ABSTRACT
Helicobacter pylori is a bacterial pathogen that can cause many gastrointestinal diseases, including ulcers and gastric cancer. A unique chemotaxis-mediated motility is critical for H. pylori to colonize in the human stomach and to establish chronic infection, but the underlying molecular mechanisms are not well understood. Here, we employ cryo-electron tomography (cryo-ET) to reveal detailed structures of the H. pylori cell envelope, including the sheathed flagella and chemotaxis arrays. Notably, H. pylori possesses a distinctive periplasmic cage-like structure with 18-fold symmetry. We propose that this structure forms a robust platform for recruiting 18 torque generators, which likely provide the higher torque needed for swimming in high-viscosity environments. We also reveal a series of key flagellar assembly intermediates, providing structural evidence that flagellar assembly is tightly coupled with the biogenesis of the membrane sheath. Finally, we determine the structure of putative chemotaxis arrays at the flagellar pole, which have implications for how the direction of flagellar rotation is regulated. Together, our pilot cryo-ET studies provide novel structural insights into the unipolar flagella of H. pylori and lay a foundation for a better understanding of the unique motility of this organism.
IMPORTANCE Helicobacter pylori is a highly motile bacterial pathogen that colonizes approximately 50% of the world's population. H. pylori can move readily within the viscous mucosal layer of the stomach. It has become increasingly clear that its unique flagella-driven motility is essential for successful gastric colonization and pathogenesis. Here, we use advanced imaging techniques to visualize novel in situ structures with unprecedented detail in intact H. pylori cells. Remarkably, H. pylori possesses multiple unipolar flagella, which are driven by one of the largest flagellar motors found in bacteria. These large motors presumably provide the higher torque needed by the bacterial pathogens to navigate in the viscous environment of the human stomach.
KEYWORDS: chemotaxis, electron microscopy, flagellar motility, nanomachine
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the human gastric mucosa and infects approximately 50% of the world's population (1). One of most unique characteristics of H. pylori is its capacity to colonize and thrive in the gastric environment of high acidity, where few other bacterial pathogens can survive. Persistent colonization by this pathogen can cause many gastrointestinal diseases, including ulcers and gastric cancer (2, 3).
H. pylori has a unique helical cell shape and fine-tuned flagellar motility (4). Compared to other motile bacteria, H. pylori cells have the unique ability to swim rapidly within the viscous mucosal layer of the stomach (5, 6). It has become increasingly clear that this unique motility is essential for its successful gastric colonization and pathogenesis (7, 8). The motility of H. pylori is driven by multiple unipolar flagella, which are enveloped within a membrane sheath. The overall organization of the H. pylori flagellum is similar to those of the model organisms Escherichia coli and Salmonella, which have been extensively studied and comprehensively reviewed (9–13). The H. pylori flagellum is composed of the following three major parts: a rotary motor embedded in the cell envelope, a filament that functions as a helical propeller, and a hook serving as a flexible joint between motor and filament. At least several dozen different proteins are involved in the assembly and function of the polar flagellum (4).
The rotary motor is the most complex part of the flagellum and can be divided into the following morphological domains: the MS ring (the base for the flagellar motor), the C ring (the switch complex regulating motor rotation), the export apparatus (multiple-protein complex located at the cytoplasmic side of the MS ring), the rod (connecting the MS ring and the hook), and the stator (membrane protein complex of MotA and MotB). The stator functions as the torque generator and is powered by proton motive force (9). A motB mutant lacking the stator complex is flagellated but nonmotile (8). The crystal structure of the cell wall anchor domain of MotB has provided molecular insights into stator-peptidoglycan interactions (14). However, the exact location and structure of the stator are not well understood, despite its essential roles in flagellar rotation and motility in H. pylori.
The C ring is the major cytoplasmic component and is important for flagellar rotation and assembly (4). The H. pylori genome encodes four flagellar switch proteins (FliG, FliM, FliN, and FliY) (15) compared to the three switch proteins (FliG, FliM, and FliN) in Salmonella and E. coli. It has been shown that each of the four switch proteins is required for wild-type levels of flagellation and function. High-resolution structures revealed the molecular details of FliM-FliG interactions (16). Presumably, multiple copies of these four proteins are involved in the assembly of the C ring, but the exact role and location of each protein remain to be defined in the context of functional flagella.
The motility of H. pylori is mediated by a chemotactic signaling system, which senses the environment to help bacteria direct their movement (17, 18). The signaling system of H. pylori is crucial for its unique ability to move away from a low-pH environment and successfully colonize the mucosal layer (4). The core components of the H. pylori chemotactic signaling system are comparable with those of E. coli (4) and include multiple chemoreceptors, a CheA kinase, a CheW coupling protein, and a CheY response regulator. Additionally, H. pylori possesses three different coupling proteins (CheV1, CheV2, and CheV3). Upon activation, the CheA histidine kinase domain undergoes autophosphorylation, which then phosphorylates the response regulator CheY. The interaction between the response regulator and the C ring proteins ultimately alters the direction of motor rotation. H. pylori has three integral membrane chemoreceptors, TlpA, TlpB, and TlpC, and one soluble chemoreceptor, TlpD. Together, they can sense arginine, bicarbonate (19), pH (20, 21), the quorum sensing molecule, autoinducer-2 (22), and energy (23). These chemotactic signaling proteins form clusters found at the cell pole of many bacterial species (24–27), and they form clusters at the flagellar pole of H. pylori as well (5, 28). However, the structure of the H. pylori clusters has not been extensively studied.
Recently, cryo-electron tomography (cryo-ET) has emerged as a powerful tool to visualize intact flagella and chemoreceptor clusters from different bacterial species, such as E. coli, Salmonella, Campylobacter jejuni, Vibrio cholerae, Borrelia burgdorferi, Leptospira interrogans, and Treponema primitia (29–36). Notably, Chen et al. imaged flagellar motors from 11 different bacteria and revealed divergent features as well as conserved core components (32). Although the basic architecture and the core components of flagellar motors from different bacterial species are highly conserved, it is becoming increasingly clear that there are significant species-specific differences (32, 35). Despite its significance for understanding the unique motility of H. pylori, the structure of its flagellar motor has not yet been reported. As a step toward understanding the flagellar assembly and unique motility of H. pylori, we employed cryo-ET to visualize intact wild-type H. pylori cells, with particular focus on flagella and chemoreceptor arrays. We were able to resolve novel features of H. pylori and, therefore, provide new insights into the cell biology of this important bacterial pathogen.
RESULTS
Ultrastructures of H. pylori cells revealed by cryo-EM.
To visualize intact H. pylori cells in near-native states, we prepared frozen-hydrated specimens from a freshly grown wild-type H. pylori culture (see Materials and Methods). At low magnification (×2,200), whole cells and the sheath flagella attached at cell poles were readily visible (Fig. 1A and B). However, most cells were too large for high-resolution cryo-electron microscopy (cryo-EM) studies, so it was necessary to only visualize bacteria embedded in thin ice, which is often less than 0.5 μm thick.
FIG 1.
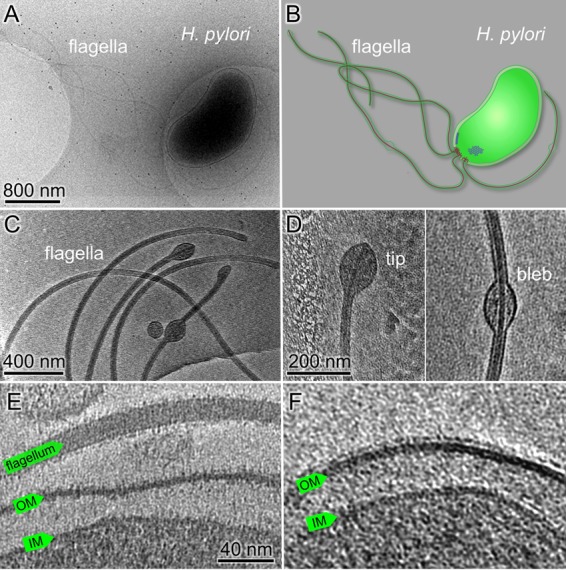
Ultrastructure of H. pylori cells revealed by cryo-ET. (A) An overall cryo-EM image of a wild-type H. pylori cell with many flagella. (B) A schematic model of the H. pylori cell with particular emphasis on the cell envelope, flagella, and chemoreceptor arrays. (C) A tomographic slice shows the sheathed flagella with membrane blebs at the tip or in the middle of the flagella. (D) Zoomed-in views of a membrane bulb at the flagellar tip (left) and a membrane bleb in the middle of a flagellum (right). (E) A tomographic slice at the cell pole shows a sheathed flagellum and the cell envelope, including the outer membrane (OM) and the inner membrane (IM). (F) Fine filamentous features that cover the outer membrane and the flagellar sheath presumably represent bacterial lipopolysaccharide (LPS).
High-magnification images of H. pylori cells revealed detailed structures of the cell envelope, including the inner and the outer membranes (Fig. 1; see also Fig. S2 in the supplemental material). The outer membrane is covered with a layer of fine filamentous features (Fig. 1E and F; Fig. S2). This layer is probably the bacterial lipopolysaccharide (LPS), which has an important role in H. pylori pathogenesis (25). The outer membrane also forms the sheath covering the flagella. Membrane blebs were commonly found along the filaments as previously described (37, 38). Unsheathed flagella were rarely seen in our reconstructions, suggesting that the membrane sheath biosynthesis is coupled with flagellar assembly.
Chemoreceptor arrays in H. pylori.
Chemotaxis signaling proteins of H. pylori form clusters at the flagellar pole (5). Here, we directly observe clusters at the flagellar poles by cryo-ET for the first time (Fig. 2). At higher magnification, the clusters appear as arrays of pillar-like densities that extend from the cytoplasmic membrane and connect with a layer of dark density at the membrane-distal ends (Fig. 2B). They share similar features with the chemoreceptor arrays reported in E. coli and many other bacterial species (24, 26). Therefore, the highly ordered array is presumably formed by the chemoreceptors together with CheA and CheW.
FIG 2.
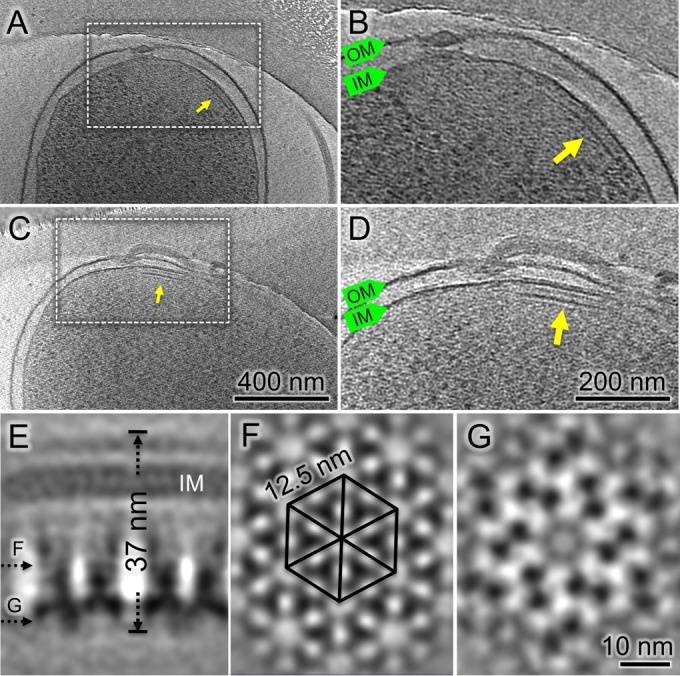
Cryo-ET of H. pylori cells reveals chemoreceptor arrays at the cell pole. (A, C) Tomographic slices show arrays at the flagellar poles. (B, D) Zoomed-in views of the arrays, highlighted by arrows. (E) A side view of the averaged structure of the array. (F) A central cross-section from the array shows a hexagonal lattice of receptors. (G) Another central cross-section from the bottom of the arrays shows the CheA densities.
To investigate the detailed organization of the arrays in intact H. pylori cells, subtomogram averaging was utilized to analyze the arrays as previously described (39). The hexagonal structure of the array emerges after multiple cycles of alignment and classification (Fig. 2F). The overall structure of the array is consistent with the universal architecture of bacterial chemoreceptor arrays (24). Specifically, there are six triangular densities in a hexagonal unit, and each of these is likely formed by a trimer of a chemoreceptor dimer. The distance between two adjacent triangular densities is 7.5 nm. The receptor trimers extend from the periplasm to the cytoplasm with a total length of 37 nm. The periplasmic domain and the transmembrane domain of the chemoreceptor trimers appear to be flexible as indicated by the less-defined densities. Similar to the chemoreceptor arrays in E. coli (39), two receptor trimers are connected at the distal end by a continuous layer of density likely formed by CheA and CheW (Fig. 2E and G). They are likely responsible for the ultrastability of the chemoreceptor arrays as shown in E. coli and other bacterial species (39, 40).
Architecture of the unipolar flagellum.
Our tomographic data from over 100 cells show that the flagella appear to cluster at cell poles. The closest distance between two adjacent flagella is ∼100 nm (see Fig. S3 in the supplemental material). The flagella are located at the cell poles where the receptor arrays are commonly observed (Fig. 2 and 3), which is consistent with the observation of their colocalization (5).
FIG 3.
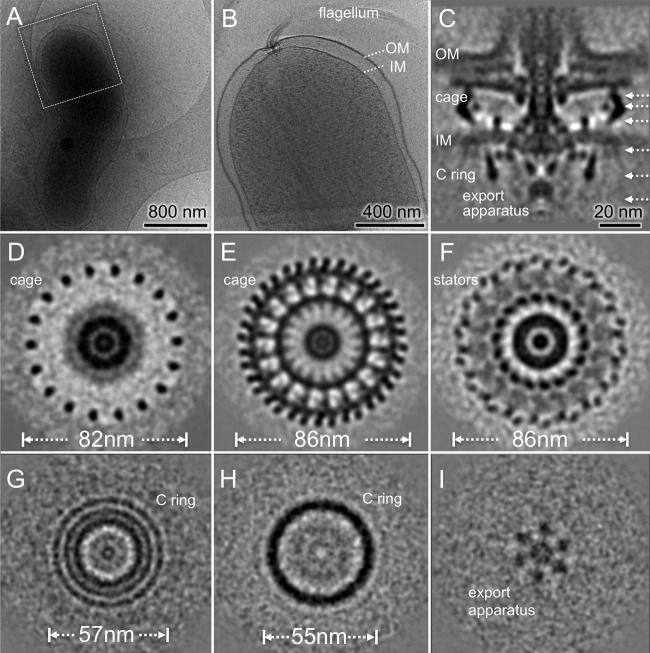
Flagellar motor structure of H. pylori revealed by cryo-ET and subtomogram averaging. (A) A typical cryo-EM image of one bacterium at low magnification. (B) A central slice of the tomogram from the cell pole outlined in panel A. Note that one flagellar motor is embedded in the cell envelope, including the outer membrane (OM) and the inner membrane (IM). (C) A central slice of the averaged structure. (D to F) Three cross sections of the flagellar motors show distinct features with 18-fold symmetry in periplasmic space. (G and H) Two cross sections from the top and the bottom of the C ring, respectively. (I) One section from the export apparatus shows a hexagonal pattern.
To determine the H. pylori motor structure in detail, we selected 365 subtomograms of motors from over 100 reconstructions of intact cells and then analyzed them by subtomogram averaging and classification (see Materials and Methods). The resulting motor structure is striking as shown in Fig. 3. First, this is one of the largest bacterial motors observed by far. It is ∼86 nm in diameter and ∼81 nm in height (from the export apparatus to the outer membrane). Second, the C ring is 57 nm in diameter, and it is considerably larger than the C ring of Salmonella (45 nm) (41). Third, there is a distinctive periplasmic feature that is ∼86 nm in diameter and at least 25 nm in height. More importantly, the 18-fold symmetry of the periplasmic feature is evident in the cross sections (Fig. 3; see also Movie S1 in the supplemental material), which has not been reported previously in any flagellar system (Fig. 3). Our symmetry analysis further confirms that the 18-fold symmetric features are present predominately in the periplasmic region but not for the C ring and other flagellar components (see Fig. S1 in the supplemental material).
Guided by recent findings on conserved cores and diverse features of bacterial flagellar motors among a wide range of bacterial species (32, 35), we divided the density map into different segments for better visualization of the H. pylori motor in three dimensions (3D) (Fig. 4; see also Movie S2 in the supplemental material). The H. pylori motor is composed of several major conserved components: the C ring, the export apparatus, the MS ring, and the rod. Around the rod, there is a large disk in a similar location of the P ring and the L ring in the Salmonella motor (32, 41) that we named the P disk. In addition, there is another smaller disk sandwiched between the P disk and the outer membrane sheath. The outer membrane covers the top portion of the motor, the hook, and the filament.
FIG 4.
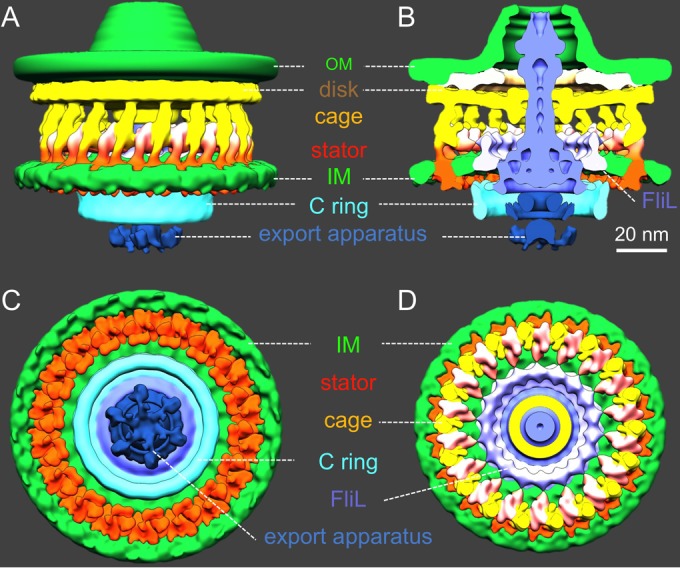
Architecture of the H. pylori motor in 3D. (A) A side view of the surface rendering of the motor shows a cage-like structure (yellow) in the periplasmic space. The outer membrane (OM) and the inner membrane (IM) are colored in green. The rotor, including the C ring, the MS ring, and the rod, is colored in blue and cyan. The export apparatus (dark blue) is located underneath the C ring. (B) A cross-section view of the motor shows the central portion of the motor: the MS ring, the rod, the C ring, and the export apparatus. A cage-like structure appears to surround the rotor. Interestingly, there is an extra ring between the cage and the rotor. We hypothesize that the putative FliL molecules form the extra ring. (C) A bottom view shows the export apparatus and the C ring and the cytoplasmic densities of the putative stators (orange). Note that the putative stator complexes form a ring around the C ring. (D) A top view of the motor after removing the outer membrane. The cage-like structure forms a large ring (orange and yellow) around the putative FliL ring.
Our intact motor structure reveals three concentric rings at the top of the C ring (Fig. 3G). The outermost ring and the middle ring are the top portion of the C ring. They are likely formed by the C-terminal domain (FliGC) and the N-terminal domain (FliGN) of FliG, respectively, as recently proposed in other flagellar systems (35). The inner ring appears to be an extension of the MS ring, which is highly conserved and is formed by multiple copies of the membrane protein FliF. As FliGN has been shown to interact with the C terminus (FliFC) of FliF (42), the inner ring is likely formed by FliFC, which directly interacts with FliGN. A similar interconnection between the C ring and the MS ring was previously shown in a spirochete flagellar motor (30), suggesting that the FliG-FliF interaction may be conserved among different bacterial species.
The export apparatus is a large multiprotein complex that is responsible for flagellar assembly and substrate export (11, 12, 35). The centrally located export apparatus is visible on the cytoplasmic side of the MS ring (Fig. 4). The export apparatus consists of at least five integral membrane proteins (FlhA, FlhB, FliP, FliQ, and FliR) and three cytoplasmic proteins (FliH, FliI, and FliJ) (11), all of which are highly conserved. The membrane proteins form a large export gate for substrate secretion, whereas the cytoplasmic proteins form a hexagonal ATPase complex promoting the export process (11). The ATPase complex appears to be linked to the bottom of the C ring. Together, our results provide direct evidence that the export apparatus in H. pylori is similar to those in other bacterial species (32, 35).
A cage-like structure in the periplasm displays 18-fold symmetry.
The central section of the H. pylori motor appears to be similar to some previously published motor structures from other species (32). However, our structure reveals, for the first time, a unique cage-like periplasmic structure with 18-fold symmetry, ∼86 nm in diameter and 25 nm in height. The cage structure completely surrounds the periplasmic portion of the motor, including the MS ring, the rod, and the P-disk. It appears to be anchored to the inner membrane and extends to the disk underneath the outer membrane. There is an additional ring between the MS ring and the cage (Fig. 3C and F and 4B and D). It is apparently embedded in the cytoplasmic membrane, presumably interacting with the MS ring (Fig. 4B). The location and structure of the additional ring are similar to those of the FliL ring found in B. burgdorferi (43, 44). FliL has been shown to play important but diverse roles in many bacterial species (43, 45–47). The exact structure and function of FliL in H. pylori remain to be elucidated.
Another striking finding is that there are 18 stud-like densities immediately underneath the cytoplasmic membrane. They form a larger ring that is ∼81 nm in diameter as shown in the bottom view (viewed from the bottom of the C ring; Fig. 4C). Each stud-like density is about 4 nm in diameter and is comparable to the stator structure observed previously with freeze fracture microscopy (48) and electron microscopy (49). Therefore, we hypothesize that the stud-like density corresponds to the cytoplasmic domain of the stator. The putative stator complex appears to interact with the C ring in the cytoplasm, and it also links to the cage-like structure in the periplasm. Based on the configuration, we propose that the cage-like structures provide binding sites to recruit 18 stator complexes, which otherwise would be dynamic and difficult to visualize (50).
Visualization of flagellar assembly intermediates.
Most of the flagellar motors that we observed seem to be associated with a long sheathed filament, while some flagellar motors lack a visible hook and filament. Presumably, the latter motors are in an early stage of assembly. To gain a better understanding of this early stage, we generated a specific class average of the H. pylori motor at this stage. As shown in Fig. 5, the motor lacks the hook and the filament. Notably, the outer membrane remains intact; however, it appears that the rod assembly pushes the outer membrane outward and induces significant deformation of the outer membrane. At this particular stage, the intact rod has been assembled without the hook, potentially representing a key assembly intermediate. Interestingly, the cage and other motor components remain similar to those observed in the intact motor. In addition, the putative stator units interconnect with the cage, suggesting that both cage and stator can assemble in the periplasm independently from the assembly of the hook and filament, which is mediated by the export apparatus (11).
FIG 5.
A flagellar motor structure lacking the hook and filament. (A) A central section of the flagellar motor structure shows the outer membrane (OM) with significant deformation. (B) A cross-section view of the motor reveals 18-fold symmetric features. A side view (C) and a cut-in view (D) of the motor. The cage-like structure surrounds the rotor. The following color scheme is the same as that in Fig. 4: yellow, cage; orange, stator; green, membranes; blue, rod and MS ring; cyan, C ring; and dark blue, export apparatus.
Flagellar assembly is a highly ordered process, which is believed to proceed in a linear fashion from the rod and hook to the distal filament (11, 12, 51). However, this sequential assembly process has not been visualized in wild-type bacteria partly because of the transient and dynamic nature of the process. The use of high-throughput cryo-ET enabled us to visualize over 300 flagella. Consequently, we were able to capture a series of intermediate structures during flagellar assembly (Fig. 6; see also Fig. S4 in the supplemental material). For example, we visualized a flagellar intermediate lacking the hook and filament, while the outer membrane appeared to be flat (Fig. 6A). We also found a motor in which the outer membrane undergoes extensive remodeling during hook assembly (Fig. 6B). In addition, some motors appear to possess an intact hook (Fig. 6C) or short filament (Fig. 6D). This series of snapshots of the dynamic process of flagellar assembly provides evidence that the assembly of the flagella is tightly coupled with the biogenesis of the outer membrane sheath.
FIG 6.
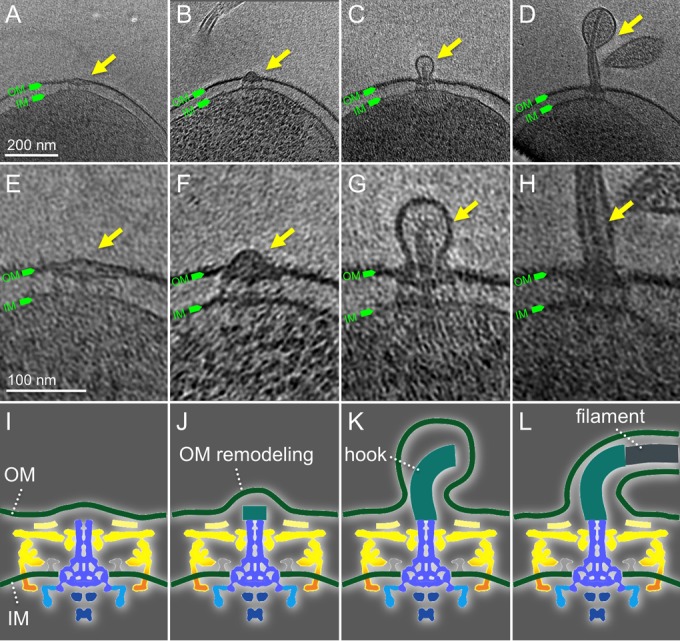
The assembly of flagella couples with the biosynthesis of the outer membrane sheath. (A to D) Representative slices of cryo-ET reconstructions show different intermediates during flagellar assembly. (E to H) The corresponding zoomed-in views of images from panels A through D. The corresponding cartoon illustrations show different stages: rod assembly (I), hook assembly (J, K), and filament assembly (L). The following color scheme is the same as that in Fig. 4: yellow, cage; orange, stator; green, membranes; blue, rod and MS ring; cyan, C ring; and dark blue, export apparatus.
DISCUSSION
H. pylori is a highly successful bacterial pathogen that infects over 50% of the world's population and has coevolved with the human host for tens of thousands of years (52). H. pylori has evolved sophisticated motility and chemotaxis strategies to colonize and survive within the hostile environment of the acidic stomach and navigate toward neutral gastric mucosa (53). To advance our understanding of these strategies, we deployed cryo-ET to visualize intact frozen-hydrated H. pylori cells and revealed novel structural features that heretofore were not visualized by any other techniques.
H. pylori possesses four different chemoreceptors that respond to different chemotactic signals (4, 54) and are known to form clusters at flagellar cell poles (5, 28). We provide direct evidence that the overall structure of the receptor array is similar to that observed in other bacteria, supporting the universal architecture of bacterial chemoreceptor arrays (24). Presumably, the conserved architecture of the arrays provides the basis for the high sensitivity and large dynamic range of chemotaxis in responding to complex chemical cues in the host environment.
Our most striking finding was the unique periplasmic cage displaying 18-fold symmetry. Periplasmic collar structures around the MS ring have been observed in many spirochete flagellar motors (29, 30, 32, 55). It was recently shown that the collar not only plays important roles for the assembly of many flagellar proteins but also has profound impacts on the cell morphology and motility of B. burgdorferi (44). In Vibrio alginolyticus, MotX and MotY form a T ring (56). While the exact function of the T ring remains to be determined, the T ring was proposed to be involved in the incorporation and/or stabilization of the stator complexes in the Vibrio motor (49). In C. jejuni, multiple proteins are responsible for the assembly of the periplasmic disk complex, and recent studies indicate that it is required for bacterial motility and stator assembly (57). The cage structure in H. pylori is strikingly different from the periplasmic features in other bacteria, yet it does have a similar location relative to the C ring and the stator. Therefore, we postulate that by virtue of its size and structure, the periplasmic cage likely provides a stable platform to recruit and stabilize more stators for higher torque generation. The precise function and protein components of the cage remain to be determined.
The stator complex consists of two conserved membrane proteins and is important for generating torque. The stator is not static as its name implies but instead is dynamic within the membrane (50). In E. coli and Salmonella, the stator is driven by proton motive force and is composed of four MotA and two MotB proteins (58). A conserved aspartic acid residue in the transmembrane segment of MotB is predicted to be the proton-binding site and plays a crucial role for torque generation and bacterial motility (59). In V. alginolyticus and Shewanella spp., the MotA/MotB homologs PomA and PomB form the stator complex, which is driven by sodium motive force (60). The stoichiometry of the stator varies significantly among different species. In E. coli, the number of stator units was reported to be 11 by using total internal reflection fluorescence microscopy (50, 61). Spirochete species, such as T. primitia and B. burgdorferi, have 16 putative stator units as visualized by cryo-ET (29, 30). Recent data have suggested that the C. jejuni motor can possess 17 stator units while Vibrio fischeri has 13 stator units (57). Our data indicate that the H. pylori motor can assemble 18 stator units, potentially allowing for the generation of higher torque than other bacterial species. This provides a plausible explanation for how H. pylori is able to move through the viscous mucus layers to establish chronic infection.
MATERIALS AND METHODS
Bacterial culture and cryo-EM sample preparation.
H. pylori strain 7.13 was derived via in vivo adaptation of clinical strain B128 (62). The strain was grown for 24 h on tryptic soy agar plates with 5% sheep blood (BD Biosciences) under microaerobic conditions. Bacterial samples were collected from plates by gently washing plates with 500 μl of sterile phosphate-buffered saline (PBS). Samples were concentrated by gently precipitating by centrifugation with a benchtop mini microcentrifuge (Corning) for 1 min and were resuspended in 100 to 300 μl of sterile PBS. Resuspended bacterial cultures were gently mixed with aliquots of 15-nm gold particles, which were used as fiducial markers later for tomogram alignment by IMOD (63). A 4-μl sample was deposited onto a freshly grow-discharged holey carbon grid. The grid was blotted with filter paper and rapidly plunged into liquid ethane using a homemade gravity-driven plunger apparatus as previously described (30). The preparation not only preserves high-resolution cellular features but also minimizes possible artifacts during conventional EM preparation, such as dehydration, fixation, and staining. However, compared to conventionally dehydrated and stained specimens, frozen-hydrated cells are extremely sensitive to the electron beam.
Cryo-ET data collection and reconstruction.
Detailed information on data collection and reconstruction has been previously described (64). Briefly, the frozen-hydrated specimens were imaged at −170°C in a cryo-electron microscope (FEI Polara) equipped with a field emission gun and a direct detection device (K2 Summit; Gatan). The microscope was initially operated at 300 kV with a magnification of ×2,200 for taking low-magnification images. The montage of those images gave an overview of bacterial cell distribution and size. More importantly, we are able to estimate the thickness of the cells based on the low-magnification overviews. To capture cryo-EM data at higher contrast and resolution, we only select cells embedded in very thin ice for further tomographic data collection at a magnification of ×9,400.
The tomographic package SerialEM (65) was utilized to collect a low-dose single-axis tilt series at ∼8 μm defocus with cumulative does of ∼100 e−/Å2 spread over 41 stacks at a range of −60° and +60° with 3° increments. Each stack contains about eight images, which were first aligned using MotionCorr (66) and were assembled into the drift-corrected stacks. TOMOAUTO was used to facilitate the alignment in IMOD (63, 67). All aligned tilt series were reconstructed into tomograms using TOMO3D (68). In total, we reconstructed 187 tomograms from the cell poles of H. pylori. Each tomographic reconstruction was 3,710 by 3,838 by 1,800 pixels and ∼100 Gb in size. The effective pixel size is 4.5 Å in original tomograms. To enhance the contrast, we also generated a reconstruction from the binned by 4 aligned tilt series using the simultaneous iterative reconstruction technique (SIRT) implemented in TOMO3D (68). Each tomogram was 928 by 960 by 450 pixels.
Subtomogram classification and averaging.
Tomography package i3 (69, 70) was utilized for subtomogram picking and averaging. The subtomogram analysis of the arrays was carried out as follows. First, we selected 30 tomograms in which the receptor arrays are reasonably large and visible. Second, we built an ellipsoid model based on the curvature of the bacterial inner membrane. A uniform distributed mesh was made on the surface of the ellipsoid model, and each point on the mesh has a coordinate and normal vector (surface normal), which were used to estimate the initial Euler angles. Third, we visually selected the arrays around the cell envelope. Only those near the marked position of the receptor arrays were selected. In total, about 3,000 subtomograms of arrays were extracted from tomograms. Fourth, the subtomograms were analyzed through multiple iterations of alignment. The final structure was derived from 677 independent subtomograms.
To determine the flagellar motor structure, we first manually select motors from tomograms. The orientation of the flagellar motor was estimated based on the coordinates of the two points on each motor (the center of the C ring and the rod). The coordinates and Euler angles of all motor subtomograms were refined through iterations of transitional, polar, and spin alignment. To identify the rotation symmetry of the cage and the stators, the subtomograms of the flagellar motor were first aligned and then classified on the stator part with a cylinder mask. The rotational symmetry of the class averages was estimated. The average structure was rotated 360° with a step size of 1° and was compared with the original image by cross correlation. The rotational cross correlation shows peaks every 20° (see Fig. S1A in the supplemental material). The Fourier transform of the cross correlation shows a peak at 18 (Fig. S1B).
Visualization of the flagellar motor.
We used UCSF Chimera (71) to generate surface rendering of the 3D maps. Watershed segmentation (72) was used to separate the major components of the motor: the disk, the MS ring, the periplasmic cage, the stator, the C-ring, and the export apparatus. The segmentation was mainly guided by prior knowledge of various bacterial flagellar motors (4, 32, 35).
Accession number(s).
The final subtomogram structure was deposited in the Electron Microscopy Data Bank under accession number EMD-8460. The motor structures were deposited in the Electron Microscopy Data Bank under accession number EMD-8459.
Supplementary Material
ACKNOWLEDGMENTS
We thank William Margolin and James Stoops for careful reading and helpful suggestions.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01AI087946), the National Institute of General Medical Sciences (R01GM107629), the NIH Center for Microbial Pathogenesis and Host Inflammatory Responses project at the University of Arkansas for Medical Sciences (COBRE grant 1P20GM103625-02), and the Welch Foundation (AU-1714) and by the McGovern Scholar Award.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00695-16.
REFERENCES
- 1.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 2.Salama NR, Hartung ML, Muller A. 2013. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol 11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishaq S, Nunn L. 2015. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench 8:S6–S14. [PMC free article] [PubMed] [Google Scholar]
- 4.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howitt MR, Lee JY, Lertsethtakarn P, Vogelmann R, Joubert LM, Ottemann KM, Amieva MR. 2011. ChePep controls Helicobacter pylori Infection of the gastric glands and chemotaxis in the Epsilonproteobacteria. mBio 2:e00098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci U S A 101:5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton KA, Morgan DR, Krakowka S. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J Med Microbiol 37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 8.Ottemann KM, Lowenthal AC. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect Immun 70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manson MD, Tedesco P, Berg HC, Harold FM, Van der Drift C. 1977. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci U S A 74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg HC. 2003. The rotary motor of bacterial flagella. Annu Rev Biochem 72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 11.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 12.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol 6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb Perspect Biol 2:a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roujeinikova A. 2008. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc Natl Acad Sci U S A 105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191:7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam KH, Lam WW, Wong JY, Chan LC, Kotaka M, Ling TK, Jin DY, Ottemann KM, Au SW. 2013. Structural basis of FliG-FliM interaction in Helicobacter pylori. Mol Microbiol 88:798–812. doi: 10.1111/mmi.12222. [DOI] [PubMed] [Google Scholar]
- 17.Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerda O, Rivas A, Toledo H. 2003. Helicobacter pylori strain ATCC 700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol Lett 224:175–181. doi: 10.1016/S0378-1097(03)00423-3. [DOI] [PubMed] [Google Scholar]
- 20.Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. 2012. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol 188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. 2011. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, Schreiber S, Suerbaum S, Aizawa S-I, Josenhans C. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol 190:3244–3255. doi: 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A 106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran AP. 1996. The role of lipopolysaccharide in Helicobacter pylori pathogenesis. Aliment Pharmacol Ther 10(Suppl):S39–S50. [DOI] [PubMed] [Google Scholar]
- 26.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. 2007. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A 104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddock JR, Shapiro L. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 28.Lertsethtakarn P, Howitt MR, Castellon J, Amieva MR, Ottemann KM. 2015. Helicobacter pylori CheZ(HP) and ChePep form a novel chemotaxis-regulatory complex distinct from the core chemotaxis signaling proteins and the flagellar motor. Mol Microbiol 97:1063–1078. doi: 10.1111/mmi.13086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. 2010. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol 169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. 2011. Structural diversity of bacterial flagellar motors. EMBO J 30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamoto A, Morimoto YV, Miyata T, Minamino T, Hughes KT, Kato T, Namba K. 2013. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep 3:3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller A, Beeby M, McDowall AW, Chow J, Jensen GJ, Clemons WM Jr. 2014. Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. Microbiologyopen 3:702–710. doi: 10.1002/mbo3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao X, Norris SJ, Liu J. 2014. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry 53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Howell JK, Bradley SD, Zheng Y, Zhou ZH, Norris SJ. 2010. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol 403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geis G, Leying H, Suerbaum S, Mai U, Opferkuch W. 1989. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J Clin Microbiol 27:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luke CJ, Kubiak E, Cockayne A, Elliott TS, Penn CW. 1990. Identification of flagellar and associated polypeptides of Helicobacter (formerly Campylobacter) pylori. FEMS Microbiol Lett 59:225–230. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. 2012. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A 109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. 2012. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A 109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas DR, Francis NR, Xu C, DeRosier DJ. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grunenfelder B, Gehrig S, Jenal U. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J Bacteriol 185:1624–1633. doi: 10.1128/JB.185.5.1624-1633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon KH, Zhao X, Manne A, Wang J, Yu Z, Liu J, Motaleb MA. 2016. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol 102:336–348. doi: 10.1111/mmi.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Partridge JD, Nieto V, Harshey RM. 2015. A new player at the flagellar motor: FliL controls both motor output and bias. mBio 6:e02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YY, Belas R. 2015. Loss of FliL alters Proteus mirabilis surface sensing and temperature-dependent swarming. J Bacteriol 197:159–173. doi: 10.1128/JB.02235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu S, Kumar A, Kojima S, Homma M. 2015. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol 98:101–110. doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
- 48.Khan S, Dapice M, Reese TS. 1988. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol 202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 49.Yonekura K, Maki-Yonekura S, Homma M. 2011. Structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J Bacteriol 193:3863–3870. doi: 10.1128/JB.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 51.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci U S A 110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomura A, Stemmermann GN, Chyou P-H, Kato I, Perez-Perez GI, Blaser MJ. 1991. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med 325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 54.Uzureau S, Godefroid M, Deschamps C, Lemaire J, De Bolle X, Letesson J. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J Bacteriol 189:6035–6047. doi: 10.1128/JB.00265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raddi G, Morado DR, Yan J, Haake DA, Yang XF, Liu J. 2012. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol 194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. 2006. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol 62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 57.Beeby M, Ribardo MA, Brennan CA, Ruby EG, Jensen GJ, Hendrixson DR. 2016. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci U S A 113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braun TF, Blair DF. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H(+) channels in the stator complex. Biochemistry 40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 59.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180:2729–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato K, Homma M. 2000. Functional reconstitution of the Na(+)-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275:5718–5722. doi: 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 61.Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Franco AT, Friedman DB, Nagy TA, Romero-Gallo J, Krishna U, Kendall A, Israel DA, Tegtmeyer N, Washington MK, Peek RM Jr. 2009. Delineation of a carcinogenic Helicobacter pylori proteome. Mol Cell Proteomics 8:1947–1958. doi: 10.1074/mcp.M900139-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 64.Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J. 2015. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci U S A 112:1047–1052. doi: 10.1073/pnas.1411610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 66.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. 2009. CTF determination and correction for low dose tomographic tilt series. J Struct Biol 168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agulleiro JI, Fernandez JJ. 2011. Fast tomographic reconstruction on multicore computers. Bioinformatics 27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 69.Winkler H, Zhu P, Liu J, Ye F, Roux KH, Taylor KA. 2009. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol 165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winkler H. 2007. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol 157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 71.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 72.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. 2010. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol 170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.