Abstract
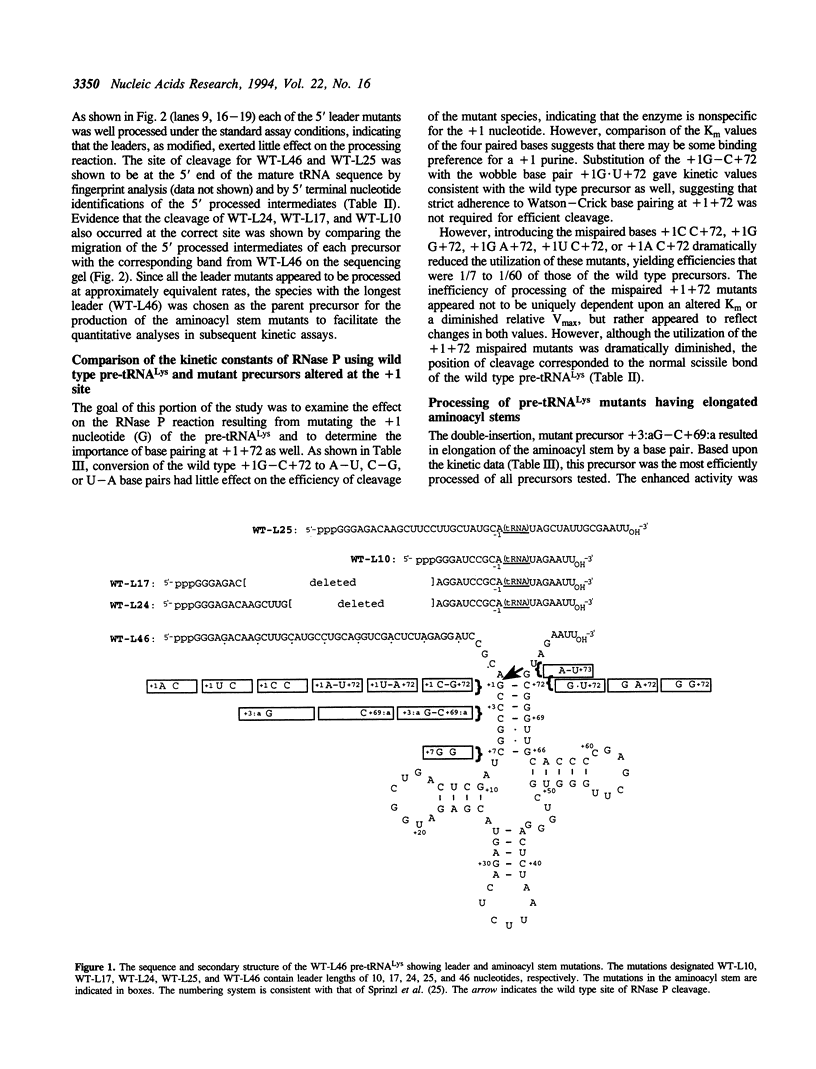
The 5' processing of rat pre-tRNA(Lys) and a series of mutant derivatives by rat cytosolic RNase P was examined. In standard, non-kinetic assays, mutant precursors synthesized in vitro with 5' leader sequences of 10, 17, 24, 25, and 46 nucleotides were processed to approximately equal levels and yielded precisely cleaved 5' processed intermediates with the normal 7-base pair aminoacyl stems. The construct containing the tRNA(Lys) with the 46-nucleotide leader was modified by PCR to give a series of pre-tRNA(Lys) mutants designed to measure the effect on processing by (1) substituting the nucleotide at the +1 position, (2) pairing and unpairing the +1 and +72 bases, (3) elongating the aminoacyl stem, and (4) disrupting the helix of the aminoacyl stem. Comparative kinetic analyses revealed that changing the wild type +1G to A, C, or U was well tolerated by the RNase P provided that compensatory changes at +72 created a base pair or a G.U noncanonical pair. Mutants with elongated aminoacyl stems that were produced either by inserting an additional base pair at +3:a + 69:a or by pairing the -1A with a +73U, were processed to yield 7-base pair aminoacyl stems, but with different efficiencies. The efficiency seen with the double insertion mutant was higher than even the wild type precursor, but the -1A-U + 73 mutant was a relatively poor substrate. Disrupting the aminoacyl stem helix by introducing a +7G G + 66 mispairing or by inserting a single G at the +3:a position dramatically reduced the processing efficiency, although the position of cleavage occurred precisely at the wild type cleavage site. However, the single insertion of a C at the +69:a position resulted in an efficiently cleaved precursor, but permitted a minor, secondary cleavage within the leader between the -6 and -5 nucleotides in addition to the dominant wild type scission.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Altman S. Ribonuclease P. Postscript. J Biol Chem. 1990 Nov 25;265(33):20053–20056. [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. RNA fingerprinting. Methods Enzymol. 1989;180:130–154. doi: 10.1016/0076-6879(89)80098-9. [DOI] [PubMed] [Google Scholar]
- Brown J. W., Haas E. S., Pace N. R. Characterization of ribonuclease P RNAs from thermophilic bacteria. Nucleic Acids Res. 1993 Feb 11;21(3):671–679. doi: 10.1093/nar/21.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara G., Calandra P., Fruscoloni P., Doria M., Tocchini-Valentini G. P. Site selection by Xenopus laevis RNAase P. Cell. 1989 Jul 14;58(1):37–45. doi: 10.1016/0092-8674(89)90400-5. [DOI] [PubMed] [Google Scholar]
- Darr S. C., Brown J. W., Pace N. R. The varieties of ribonuclease P. Trends Biochem Sci. 1992 May;17(5):178–182. doi: 10.1016/0968-0004(92)90262-8. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. Processing of tRNA in prokaryotes and eukaryotes. CRC Crit Rev Biochem. 1984;17(1):45–71. doi: 10.3109/10409238409110269. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drainas D., Zimmerly S., Willis I., Söll D. Substrate structural requirements of Schizosaccharomyces pombe RNase P. FEBS Lett. 1989 Jul 17;251(1-2):84–88. doi: 10.1016/0014-5793(89)81433-4. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Gegenheimer P., Abelson J. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem. 1985 Jan 25;260(2):1271–1279. [PubMed] [Google Scholar]
- Hanic-Joyce P. J., Gray M. W. Processing of transfer RNA precursors in a wheat mitochondrial extract. J Biol Chem. 1990 Aug 15;265(23):13782–13791. [PubMed] [Google Scholar]
- Holm P. S., Krupp G. The acceptor stem in pre-tRNAs determines the cleavage specificity of RNase P. Nucleic Acids Res. 1992 Feb 11;20(3):421–423. doi: 10.1093/nar/20.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayanthi G. P., Van Tuyle G. C. Characterization of ribonuclease P isolated from rat liver cytosol. Arch Biochem Biophys. 1992 Jul;296(1):264–270. doi: 10.1016/0003-9861(92)90571-d. [DOI] [PubMed] [Google Scholar]
- Kirsebom L. A., Svärd S. G. The kinetics and specificity of cleavage by RNase P is mainly dependent on the structure of the amino acid acceptor stem. Nucleic Acids Res. 1992 Feb 11;20(3):425–432. doi: 10.1093/nar/20.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manam S., Van Tuyle G. C. Separation and characterization of 5'- and 3'-tRNA processing nucleases from rat liver mitochondria. J Biol Chem. 1987 Jul 25;262(21):10272–10279. [PubMed] [Google Scholar]
- Marchfelder A., Brennicke A. Plant mitochondrial RNase P and E. coli RNase P have different substrate specificities. Biochem Mol Biol Int. 1993 Mar;29(4):621–633. [PubMed] [Google Scholar]
- Marchfelder A., Schuster W., Brennicke A. In vitro processing of mitochondrial and plastid derived tRNA precursors in a plant mitochondrial extract. Nucleic Acids Res. 1990 Mar 25;18(6):1401–1406. doi: 10.1093/nar/18.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Guerrier-Takada C., Altman S. Model substrates for an RNA enzyme. Science. 1987 Oct 23;238(4826):527–530. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Nemoto F., Okazaki T., Mizushima H., Müller W. E., Kuchino Y. Nucleotide sequence of the human tRNA(UUGGln) gene. Nucleic Acids Res. 1991 May 25;19(10):2779–2779. doi: 10.1093/nar/19.10.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols M., Söll D., Willis I. Yeast RNase P: catalytic activity and substrate binding are separate functions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1379–1383. doi: 10.1073/pnas.85.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson S. V., Magnusson G. Sealing of gaps in duplex DNA by T4 DNA ligase. Nucleic Acids Res. 1982 Mar 11;10(5):1425–1437. doi: 10.1093/nar/10.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Smith D. Ribonuclease P: function and variation. J Biol Chem. 1990 Mar 5;265(7):3587–3590. [PubMed] [Google Scholar]
- Pirtle R. M., Pirtle I. L., Inouye M. Messenger ribonucleic acid of the lipoprotein of the Escherichia coli outer membrane. I. Nucleotide sequence at the 3' terminus and sequences of oligonucleotides derived from complete digests of the mRNA. J Biol Chem. 1980 Jan 10;255(1):199–209. [PubMed] [Google Scholar]
- Randerath K., Gupta R. C., Randerath E. 3H and 32P derivative methods for base composition and sequence analysis of RNA. Methods Enzymol. 1980;65(1):638–680. doi: 10.1016/s0076-6879(80)65065-4. [DOI] [PubMed] [Google Scholar]
- Roy K. L., Cooke H., Buckland R. Nucleotide sequence of a segment of human DNA containing the three tRNA genes. Nucleic Acids Res. 1982 Nov 25;10(22):7313–7322. doi: 10.1093/nar/10.22.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T., Nishizawa R., Matsuda K., Taya Y., Nishimura S. A rat tRNA gene cluster containing the genes for tRNAPro and tRNALys. Analysis of nucleotide sequences of the genes and the surrounding regions. Nucleic Acids Res. 1982 Oct 25;10(20):6411–6419. doi: 10.1093/nar/10.20.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svärd S. G., Kirsebom L. A. Several regions of a tRNA precursor determine the Escherichia coli RNase P cleavage site. J Mol Biol. 1992 Oct 20;227(4):1019–1031. doi: 10.1016/0022-2836(92)90518-o. [DOI] [PubMed] [Google Scholar]
- Thurlow D. L., Shilowski D., Marsh T. L. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991 Feb 25;19(4):885–891. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILKINSON G. N. Statistical estimations in enzyme kinetics. Biochem J. 1961 Aug;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]