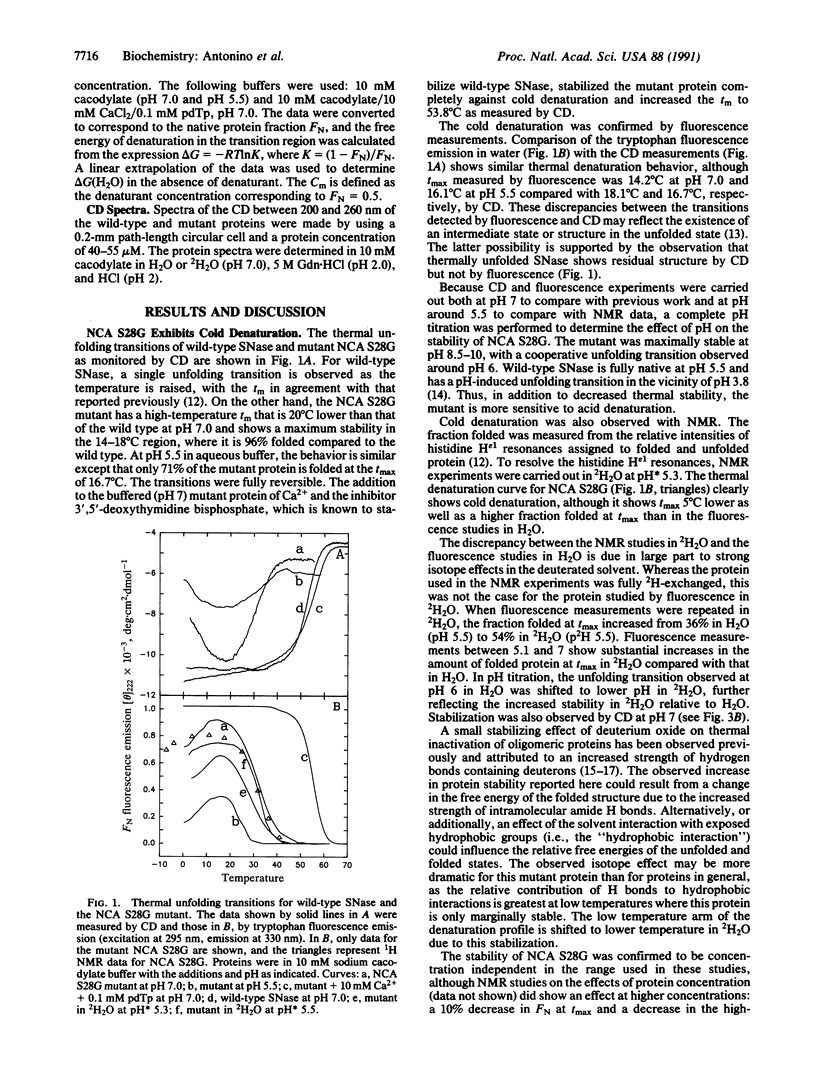
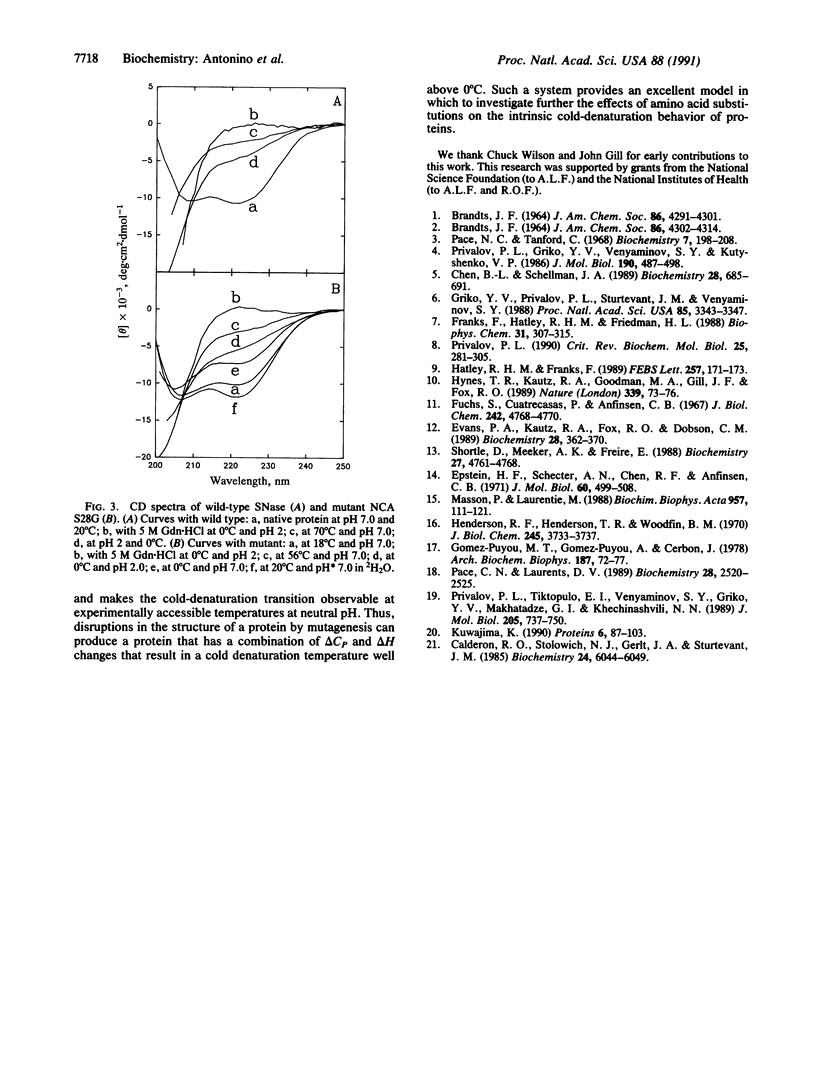
Abstract
Cold denaturation is now recognized as a general property of proteins but has been observed only under destabilizing conditions, such as moderate denaturant concentration or low pH. By destabilizing the protein using site-directed mutagenesis, we have observed cold denaturation at pH 7.0 in the absence of denaturants in a mutant of staphylococcal nuclease, which we call NCA S28G for a hybrid protein between staphylococcal nuclease and concanavalin A in which there is the point mutation Ser-28----Gly. The temperature of maximum stability (tmax) as determined by circular dichroism (CD) was 18.1 degrees C, and the midpoints of the thermal unfolding transitions (tm) were 0.6 degrees C and 30.0 degrees C. These values may be compared with the tm of 52.5 degrees C for wild-type staphylococcal nuclease, for which no cold denaturation was observed under these conditions. When the stability of the mutant was examined in 2H2O by NMR, CD, or fluorescence, a substantial increase in the amount of folded protein at the tmax was noted as well as a decrease in tmax, reflecting increased stability.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calderon R. O., Stolowich N. J., Gerlt J. A., Sturtevant J. M. Thermal denaturation of staphylococcal nuclease. Biochemistry. 1985 Oct 22;24(22):6044–6049. doi: 10.1021/bi00343a004. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Schellman J. A. Low-temperature unfolding of a mutant of phage T4 lysozyme. 1. Equilibrium studies. Biochemistry. 1989 Jan 24;28(2):685–691. doi: 10.1021/bi00428a041. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Schechter A. N., Chen R. F., Anfinsen C. B. Folding of staphylococcal nuclease: kinetic studies of two processes in acid renaturation. J Mol Biol. 1971 Sep 28;60(3):499–508. doi: 10.1016/0022-2836(71)90184-7. [DOI] [PubMed] [Google Scholar]
- Evans P. A., Kautz R. A., Fox R. O., Dobson C. M. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry. 1989 Jan 10;28(1):362–370. doi: 10.1021/bi00427a050. [DOI] [PubMed] [Google Scholar]
- Franks F., Hatley R. H., Friedman H. L. The thermodynamics of protein stability. Cold destabilization as a general phenomenon. Biophys Chem. 1988 Sep;31(3):307–315. doi: 10.1016/0301-4622(88)80037-1. [DOI] [PubMed] [Google Scholar]
- Fuchs S., Cuatrecasas P., Anfinsen C. B. An improved method for the purification of staphylococcal nuclease. J Biol Chem. 1967 Oct 25;242(20):4768–4770. [PubMed] [Google Scholar]
- Griko Y. V., Privalov P. L., Sturtevant J. M., Venyaminov SYu Cold denaturation of staphylococcal nuclease. Proc Natl Acad Sci U S A. 1988 May;85(10):3343–3347. doi: 10.1073/pnas.85.10.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatley R. H., Franks F. The cold-induced denaturation of lactate dehydrogenase at sub-zero temperatures in the absence of perturbants. FEBS Lett. 1989 Oct 23;257(1):171–173. doi: 10.1016/0014-5793(89)81813-7. [DOI] [PubMed] [Google Scholar]
- Hynes T. R., Kautz R. A., Goodman M. A., Gill J. F., Fox R. O. Transfer of a beta-turn structure to a new protein context. Nature. 1989 May 4;339(6219):73–76. doi: 10.1038/339073a0. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Masson P., Laurentie M. Stability of butyrylcholinesterase: thermal inactivation in water and deuterium oxide. Biochim Biophys Acta. 1988 Nov 2;957(1):111–121. doi: 10.1016/0167-4838(88)90163-x. [DOI] [PubMed] [Google Scholar]
- Pace C. N., Laurents D. V. A new method for determining the heat capacity change for protein folding. Biochemistry. 1989 Mar 21;28(6):2520–2525. doi: 10.1021/bi00432a026. [DOI] [PubMed] [Google Scholar]
- Pace N. C., Tanford C. Thermodynamics of the unfolding of beta-lactoglobulin A in aqueous urea solutions between 5 and 55 degrees. Biochemistry. 1968 Jan;7(1):198–208. doi: 10.1021/bi00841a025. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Cold denaturation of proteins. Crit Rev Biochem Mol Biol. 1990;25(4):281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Griko YuV, Venyaminov SYu, Kutyshenko V. P. Cold denaturation of myoglobin. J Mol Biol. 1986 Aug 5;190(3):487–498. doi: 10.1016/0022-2836(86)90017-3. [DOI] [PubMed] [Google Scholar]
- Privalov P. L., Tiktopulo E. I., Venyaminov SYu, Griko YuV, Makhatadze G. I., Khechinashvili N. N. Heat capacity and conformation of proteins in the denatured state. J Mol Biol. 1989 Feb 20;205(4):737–750. doi: 10.1016/0022-2836(89)90318-5. [DOI] [PubMed] [Google Scholar]
- Shortle D., Meeker A. K., Freire E. Stability mutants of staphylococcal nuclease: large compensating enthalpy-entropy changes for the reversible denaturation reaction. Biochemistry. 1988 Jun 28;27(13):4761–4768. doi: 10.1021/bi00413a027. [DOI] [PubMed] [Google Scholar]
- Woodfin B. M., Henderson R. F., Henderson T. R. Effects of D2O on the association-dissociation equilibrium in subunit proteins. J Biol Chem. 1970 Aug 10;245(15):3733–3737. [PubMed] [Google Scholar]
- de Gómez-Puyou M. T., Gómez-Puyou A., Cerbón J. Increased conformational stability of mitochondrial soluble ATPase (F1) by substitution of H2O for D2O. Arch Biochem Biophys. 1978 Apr 15;187(1):72–77. doi: 10.1016/0003-9861(78)90007-3. [DOI] [PubMed] [Google Scholar]


