Abstract
HIV-1-specific broadly neutralizing antibodies (bNAbs) can protect rhesus monkeys against simian-human immunodeficiency virus (SHIV) challenge. However, the site of antibody interception of virus and the mechanism of antibody-mediated protection remain unclear. We administered a fully protective dose of the bNAb PGT121 to rhesus monkeys and challenged them intravaginally with SHIV-SF162P3. In PGT121 treated animals, we detected low levels of viral RNA and viral DNA in distal tissues for several days following challenge. Viral RNA positive tissues showed transcriptomic changes indicative of innate immune activation, and cells from these tissues initiated infection following adoptive transfer into naïve hosts. These data demonstrate that bNAb mediated protection against a mucosal virus challenge can involve clearance of infectious virus in distal tissues.
Multiple bNAbs targeting HIV-1 Env have demonstrated complete protection against SHIV challenges in rhesus monkeys (1–6) and are currently being evaluated as a strategy to prevent HIV-1 acquisition in humans. However, the anatomic sites and mechanisms of antibody-mediated protection have not been fully elucidated. In particular, it remains unclear whether bNAbs completely block virus at the local portal of entry following mucosal virus challenge. To address this question, we performed comprehensive necropsies following intravaginal SHIV-SF162P3 challenge of rhesus monkeys that received a fully protective dose of the potent V3 glycan-dependent bNAb PGT121 (7).
We verified the protective efficacy of PGT121 against intravaginal challenge with SHIV-SF162P3 (8–10) in a preliminary study in 12 female rhesus monkeys (M. mulatta). Consistent with previously published data (1), an intravenous infusion of 2 mg/kg PGT121 afforded complete protection against intravaginal challenge with 5×104 TCID50 SHIV-SF162P3, as evidenced by no detectable plasma viral RNA for over 6 months following challenge (Fig. S1). To evaluate the mechanism of this observed protection, 24 female rhesus monkeys received 2 mg/kg PGT121 (N=12) or an isotype matched sham control antibody (N=12) by the intravenous route on day −1 and were challenged intravaginally with 5×104 TCID50 SHIV-SF162P3 on day 0. Serum PGT121 levels were 20–50 μg/ml on the day of challenge in all animals. We performed serial necropsies on day 1 (N=4), day 3 (N=4), day 7 (N=10), and day 10 (N=6) following challenge for comprehensive assessments of virologic, immunologic, and transcriptomic profiles in multiple tissues in each animal (11).
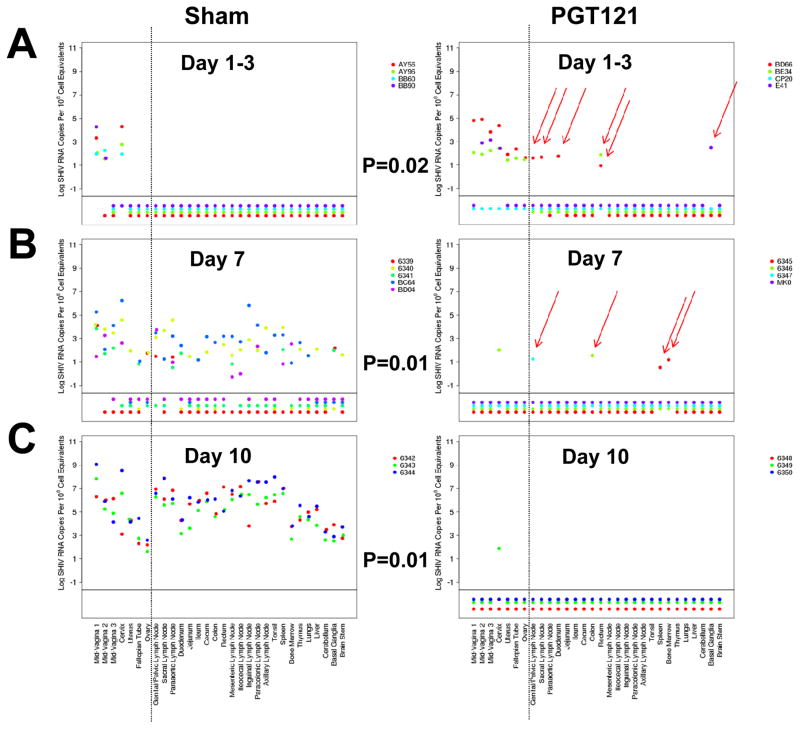
Tissue viral RNA levels were quantitated using an ultrasensitive nested RT-PCR assay (12) assessing 30 independent tissues from each animal from the female reproductive tract, draining lymph nodes, gastrointestinal tract, distal lymph nodes, tonsil, spleen, bone marrow, thymus, lung, liver, and central nervous system. In 75% (3 of 4) of PGT121 treated animals on day 1 and day 3 following SHIV challenge, we observed low levels of viral RNA in at least one tissue distal to the female reproductive tract, primarily in draining lymph nodes and gastrointestinal tissue (Fig. 1A). Viral RNA was observed more frequently in PGT121 treated animals than in sham controls at these timepoints (Fig. 1A; P=0.02, two-sided Fisher’s exact test), suggesting that the antibody may have facilitated translocation of virus across the mucosal barrier. On day 7, viral RNA distal to the female reproductive tract was still detected sporadically in 75% (3 of 4) of PGT121 treated animals, but was not detected in plasma. Viral RNA was detected far more extensively in sham controls than in PGT121 treated animals on day 7 (Fig. 1B; P=0.01). On day 10, viral RNA was not detected in any PGT121 treated animals at distal sites but was present at high levels in all tissues in the sham controls, as expected (13–15) (Fig. 1C; P=0.01).
Figure 1. Viral RNA in tissues following SHIV-SF162P3 challenge.
Tissue viral RNA (log RNA copies/108 cell equivalents) across multiple tissues at necropsy in PGT121 treated animals and sham controls on (A) day 1 (AY56, AY96, BD66, BE34) and day 3 (BB60, BB90, CP20, E41), (B) day 7, and (C) day 10 following SHIV-SF162P3 challenge. Colors on each plot reflect individual animals. Values plotted below the horizontal line indicate samples for which viral RNA was not measured above the threshold of detection. Values to the right of the vertical line reflect samples distal to the female reproductive tract. Red arrows highlight viral RNA positive distal tissues in the PGT121 treated animals. P-values reflect Fisher’s exact tests. Data are presented based on calculations normalized as log viral RNA copies/108 diploid genome equivalents of DNA; for this study the majority of the specimens analyzed contained 106–107 cell equivalents.
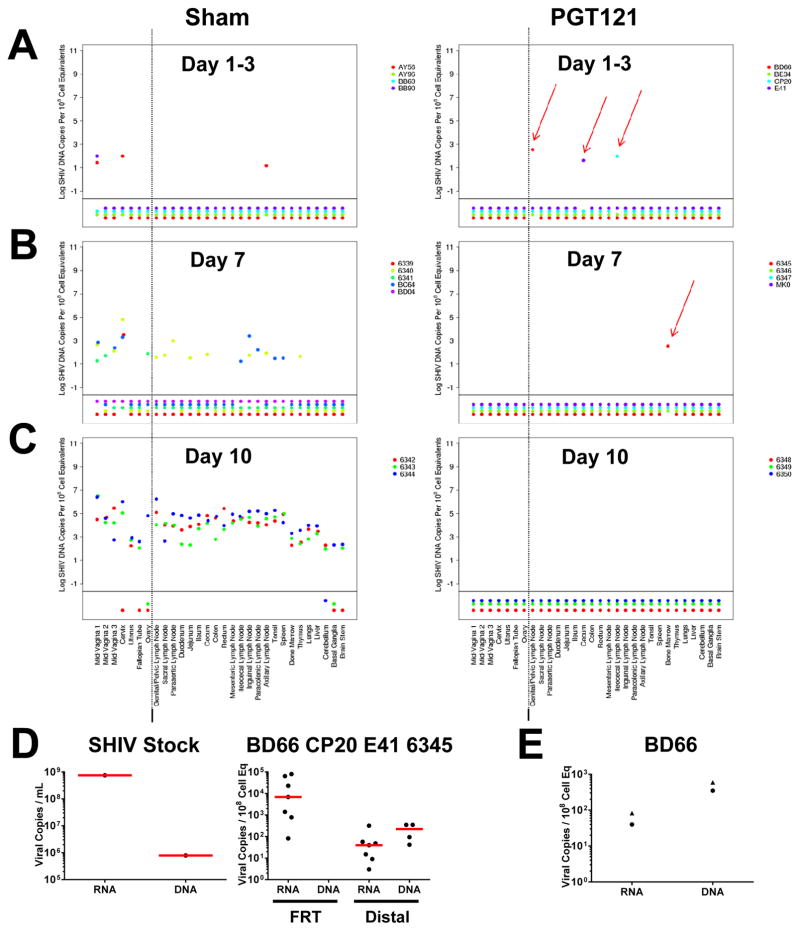
Viral DNA was also observed sporadically and at a declining frequency in PGT121 treated animals on days 1, 3, and 7 following challenge (Fig. 2A–C). In contrast, viral DNA was detected at increasing magnitude and frequency over time in the sham controls, as expected. Taken together, these data show that low but declining levels of viral RNA and viral DNA were detectable in distal tissues in PGT121 treated animals for approximately 7 days following SHIV-SF162P3 challenge but were undetectable by day 10 (Figs. 1–2, S2).
Figure 2. Viral DNA in tissues following SHIV-SF162P3 challenge.
Tissue viral DNA (log DNA copies/108 cell equivalents) across multiple tissues at necropsy in PGT121 treated animals and sham controls on (A) day 1–3, (B) day 7, and (C) day 10 following SHIV-SF162P3 challenge. Colors on each plot reflect individual animals. Values plotted below the horizontal line indicate samples for which viral DNA was not measured above the threshold of detection. Values to the right of the vertical line indicate samples distal to the female reproductive tract. Red arrows highlight viral DNA positive distal tissues in the PGT121 treated animals. Data are presented based on calculations normalized as log viral DNA copies/108 diploid genome equivalents of DNA; for this study the majority of the specimens analyzed contained 106–107 cell equivalents. (D) Viral RNA and viral DNA in the SHIV-SF162P3 stock (left) and in female reproductive tract (FRT) and distal tissues in the 4 animals that were viral DNA positive on days 1–7 (BD66, CP20, E41, 6345) (right). (E) Viral RNA and viral DNA in total cells (circles) and purified CD4+ T lymphocytes (triangles) from BD66 genital/pelvic lymph node on day 1. Data are based on analysis of 2.7×106 purified CD4+ T lymphocytes.
In our SHIV-SF162P3 challenge stock, the level of viral gag RNA (7.5×108 copies/mL) was approximately 3 logs greater than the corresponding level of viral gag DNA (7.9×105 copies/mL) (Fig. 2D). In the 4 animals that exhibited viral DNA in distal tissues on days 1–7 (BD66, CP20, E41, 6345), the median level of viral RNA (6.9×103 copies/108 cell equivalents) was similarly over 3 logs higher than the median level of viral DNA (<10 copies/108 cell equivalents) at the site of inoculation in the female reproductive tract, consistent with the notion that this virus largely represented the input challenge stock. In contrast, in distal tissues, the median level of viral RNA (4.0×101 copies/108 cell equivalents) was slightly lower than the median level of viral DNA (2.2×102 copies/108 cell equivalents) (Fig. 2D). Moreover, in the day 1 genital/pelvic lymph node from monkey BD66, viral RNA and viral DNA was detected in purified (>99%) CD4+ T lymphocytes (Fig. 2E). These data suggest that the viral DNA detected in distal tissues reflected a limited degree of new virus replication at the distal sites.
We next assessed Gag-specific CD8+ and CD4+ T lymphocyte responses in multiple tissues from each animal by multiparameter intracellular cytokine staining assays (16, 17). CD8+ T lymphocyte responses were not detected in any tissue at any timepoint in the PGT121 treated animals, but these responses were observed in the female genital tract in all sham controls on day 7 and day 10 (Figs. S3–S5). The lack of detectable T lymphocyte responses in the PGT121 treated animals indicates insufficient antigen exposure to induce measurable cellular immune responses and suggests that virus-specific T cell responses were likely not responsible for the clearance of virus in distal tissues at these early timepoints.
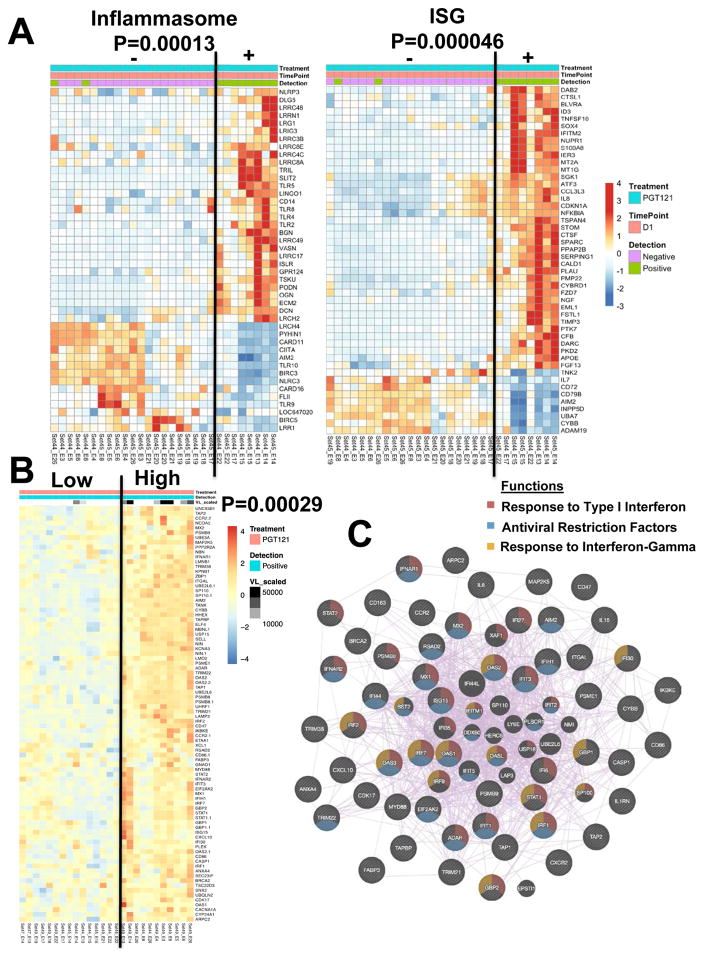
We next evaluated gene expression profiles in multiple tissues in each animal at necropsy (11, 18). A multidimensional scaling plot revealed differences between PGT121 treated animals and controls for all tissues analyzed (Fig. S6). We compared transcriptomic profiles in viral RNA positive compared with viral RNA negative tissues in the PGT121 treated animals. Inflammasome-related genes (P=0.00013; NLRP3, TLR) and interferon-stimulated genes (P=0.000046; IFITM2, IL8, S100A8) were upregulated specifically in viral RNA positive local and distal tissues from animals that received PGT121 (Figs. 3A, S7). Pathway analysis (19) confirmed upregulation of several pathways associated with host response to viruses (interferon alpha, interferon beta) and cellular activation (hypoxia, TNF signaling via NFKB; JAK STAT signaling) in these tissues (Fig. S8). Moreover, a linear regression analysis revealed a transcriptomic signature that correlated with viral RNA levels in PGT121 treated animals necropsied on days 1, 3, and 7 (Fig. 3B; P=0.00029). Network inference analysis (20, 21) revealed that this signature involved activation of innate immunity and antiviral pathways, including type 1 interferon signaling (FDR=1.64×10−46), host response to viral infection (FDR=2.85×10−29), regulation of viral replication (FDR=2.49×10−17), and response to interferon-γ (FDR=1.91×10−16) (Figs. 3C, S9).
Figure 3. Transcriptomic signature in viral RNA positive tissues from PGT121 treated animals.
(A) Heatmaps reveal inflammasome and interferon-stimulated gene (ISG) signatures in viral RNA positive compared with viral RNA negative local and distal tissues from PGT121 treated tissues on day 1 following SHIV-SF162P3 challenge. Gene expression is represented by standardized expression (Z-score) with P < 0.05. Red and blue correspond to upregulated and downregulated genes, respectively. P-values represent the significance of the two clusters using Fisher’s exact tests. 29 samples from 2 monkeys (BD66 and BE34) are represented, reflecting 10 viral RNA positive samples and 19 viral RNA negative samples. (B) Linear regression analysis of viral RNA as a continuous variable and gene expression (LIMMA t-test with P < 0.05) in viral RNA positive tissues from PGT121 treated animals reveals a transcriptomic signature that correlates with levels of viral RNA. Heatmap represents the top positively correlated genes that were part of the full regression signature. 26 samples from 4 monkeys were included in this analysis, reflecting 10 samples from day 1, 2 samples from day 3, and 14 samples from day 7. (C) Network of genes that positively correlated with levels of viral RNA in tissues from PGT121 treated animals. Nodes represent genes and purple lines represent co-expression between genes.
We also observed an increase in the number of differentially expressed total and interferon-stimulated genes over the first 7 days in viral RNA positive tissues from PGT121 treated animals (Fig. S10). By day 7, upregulation of classic antiviral restriction factors such as IRF7, TRIM5, APOBEC, and IFITM was evident in these tissues, suggesting an evolving antiviral innate response. The late expression of these antiviral restriction factors is consistent with our previous findings in SIVmac251 infection (11). The upregulated genes in viral RNA positive tissues from PGT121 treated animals also partially overlapped with a previously defined transcriptomic signature for viral replication (22) (Fig. S11).
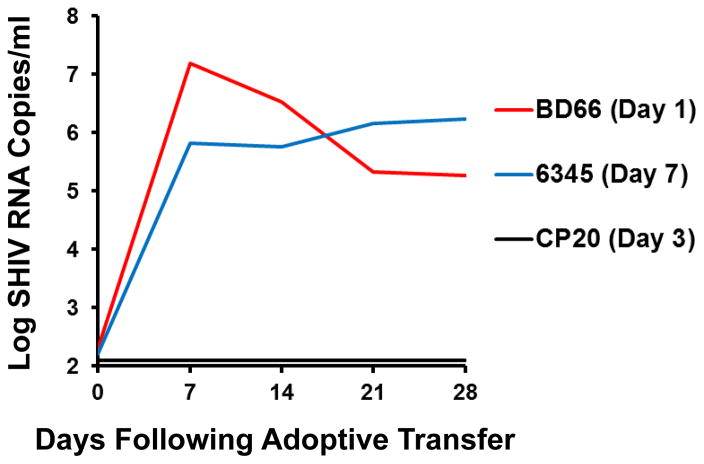
To investigate whether the low levels of detectable virus in distal tissues in PGT121 treated animals were infectious, we adoptively transferred 30 million cells from the following animals and tissues into naïve hosts by the intravenous route: BD66 (genital/pelvic lymph node; day 1), CP20 (ileocecal lymph node; day 3), and 6345 (spleen; day 7). Tissues from BD66 and 6345 were positive for both viral RNA and viral DNA, whereas tissues from CP20 were negative for viral RNA but positive for viral DNA. Adoptive transfer of cells from BD66 and 6345 into naïve hosts efficiently transferred infection, resulting in 5.8–7.1 log RNA copies/ml of replicating virus by day 7 after adoptive transfer (Fig. 4), demonstrating that the viral RNA and viral DNA detected in these samples included replication-competent infectious virus when removed from the host milieu of antibody and innate factors. Cells from CP20 did not detectably transfer infection, suggesting that the amount of virus in this sample was too low or that the viral DNA in this sample was defective and noninfectious.
Figure 4. Adoptive transfer studies.
Adoptive transfer of 30 million cells into naïve hosts by the intravenous route from tissues from BD66 (genital/pelvic lymph node; day 1), CP20 (ileocecal lymph node; day 3), and 6345 (spleen; day 7). Plasma viral RNA copies/ml are shown following adoptive transfer.
In summary, we detected low levels of viral RNA and/or viral DNA in at least one tissue distal to the female reproductive tract in 87.5% (7 of 8) of monkeys on days 1, 3, and 7 following intravaginal SHIV-SF162P3 challenge, despite receiving a fully protective dose of PGT121 (Fig. S12). Virus in distal tissues triggered transcriptomic responses involving activation of innate immunity and antiviral pathways, and a subset of these samples proved infectious following adoptive transfer into naïve hosts. These data demonstrate that PGT121 did not completely block the challenge virus at the mucosal portal of entry. Instead, some virions appeared to transit to distal tissues, where they were progressively cleared over a period of approximately 7 days, potentially involving Fc effector mechanisms such as antibody-dependent cellular cytotoxicity. The viral RNA to viral DNA ratio at distal sites (Fig. 2D), the presence of viral RNA and viral DNA in purified CD4+ T lymphocytes (Fig. 2E), and the transcriptomics signatures (Fig. S11) suggested a limited degree of new virus replication at distal sites.
Although our studies were limited to PGT121, it is likely that other bNAbs as well as vaccine-elicited antibodies that afford protection (9) also involve viral clearance in distal tissues rather than complete blockade of virus at the mucosal surface. These data emphasize the potential importance of antibodies to clear virally infected cells in tissues to block the establishment of chronic infection (9, 23–25). These data are also consistent with previous findings from our laboratory and others that the virus can disseminate rapidly and trigger inflammatory responses in host tissues following mucosal challenge (11, 26–28). Moreover, our findings are consistent with the observation that bNAbs may be able to abort initial infection in infant rhesus monkeys when administered therapeutically after SHIV challenge (29). The possibility that PGT121 may have actually facilitated virus translocation across the mucosal barrier, potentially via immune complexes captured on Fc receptor-bearing cells, warrants further investigation.
Our findings have important implications for the development of vaccines, passively transferred antibodies, and other interventions to block HIV-1 infection. In particular, our data suggest that a systemic component of an antiviral intervention may be necessary for optimal efficacy to clear early foci of disseminated virus in distal tissues. It is noteworthy that high concentrations of topically applied bNAbs are required to protect against mucosal SHIV challenge, whereas much lower serum concentrations of intravenously administered bNAbs are required for protection (30, 31). Our data also show that the initial virus that is seeded in distal tissues may still be vulnerable to immune-mediated elimination prior to the establishment of a permanent viral reservoir. Further exploitation of this vulnerability may lead to improved HIV-1 eradication as well as improved HIV-1 prevention strategies.
Supplementary Material
Acknowledgments
We thank A. Schultz, Q. Dang, M. Pensiero, M. Ferguson, F. Stephens, J. Nkolola, K. Smith, J. Smith, S. Blackmore, L. Parenteau, S. Mathew, S. Levin, C. Shaver, and Z. Pippin for generous advice, assistance, and reagents. The SIVmac239 peptides were obtained from the NIH AIDS Research and Reference Reagent Program. The data presented in this paper are tabulated in the main paper and in the supplementary materials. The authors declare no competing financial interests. We acknowledge support from the National Institutes of Health (AI060354, AI095985, AI096040, AI100645, AI100663, AI124377, HHSN261200800001E) and the Ragon Institute of MGH, MIT, and Harvard. Transcriptomics data can be accessed at GEO (GSE83702) and NCBI (17948367).
References
- 1.Moldt B, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012 Nov 13;109:18921. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shingai M, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014 Sep 22;211:2061. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mascola JR, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J Virol. 1999 May;73:4009. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000 Feb;6:207. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 5.Baba TW, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nature medicine. 2000 Feb;6:200. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 6.Hessell AJ, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009 Aug;15:951. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011 Sep 22;477:466. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch DH, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013 Oct 24;155:531. doi: 10.1016/j.cell.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barouch DH, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science. 2015 Jul 17;349:320. doi: 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch DH, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013 Nov 14;503:224. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch DH, et al. Rapid Inflammasome Activation following Mucosal SIV Infection of Rhesus Monkeys. Cell. 2016 Apr 21;165:656. doi: 10.1016/j.cell.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SG, et al. Immune clearance of highly pathogenic SIV infection. Nature. 2013 Oct 3;502:100. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005 Apr 28;434:1148. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 14.Mattapallil JJ, et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005 Apr 28;434:1093. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 15.Brenchley JM, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004 Sep 20;200:749. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barouch DH, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012 Feb 2;482:89. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, et al. Durable mucosal simian immunodeficiency virus-specific effector memory T lymphocyte responses elicited by recombinant adenovirus vectors in rhesus monkeys. J Virol. 2011 Nov;85:11007. doi: 10.1128/JVI.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukazawa Y, et al. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med. 2012 Nov;18:1673. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102:15545. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mostafavi S, Morris Q. Fast integration of heterogeneous data sources for predicting gene function with limited annotation. Bioinformatics. 2010 Jul 15;26:1759. doi: 10.1093/bioinformatics/btq262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome biology. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011 Apr 28;472:481. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007 Sep 6;449:101. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 24.Bournazos S, et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014 Sep 11;158:1243. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung AW, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015 Nov 5;163:988. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller CJ, et al. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005 Jul;79:9217. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro Dos Santos P, et al. Rapid dissemination of SIV follows multisite entry after rectal inoculation. PLoS ONE. 2011;6:e19493. doi: 10.1371/journal.pone.0019493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Gardner MB, Miller CJ. Simian immunodeficiency virus rapidly penetrates the cervicovaginal mucosa after intravaginal inoculation and infects intraepithelial dendritic cells. J Virol. 2000 Jul;74:6087. doi: 10.1128/jvi.74.13.6087-6095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hessell AJ, et al. Early short-term treatment with neutralizing human monoclonal antibodies halts SHIV infection in infant macaques. Nat Med. 2016 Apr;22:362. doi: 10.1038/nm.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burton DR, et al. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A. 2011 Jul 5;108:11181. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veazey RS, et al. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nature medicine. 2003 Mar;9:343. doi: 10.1038/nm833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.