Abstract
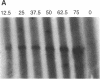
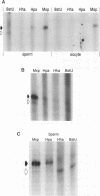
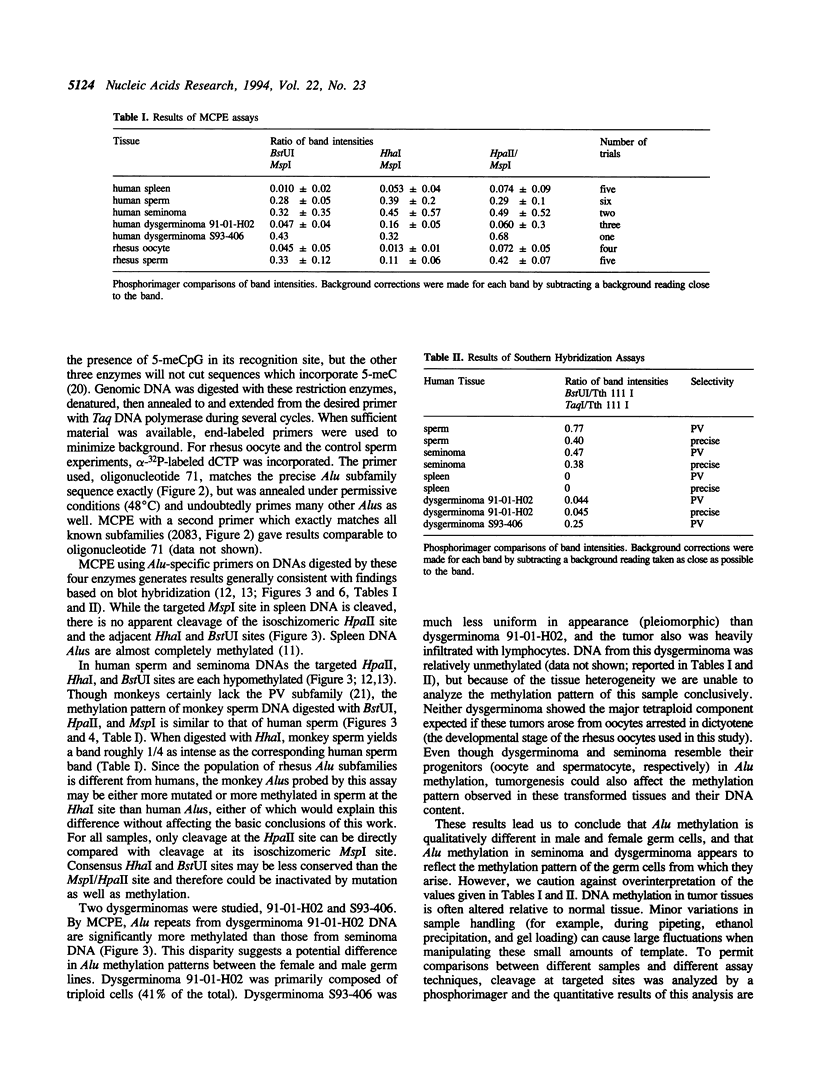
A significant fraction of Alu repeats in human sperm DNA, previously found to be unmethylated, is nearly completely methylated in DNA from many somatic tissues. A similar fraction of unmethylated Alus is observed here in sperm DNA from rhesus monkey. However, Alus are almost completely methylated at the restriction sites tested in monkey follicular oocyte DNA. The Alu methylation patterns in mature male and female monkey germ cells are consistent with Alu methylation in human germ cell tumors. Alu sequences are hypomethylated in seminoma DNAs and more methylated in a human ovarian dysgerminoma. These results contrast with methylation patterns reported for germ cell single-copy, CpG island, satellite, and L1 sequences. The function of Alu repeats is not known, but differential methylation of Alu repeats in the male and female germ lines suggests that they may serve as markers for genomic imprinting or in maintaining differences in male and female meiosis.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlow D. P. Imprinting: a gamete's point of view. Trends Genet. 1994 Jun;10(6):194–199. doi: 10.1016/0168-9525(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Batzer M. A., Hutchison C. A., 3rd, Edgell M. H. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992 Sep;8(9):307–311. doi: 10.1016/0168-9525(92)90262-3. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa M. A., Wang R. Y., Kuo K. C., Gehrke C. W., Ehrlich M. The 5-methylcytosine content of highly repeated sequences in human DNA. Nucleic Acids Res. 1983 May 25;11(10):3087–3095. doi: 10.1093/nar/11.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann-Blumberg U., Hintz M. F., Gatewood J. M., Schmid C. W. Developmental differences in methylation of human Alu repeats. Mol Cell Biol. 1993 Aug;13(8):4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett S. K., Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991 Sep;113(1):119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Jurka J., Milosavljevic A. Reconstruction and analysis of human Alu genes. J Mol Evol. 1991 Feb;32(2):105–121. doi: 10.1007/BF02515383. [DOI] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992 May;6(5):705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- Kochanek S., Renz D., Doerfler W. DNA methylation in the Alu sequences of diploid and haploid primary human cells. EMBO J. 1993 Mar;12(3):1141–1151. doi: 10.1002/j.1460-2075.1993.tb05755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeflang E. P., Liu W. M., Chesnokov I. N., Schmid C. W. Phylogenetic isolation of a human Alu founder gene: drift to new subfamily identity [corrected]. J Mol Evol. 1993 Dec;37(6):559–565. doi: 10.1007/BF00182741. [DOI] [PubMed] [Google Scholar]
- Li E., Beard C., Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993 Nov 25;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Liu W. M., Maraia R. J., Rubin C. M., Schmid C. W. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res. 1994 Mar 25;22(6):1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. M., Schmid C. W. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993 Mar 25;21(6):1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Nelson M., Raschke E., McClelland M. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1993 Jul 1;21(13):3139–3154. doi: 10.1093/nar/21.13.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J. P., Clark H. J., Chapman V. M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987 Dec;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Sanford J., Forrester L., Chapman V., Chandley A., Hastie N. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 1984 Mar 26;12(6):2823–2836. doi: 10.1093/nar/12.6.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C. W. Human Alu subfamilies and their methylation revealed by blot hybridization. Nucleic Acids Res. 1991 Oct 25;19(20):5613–5617. doi: 10.1093/nar/19.20.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid C., Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992 Dec;2(6):874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Shemer R., Kafri T., O'Connell A., Eisenberg S., Breslow J. L., Razin A. Methylation changes in the apolipoprotein AI gene during embryonic development of the mouse. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11300–11304. doi: 10.1073/pnas.88.24.11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M. A. Genomic imprinting. Silence of the genes. Nature. 1993 Nov 25;366(6453):302–303. doi: 10.1038/366302a0. [DOI] [PubMed] [Google Scholar]
- Teplitz R. L., Butler B. B., Tesluk H., Min B. H., Russell L. A., Jensen H. M., Hill L. R. Quantitative DNA patterns in human preneoplastic breast lesions. Anal Quant Cytol Histol. 1990 Apr;12(2):98–102. [PubMed] [Google Scholar]
- Varmuza S., Mann M. Genomic imprinting--defusing the ovarian time bomb. Trends Genet. 1994 Apr;10(4):118–123. doi: 10.1016/0168-9525(94)90212-7. [DOI] [PubMed] [Google Scholar]