then reacting with the guanine residues of either dsDNA or ssDNA.
Full text
PDF

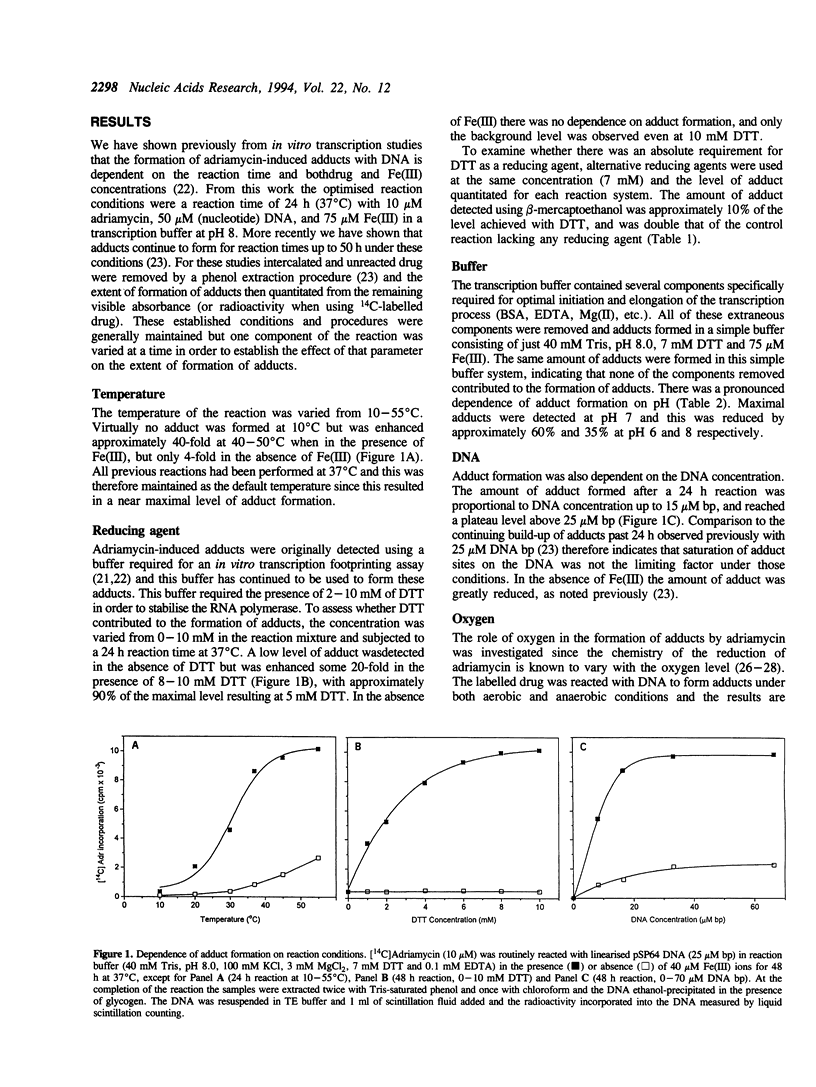

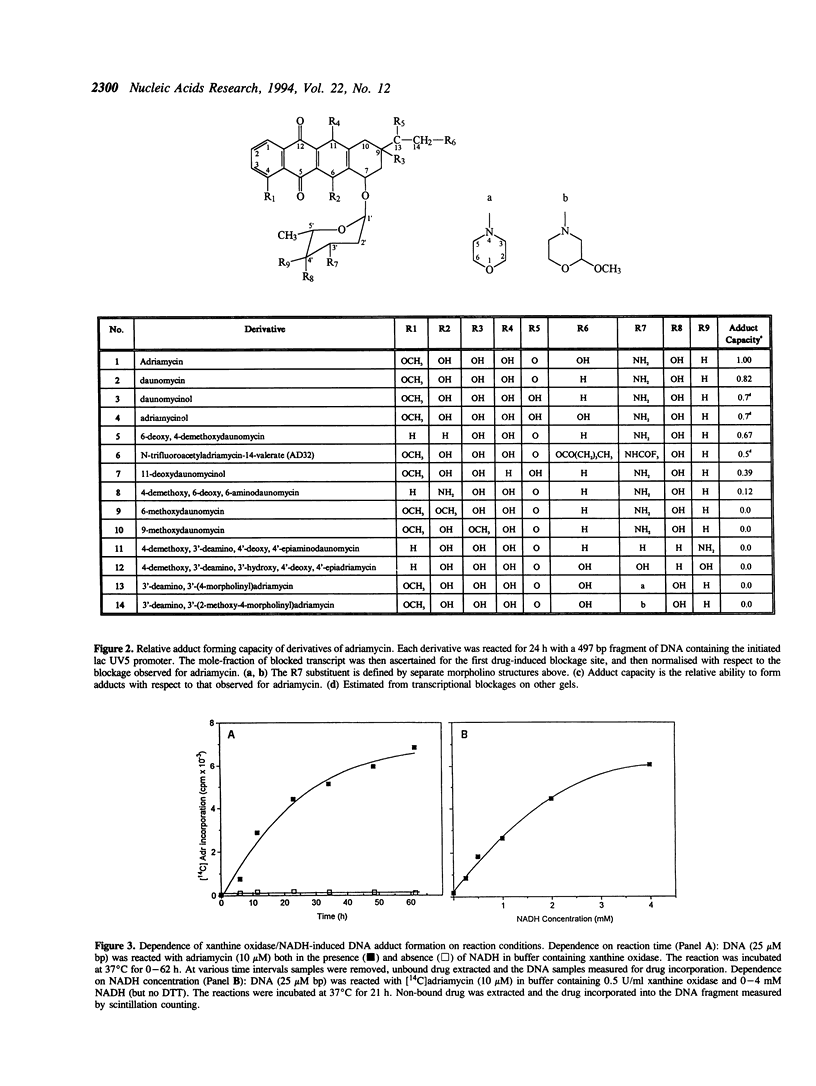

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartoszek A., Wolf C. R. Enhancement of doxorubicin toxicity following activation by NADPH cytochrome P450 reductase. Biochem Pharmacol. 1992 Apr 1;43(7):1449–1457. doi: 10.1016/0006-2952(92)90201-s. [DOI] [PubMed] [Google Scholar]
- Bartoszek A., Wolf C. R. Enhancement of doxorubicin toxicity following activation by NADPH cytochrome P450 reductase. Biochem Pharmacol. 1992 Apr 1;43(7):1449–1457. doi: 10.1016/0006-2952(92)90201-s. [DOI] [PubMed] [Google Scholar]
- Bates D. A., Winterbourn C. C. Reactions of Adriamycin with haemoglobin. Superoxide dismutase indirectly inhibits reactions of the Adriamycin semiquinone. Biochem J. 1982 Apr 1;203(1):155–160. doi: 10.1042/bj2030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binaschi M., Capranico G., De Isabella P., Mariani M., Supino R., Tinelli S., Zunino F. Comparison of DNA cleavage induced by etoposide and doxorubicin in two human small-cell lung cancer lines with different sensitivities to topoisomerase II inhibitors. Int J Cancer. 1990 Feb 15;45(2):347–352. doi: 10.1002/ijc.2910450223. [DOI] [PubMed] [Google Scholar]
- CLELAND W. W. DITHIOTHREITOL, A NEW PROTECTIVE REAGENT FOR SH GROUPS. Biochemistry. 1964 Apr;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- Capranico G., Zunino F. DNA topoisomerase-trapping antitumour drugs. Eur J Cancer. 1992;28A(12):2055–2060. doi: 10.1016/0959-8049(92)90255-z. [DOI] [PubMed] [Google Scholar]
- Cullinane C., Phillips D. R. Induction of stable transcriptional blockage sites by adriamycin: GpC specificity of apparent adriamycin-DNA adducts and dependence on iron(III) ions. Biochemistry. 1990 Jun 12;29(23):5638–5646. doi: 10.1021/bi00475a032. [DOI] [PubMed] [Google Scholar]
- Cummings J., Anderson L., Willmott N., Smyth J. F. The molecular pharmacology of doxorubicin in vivo. Eur J Cancer. 1991;27(5):532–535. doi: 10.1016/0277-5379(91)90209-v. [DOI] [PubMed] [Google Scholar]
- Cummings J., Bartoszek A., Smyth J. F. Determination of covalent binding to intact DNA, RNA, and oligonucleotides by intercalating anticancer drugs using high-performance liquid chromatography. Studies with doxorubicin and NADPH cytochrome P-450 reductase. Anal Biochem. 1991 Apr;194(1):146–155. doi: 10.1016/0003-2697(91)90162-m. [DOI] [PubMed] [Google Scholar]
- Cummings J., Willmott N., Hoey B. M., Marley E. S., Smyth J. F. The consequences of doxorubicin quinone reduction in vivo in tumour tissue. Biochem Pharmacol. 1992 Dec 1;44(11):2165–2174. doi: 10.1016/0006-2952(92)90343-h. [DOI] [PubMed] [Google Scholar]
- Del Bino G., Darzynkiewicz Z. Camptothecin, teniposide, or 4'-(9-acridinylamino)-3-methanesulfon-m-anisidide, but not mitoxantrone or doxorubicin, induces degradation of nuclear DNA in the S phase of HL-60 cells. Cancer Res. 1991 Feb 15;51(4):1165–1169. [PubMed] [Google Scholar]
- Demant E. J., Nørskov-Lauritsen N. Binding of transferrin-iron by adriamycin at acidic pH. FEBS Lett. 1986 Feb 17;196(2):321–324. doi: 10.1016/0014-5793(86)80271-x. [DOI] [PubMed] [Google Scholar]
- Dikalov S. I., Rumyantseva G. V., Piskunov A. V., Weiner L. M. Role of quinone-iron(III) interaction in NADPH-dependent enzymatic generation of hydroxyl radicals. Biochemistry. 1992 Sep 22;31(37):8947–8953. doi: 10.1021/bi00152a034. [DOI] [PubMed] [Google Scholar]
- Fisher J., Abdella B. R., McLane K. E. Anthracycline antibiotic reduction by spinach ferredoxin-NADP+ reductase and ferredoxin. Biochemistry. 1985 Jul 2;24(14):3562–3571. doi: 10.1021/bi00335a026. [DOI] [PubMed] [Google Scholar]
- Gelvan D., Samuni A. Reappraisal of the association between adriamycin and iron. Cancer Res. 1988 Oct 15;48(20):5645–5649. [PubMed] [Google Scholar]
- Gewirtz D. A. Does bulk damage to DNA explain the cytostatic and cytotoxic effects of topoisomerase II inhibitors? Biochem Pharmacol. 1991 Nov 27;42(12):2253–2258. doi: 10.1016/0006-2952(91)90227-v. [DOI] [PubMed] [Google Scholar]
- Gianni L., Viganò L., Lanzi C., Niggeler M., Malatesta V. Role of daunosamine and hydroxyacetyl side chain in reaction with iron and lipid peroxidation by anthracyclines. J Natl Cancer Inst. 1988 Sep 21;80(14):1104–1111. doi: 10.1093/jnci/80.14.1104. [DOI] [PubMed] [Google Scholar]
- Kappus H. Overview of enzyme systems involved in bio-reduction of drugs and in redox cycling. Biochem Pharmacol. 1986 Jan 1;35(1):1–6. doi: 10.1016/0006-2952(86)90544-7. [DOI] [PubMed] [Google Scholar]
- Land E. J., Mukherjee T., Swallow A. J., Bruce J. M. One-electron reduction of adriamycin: properties of the semiquinone. Arch Biochem Biophys. 1983 Aug;225(1):116–121. doi: 10.1016/0003-9861(83)90013-9. [DOI] [PubMed] [Google Scholar]
- May P. M., Williams G. K., Williams D. R. Solution chemistry studies of adriamycin--iron complexes present in vivo. Eur J Cancer. 1980 Sep;16(9):1275–1276. doi: 10.1016/0014-2964(80)90189-9. [DOI] [PubMed] [Google Scholar]
- Myers C. E. Anthracyclines. Cancer Chemother Biol Response Modif. 1992;13:45–52. [PubMed] [Google Scholar]
- Myers C. E., Gianni L., Simone C. B., Klecker R., Greene R. Oxidative destruction of erythrocyte ghost membranes catalyzed by the doxorubicin-iron complex. Biochemistry. 1982 Apr 13;21(8):1707–1712. doi: 10.1021/bi00537a001. [DOI] [PubMed] [Google Scholar]
- Myers C., Gianni L., Zweier J., Muindi J., Sinha B. K., Eliot H. Role of iron in adriamycin biochemistry. Fed Proc. 1986 Nov;45(12):2792–2797. [PubMed] [Google Scholar]
- Netto L. E., Ferreira A. M., Augusto O. Iron(III) binding in DNA solutions: complex formation and catalytic activity in the oxidation of hydrazine derivatives. Chem Biol Interact. 1991;79(1):1–14. doi: 10.1016/0009-2797(91)90048-c. [DOI] [PubMed] [Google Scholar]
- Pan S. S., Bachur N. R. Xanthine oxidase catalyzed reductive cleavage of anthracycline antibiotics and free radical formation. Mol Pharmacol. 1980 Jan;17(1):95–99. [PubMed] [Google Scholar]
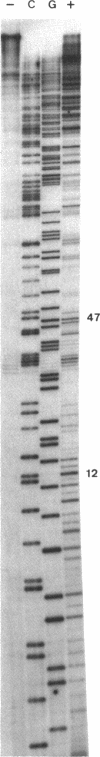
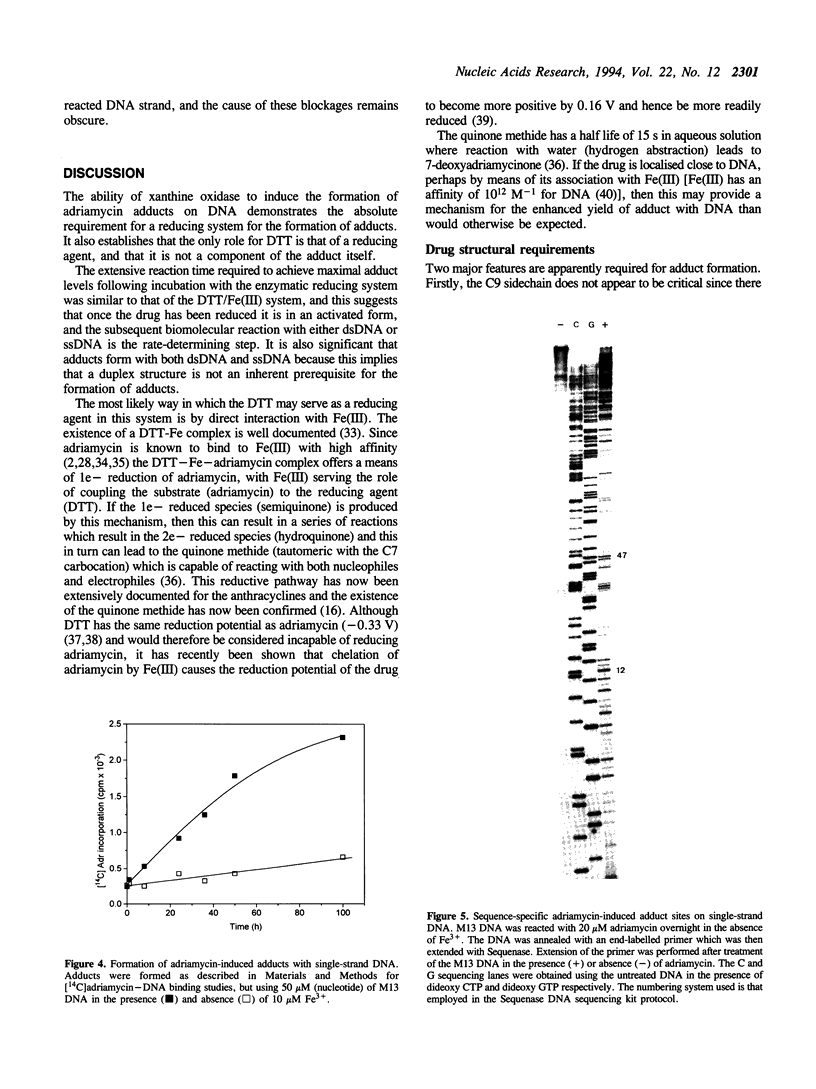
- Phillips D. R., White R. J., Cullinane C. DNA sequence-specific adducts of adriamycin and mitomycin C. FEBS Lett. 1989 Mar 27;246(1-2):233–240. doi: 10.1016/0014-5793(89)80289-3. [DOI] [PubMed] [Google Scholar]
- Schneider E., Hsiang Y. H., Liu L. F. DNA topoisomerases as anticancer drug targets. Adv Pharmacol. 1990;21:149–183. doi: 10.1016/s1054-3589(08)60342-7. [DOI] [PubMed] [Google Scholar]
- Sinha B. K. Free radicals in anticancer drug pharmacology. Chem Biol Interact. 1989;69(4):293–317. doi: 10.1016/0009-2797(89)90117-8. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Sik R. H. Binding of [14C]-adriamycin to cellular macromolecules in vivo. Biochem Pharmacol. 1980 Jun 15;29(12):1867–1868. doi: 10.1016/0006-2952(80)90156-2. [DOI] [PubMed] [Google Scholar]
- Sinha B. K., Trush M. A., Kennedy K. A., Mimnaugh E. G. Enzymatic activation and binding of adriamycin to nuclear DNA. Cancer Res. 1984 Jul;44(7):2892–2896. [PubMed] [Google Scholar]
- Skorobogaty A., White R. J., Phillips D. R., Reiss J. A. Elucidation of the DNA sequence preferences of daunomycin. Drug Des Deliv. 1988 Jul;3(2):125–151. [PubMed] [Google Scholar]
- Tomasz M., Chowdary D., Lipman R., Shimotakahara S., Veiro D., Walker V., Verdine G. L. Reaction of DNA with chemically or enzymatically activated mitomycin C: isolation and structure of the major covalent adduct. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6702–6706. doi: 10.1073/pnas.83.18.6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vile G. F., Winterbourn C. C. High-affinity iron binding by xanthine oxidase. J Free Radic Biol Med. 1986;2(5-6):393–396. doi: 10.1016/s0748-5514(86)80041-1. [DOI] [PubMed] [Google Scholar]
- Weiss R. B. The anthracyclines: will we ever find a better doxorubicin? Semin Oncol. 1992 Dec;19(6):670–686. [PubMed] [Google Scholar]
- Zwelling L. A., Bales E., Altschuler E., Mayes J. Circumvention of resistance by doxorubicin, but not by idarubicin, in a human leukemia cell line containing an intercalator-resistant form of topoisomerase II: evidence for a non-topoisomerase II-mediated mechanism of doxorubicin cytotoxicity. Biochem Pharmacol. 1993 Jan 26;45(2):516–520. doi: 10.1016/0006-2952(93)90091-a. [DOI] [PubMed] [Google Scholar]