Abstract
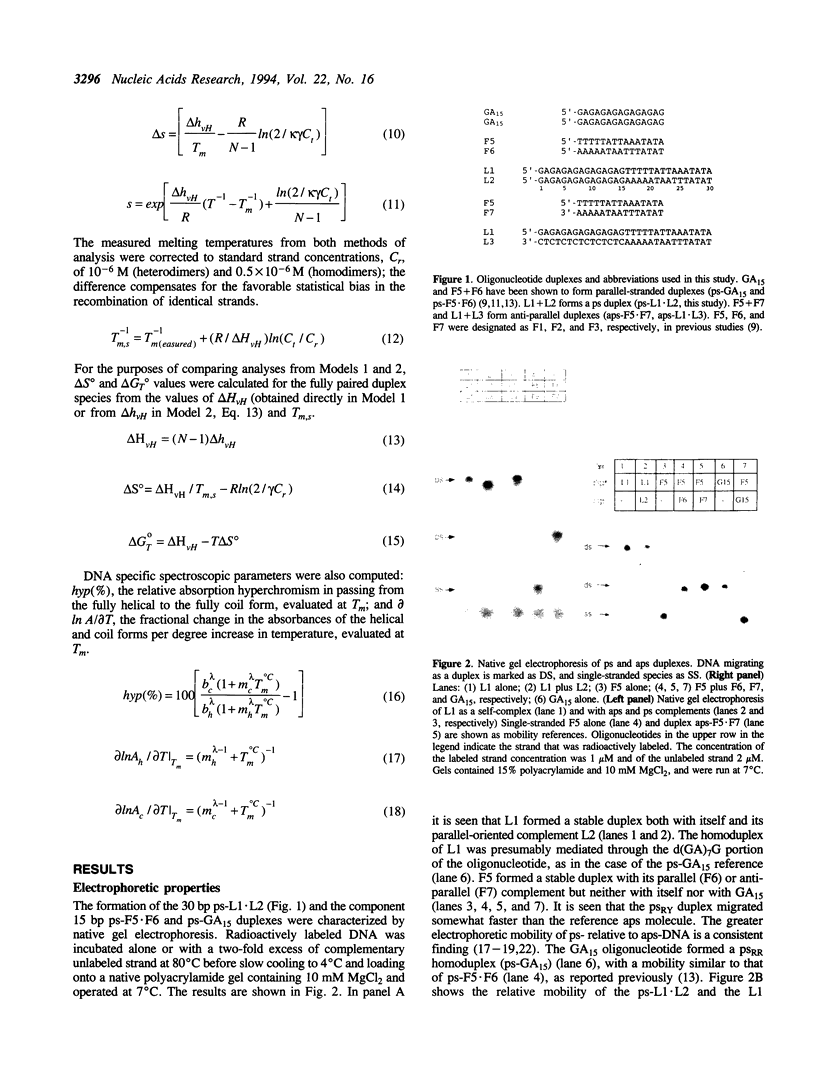
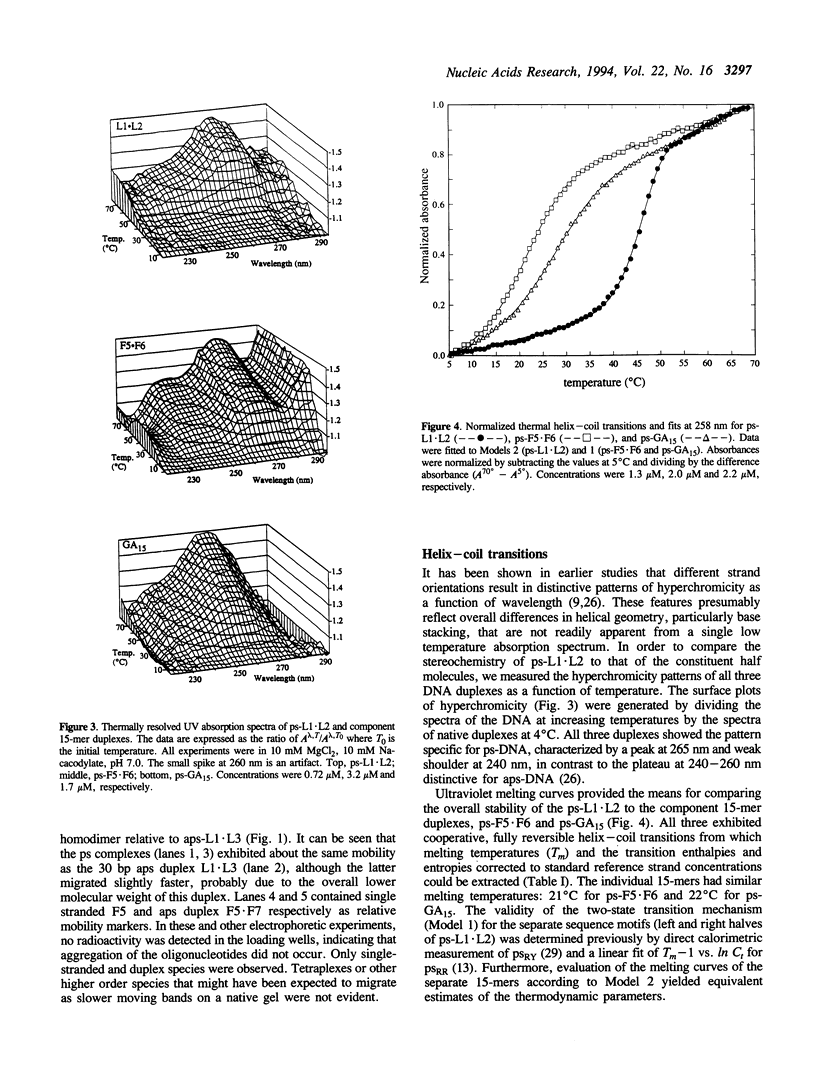
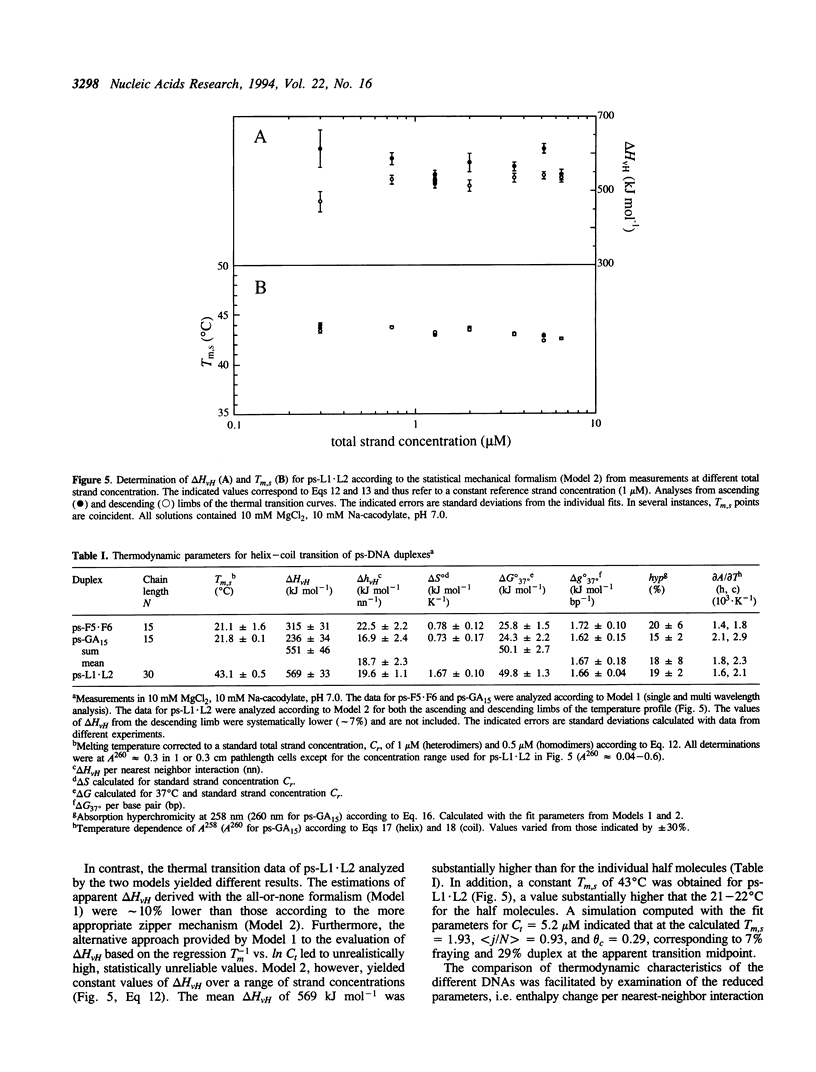
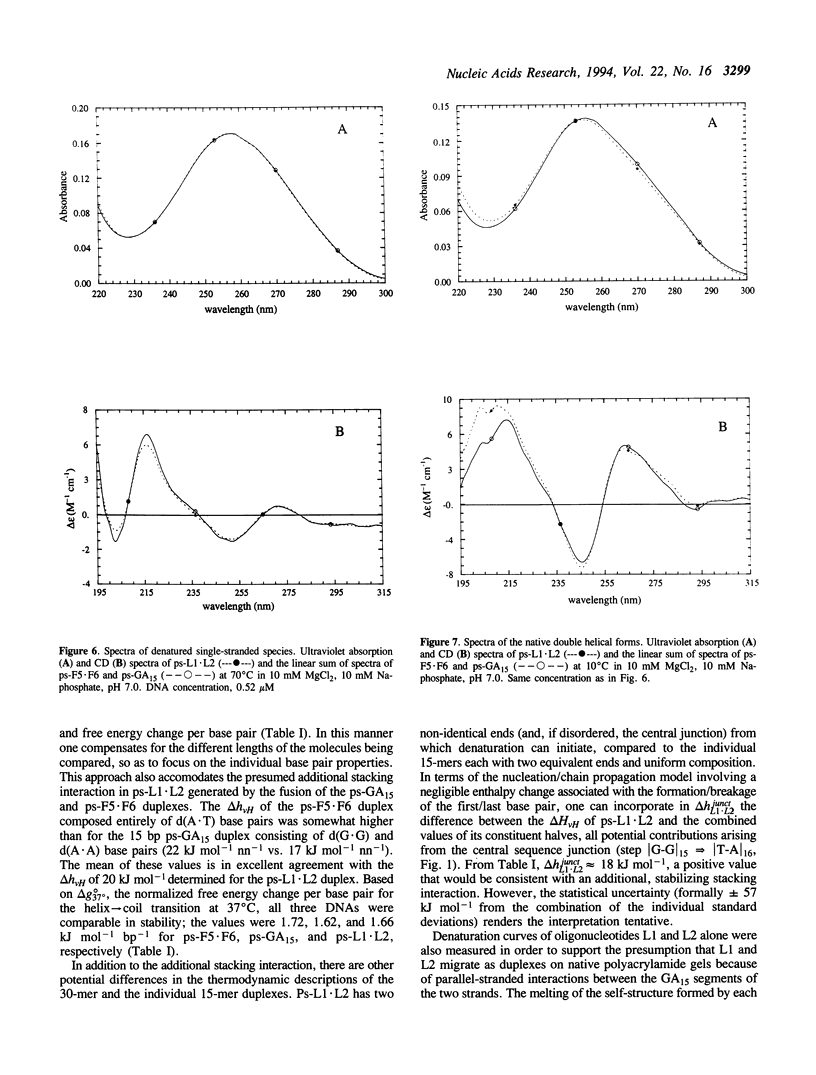
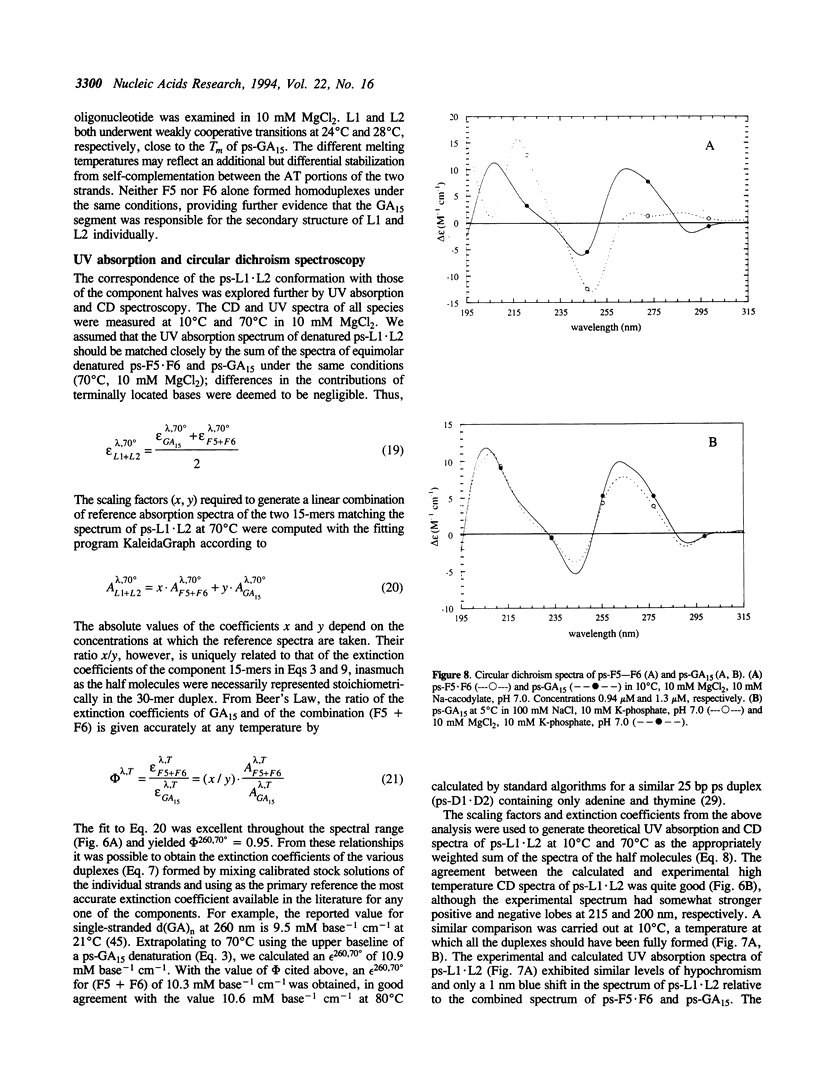
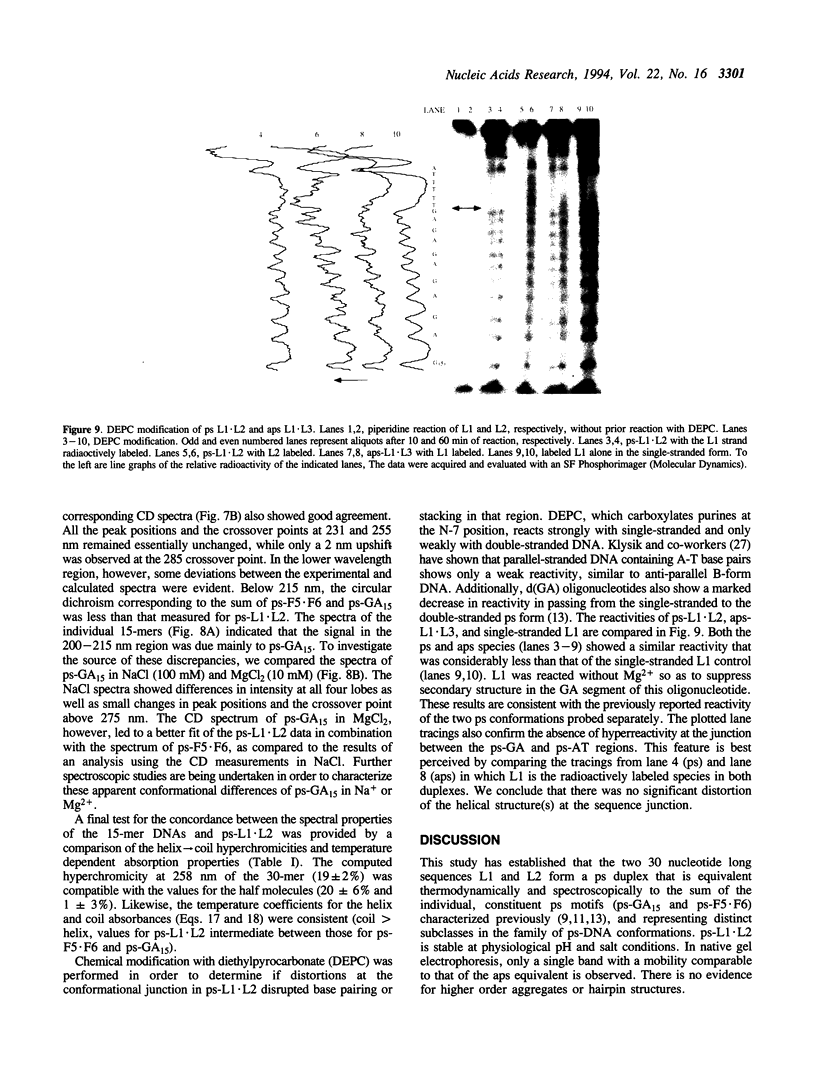
A 30 base pair parallel-stranded (ps) duplex ps-L1.L2 composed of two adjoined purine-purine and purine-pyrimidine sequence blocks has been characterized thermodynamically and spectroscopically. The 5'-terminal 15 residues in both strands ('left-half') consisted of the alternating d(GA)7G sequence that forms a ps homoduplex secondary structure stabilized by d(G.G) and d(A.A) base pairs. The 3'-terminal 15 positions of the sequence ('right-half') were combinations of A and T with complementary reverse Watson-Crick d(A.T) base pairing between the two strands. The characteristics of the full length duplex were compared to those of the constituent left and right halves in order to determine the compatibility of the two ps helical forms. The thermal denaturation curves and hyperchromicity profiles of all three duplexes determined by UV absorption spectroscopy were characteristic of ps-DNA, in accordance with previous studies. The thermodynamic properties of the 30 bp duplex corresponded within experimental error to the linear combination of the two 15-mers. Thus, the Tm and delta HvH of ps-L1.L2 in 10 mM MgCl2, derived from analyses according to a statistical mechanical formulation for the helix-coil transition, were 43 degrees C and 569 kJ mol-1, compared to 21 degrees C, 315 kJ mol-1 (ps-F5.F6) and 22 degrees C, 236 kJ mol-1 (ps-GA15). The UV absorption and CD spectra of ps-L1.L2 and the individual 15-mer ps motifs were also compared quantitatively. The sums of the two constituent native spectra (left+right halves) accurately matched that of the 30 bp duplex, with only small deviations in the 195-215 nm (CD) and 220-240 nm (absorption) regions. Based on analysis by native gel electrophoresis, the sequences studied formed duplex structures exclusively; there were no indications of higher order species. Chemical modification with diethyl pyrocarbonate showed no hyperreactivity of the junctional bases, indicating a smooth transition between the two parallel-stranded conformations. We conclude that under given salt conditions, oligonucleotides with normal primary chemical structures can readily form a parallel-stranded double helix based on blocks of very disparate non-canonical purine-purine and purine-pyrimidine base pairs and without perceptible destabilization at the junction. There are biological implications of these findings in relation to genetic structure and expression.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Murchie A. I., Lilley D. M. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T). Nature. 1992 Nov 19;360(6401):280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- Applequist J., Damle V. Theory of the effects of concentration and chain length on helix-coil equilibria in two-stranded nucleic acids. J Chem Phys. 1963 Nov 15;39(10):2719–2721. doi: 10.1063/1.1734089. [DOI] [PubMed] [Google Scholar]
- Awang G., Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993 Oct 26;32(42):11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinnaya N. G., Fresco J. R. Single-stranded nucleic acid helical secondary structure stabilized by ionic bonds: d(A(+)-G)10. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9242–9246. doi: 10.1073/pnas.89.19.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche H., Akhebat A., Taillandier E., Rippe K., Jovin T. M. Structure and drug interactions of parallel-stranded DNA studied by infrared spectroscopy and fluorescence. Nucleic Acids Res. 1993 Nov 11;21(22):5085–5091. doi: 10.1093/nar/21.22.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., Pon R. T., van de Sande J. H. Length-dependent formation of parallel-stranded DNA in alternating AT segments. Biochemistry. 1990 Oct 9;29(40):9426–9432. doi: 10.1021/bi00492a017. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Kalisch B. W., van de Sande J. H. Relative stability of parallel- and antiparallel-stranded duplex DNA. Biochemistry. 1988 Nov 1;27(22):8302–8306. doi: 10.1021/bi00422a002. [DOI] [PubMed] [Google Scholar]
- Huertas D., Bellsolell L., Casasnovas J. M., Coll M., Azorín F. Alternating d(GA)n DNA sequences form antiparallel stranded homoduplexes stabilized by the formation of G.A base pairs. EMBO J. 1993 Oct;12(10):4029–4038. doi: 10.1002/j.1460-2075.1993.tb06081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il'ychova I. A., Lysov YuP, Chernyi A. A., Shchyolkina A. K., Gottikh B. P., Florentiev V. A. Parallel double helices of DNA. Conformational analysis of regular helices with the second order symmetry axis. J Biomol Struct Dyn. 1990 Feb;7(4):879–897. doi: 10.1080/07391102.1990.10508530. [DOI] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Reactivity of parallel-stranded DNA to chemical modification reagents. Biochemistry. 1990 Oct 23;29(42):9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- Kung H. J., Hu S., Bender W., Bailey J. M., Davidson N., Nicolson M. O., McAllister R. M. RD-114, baboon, and woolly monkey viral RNA's compared in size and structure. Cell. 1976 Apr;7(4):609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- LANGRIDGE R., RICH A. Molecular structure of helical polycytidylic acid. Nature. 1963 May 25;198:725–728. doi: 10.1038/198725a0. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. The stability of polypurine tetraplexes in the presence of mono- and divalent cations. Nucleic Acids Res. 1990 Oct 25;18(20):6057–6060. doi: 10.1093/nar/18.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longfellow C. E., Kierzek R., Turner D. H. Thermodynamic and spectroscopic study of bulge loops in oligoribonucleotides. Biochemistry. 1990 Jan 9;29(1):278–285. doi: 10.1021/bi00453a038. [DOI] [PubMed] [Google Scholar]
- Luo J., Sarma M. H., Yuan R. D., Sarma R. H. NMR study of self-paired parallel duplex of d(AAAAACCCCC) in solution. FEBS Lett. 1992 Jul 20;306(2-3):223–228. doi: 10.1016/0014-5793(92)81005-7. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Breslauer K. J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987 Sep;26(9):1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- Otto C., Thomas G. A., Rippe K., Jovin T. M., Peticolas W. L. The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson-Crick. Biochemistry. 1991 Mar 26;30(12):3062–3069. doi: 10.1021/bi00226a012. [DOI] [PubMed] [Google Scholar]
- Pattabiraman N. Can the double helix be parallel? Biopolymers. 1986 Sep;25(9):1603–1606. doi: 10.1002/bip.360250903. [DOI] [PubMed] [Google Scholar]
- Petersheim M., Turner D. H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983 Jan 18;22(2):256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Ramsing N. B., Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988 Jul 25;16(14A):6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsing N. B., Rippe K., Jovin T. M. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989 Nov 28;28(24):9528–9535. doi: 10.1021/bi00450a042. [DOI] [PubMed] [Google Scholar]
- Rippe K., Fritsch V., Westhof E., Jovin T. M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992 Oct;11(10):3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Parallel-stranded duplex DNA. Methods Enzymol. 1992;211:199–220. doi: 10.1016/0076-6879(92)11013-9. [DOI] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Substrate properties of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9542–9549. doi: 10.1021/bi00450a044. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Klement R., Jovin T. M. A parallel stranded linear DNA duplex incorporating dG.dC base pairs. J Biomol Struct Dyn. 1990 Jun;7(6):1199–1209. doi: 10.1080/07391102.1990.10508559. [DOI] [PubMed] [Google Scholar]
- Robinson H., van der Marel G. A., van Boom J. H., Wang A. H. Unusual DNA conformation at low pH revealed by NMR: parallel-stranded DNA duplex with homo base pairs. Biochemistry. 1992 Nov 3;31(43):10510–10517. doi: 10.1021/bi00158a014. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Luo J., Umemoto K., Yuan R. D., Sarma R. H. Tetraplex formation of d(GGGGGTTTTT): 1H NMR study in solution. J Biomol Struct Dyn. 1992 Jun;9(6):1131–1153. doi: 10.1080/07391102.1992.10507984. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by d(TA) oligomers. II. Analysis of the helix-coli transitions of linear and circular oligomers. J Mol Biol. 1970 Feb 28;48(1):145–171. doi: 10.1016/0022-2836(70)90225-1. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988 Jul 28;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Shchyolkina A. K., Lysov YuP, Il'ichova I. A., Chernyi A. A., Golova YuB, Chernov B. K., Gottikh B. P., Florentiev V. L. Parallel stranded DNA with AT base pairing. FEBS Lett. 1989 Feb 13;244(1):39–42. doi: 10.1016/0014-5793(89)81157-3. [DOI] [PubMed] [Google Scholar]
- Sundquist W. I., Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov N. A., Chernov B. K., Golova Y. B., Nechipurenko Y. D. Parallel DNA: generation of a duplex between two Drosophila sequences in vitro. FEBS Lett. 1989 Nov 6;257(2):415–418. doi: 10.1016/0014-5793(89)81585-6. [DOI] [PubMed] [Google Scholar]
- Tchurikov N. A., Shchyolkina A. K., Borissova O. F., Chernov B. K. Southern molecular hybridization experiments with parallel complementary DNA probes. FEBS Lett. 1992 Feb 10;297(3):233–236. doi: 10.1016/0014-5793(92)80545-r. [DOI] [PubMed] [Google Scholar]
- Zhou N., Germann M. W., van de Sande J. H., Pattabiraman N., Vogel H. J. Solution structure of the parallel-stranded hairpin d(T8<text text>C4A8) as determined by two-dimensional NMR. Biochemistry. 1993 Jan 19;32(2):646–656. doi: 10.1021/bi00053a033. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Cohen G. H., Davies D. R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol. 1975 Feb 25;92(2):181–192. doi: 10.1016/0022-2836(75)90222-3. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Ramsing N. B., Germann M. W., Elhorst W., Kalisch B. W., von Kitzing E., Pon R. T., Clegg R. C., Jovin T. M. Parallel stranded DNA. Science. 1988 Jul 29;241(4865):551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]




