Abstract
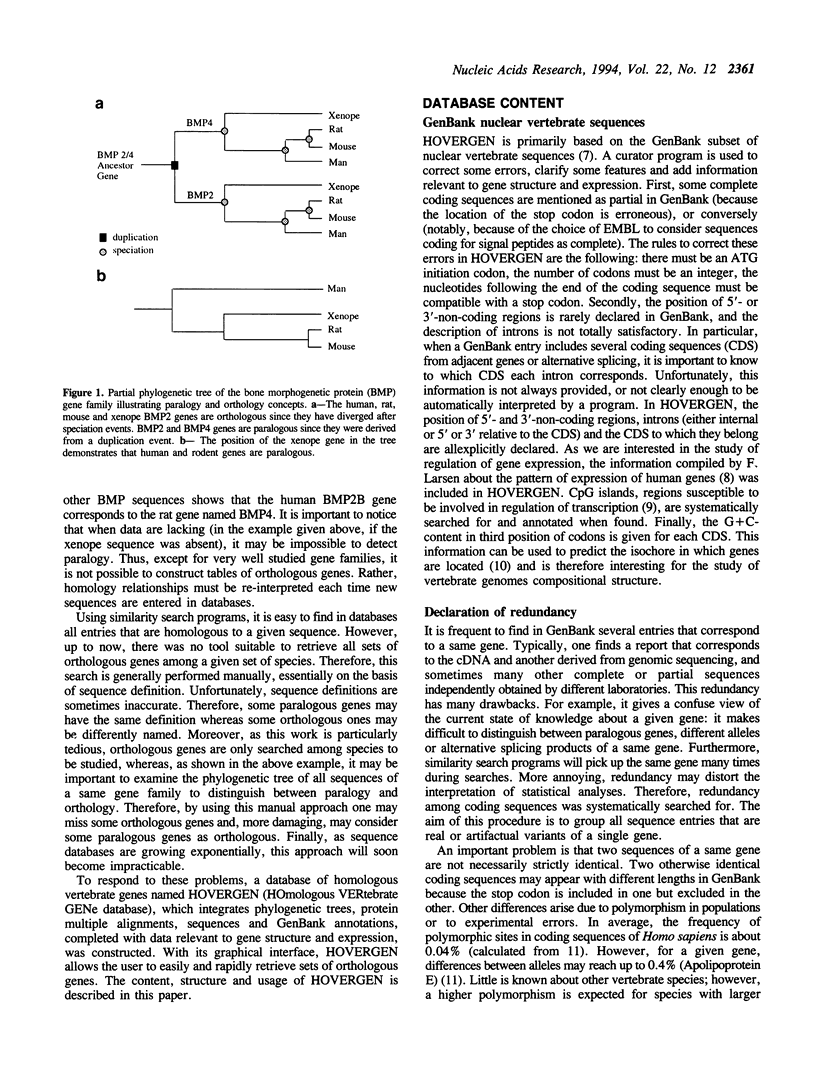
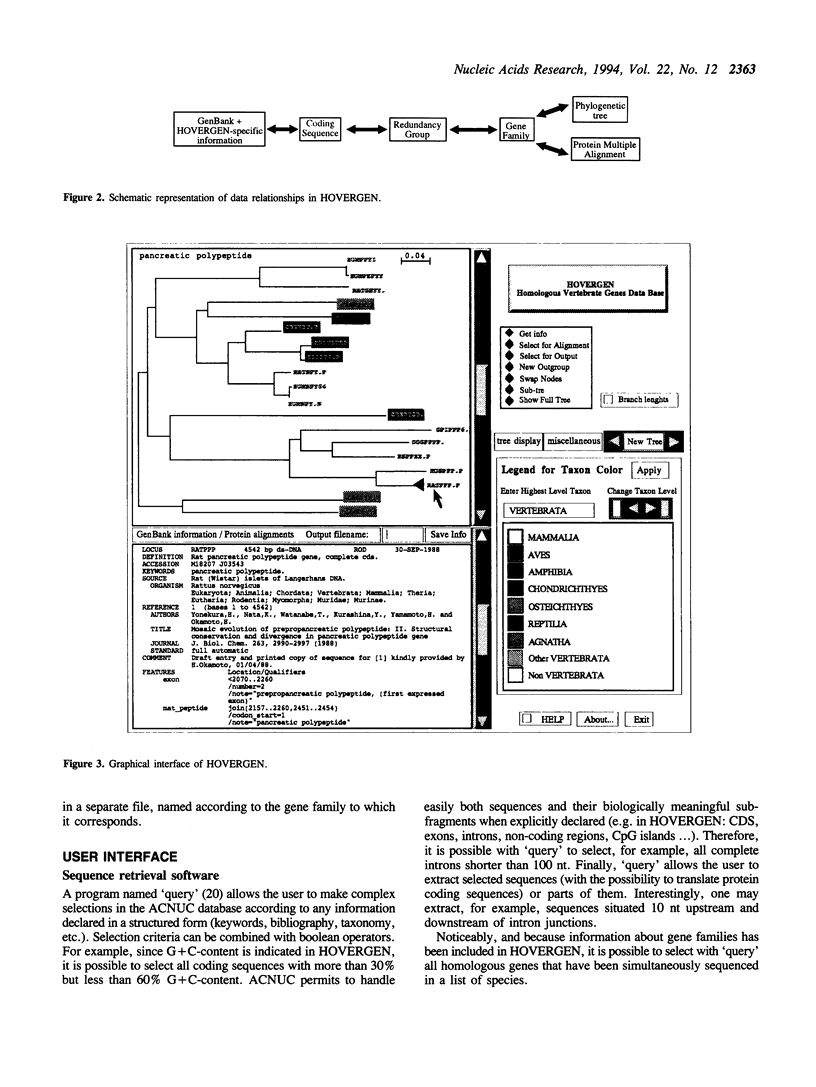
Comparison of homologous genes is a major step for many studies related to genome structure, function or evolution. Similarity search programs easily find genes homologous to a given sequence. However, only very tedious manual procedures allow the retrieval of all sets of homologous genes sequenced for a given set of species. Moreover, this search often generates errors due to the complexity of data to be managed simultaneously: phylogenetic trees, alignments, taxonomy, sequences and related information. HOVERGEN helps to solve these problems by integrating all this information. HOVERGEN corresponds to GenBank sequences from all vertebrate species, with some data corrected, clarified, or completed, notably to address the problem of redundancy. Coding sequences have been classified in gene families. Protein multiple alignments and phylogenetic trees have been calculated for each family. Sequences and related information have been structured in an ACNUC database which permits complex selections. A graphical interface has been developed to visualize and edit trees. Genes are displayed in color, according to their taxonomy. Users have directly access to all information attached to sequences and to multiple alignments simply by clicking on genes. This graphical tool gives thus a rapid and simple access to all data necessary to interpret homology relationships between genes. HOVERGEN allows the user to easily select sets of homologous vertebrate genes, and thus is particularly useful for comparative sequence analysis, or molecular evolution studies.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barker W. C., George D. G., Hunt L. T. Protein sequence database. Methods Enzymol. 1990;183:31–49. doi: 10.1016/0076-6879(90)83005-t. [DOI] [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992 Jul 10;70(1):5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bloch K. D., Friedrich S. P., Lee M. E., Eddy R. L., Shows T. B., Quertermous T. Structural organization and chromosomal assignment of the gene encoding endothelin. J Biol Chem. 1989 Jun 25;264(18):10851–10857. [PubMed] [Google Scholar]
- Burks C., Cassidy M., Cinkosky M. J., Cumella K. E., Gilna P., Hayden J. E., Keen G. M., Kelley T. A., Kelly M., Kristofferson D. GenBank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2221–2225. doi: 10.1093/nar/19.suppl.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F., Galas D. A new five-year plan for the U.S. Human Genome Project. Science. 1993 Oct 1;262(5130):43–46. doi: 10.1126/science.8211127. [DOI] [PubMed] [Google Scholar]
- Duret L., Dorkeld F., Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993 May 25;21(10):2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M., Czelusniak J., Koop B. F., Tagle D. A., Slightom J. L. Globins: a case study in molecular phylogeny. Cold Spring Harb Symp Quant Biol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C., Attimonelli M., Lanave C., di Paola G. ACNUC--a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput Appl Biosci. 1985 Sep;1(3):167–172. doi: 10.1093/bioinformatics/1.3.167. [DOI] [PubMed] [Google Scholar]
- Green P., Lipman D., Hillier L., Waterston R., States D., Claverie J. M. Ancient conserved regions in new gene sequences and the protein databases. Science. 1993 Mar 19;259(5102):1711–1716. doi: 10.1126/science.8456298. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Inoue A., Yanagisawa M., Kimura S., Kasuya Y., Miyauchi T., Goto K., Masaki T. The human endothelin family: three structurally and pharmacologically distinct isopeptides predicted by three separate genes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2863–2867. doi: 10.1073/pnas.86.8.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz S. A. Sequence errors described in GenBank: a means to determine the accuracy of DNA sequence interpretation. Nucleic Acids Res. 1989 May 25;17(10):3951–3957. doi: 10.1093/nar/17.10.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen T., Lopez R., Prydz H. An estimate of the sequencing error frequency in the DNA sequence databases. DNA Seq. 1992;2(6):343–346. doi: 10.3109/10425179209020815. [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992 Aug;13(4):1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Li W. H., Sadler L. A. Low nucleotide diversity in man. Genetics. 1991 Oct;129(2):513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud D., Bernardi G. Compositional properties of coding sequences and mammalian phylogeny. J Mol Evol. 1993 Aug;37(2):109–116. doi: 10.1007/BF02407345. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D., D'Onofrio G., Aïssani B., Macaya G., Gautier C., Bernardi G. The distribution of genes in the human genome. Gene. 1991 Apr;100:181–187. doi: 10.1016/0378-1119(91)90364-h. [DOI] [PubMed] [Google Scholar]
- Mouchiroud D., Gautier C. Codon usage changes and sequence dissimilarity between human and rat. J Mol Evol. 1990 Aug;31(2):81–91. doi: 10.1007/BF02109477. [DOI] [PubMed] [Google Scholar]
- O'hUigin C., Li W. H. The molecular clock ticks regularly in muroid rodents and hamsters. J Mol Evol. 1992 Nov;35(5):377–384. doi: 10.1007/BF00171816. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J Mol Evol. 1993 Oct;37(4):441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]



