Abstract
Volatile organic compounds (VOCs) produced by various bacteria have significant potential to enhance plant growth and to control phytopathogens. Six of the most effective antagonistic Bacillus spp. were used in this study against Ralstonia solanacearum (Rsc) TBBS1, the causal agent of bacterial wilt disease in tobacco. Bacillus amyloliquefaciens FZB42 and Bacillus artrophaeus LSSC22 had the strongest inhibitory effect against Rsc. Thirteen VOCs produced by FZB42 and 10 by LSSC22 were identified using gas chromatography-mass spectrometry analysis. Benzaldehyde, 1,2-benzisothiazol-3(2 H)-one and 1,3-butadiene significantly inhibited the colony size, cell viability, and motility of pathogens and negatively influenced chemotaxis. Transmission and scanning electron microscopy revealed severe morphological and ultra-structural changes in cells of Rsc. Furthermore, VOCs altered the transcriptional expression level of PhcA (a global virulence regulator), type III secretion system (T3SS), type IV secretion system (T4SS), extracellular polysaccharides and chemotaxis-related genes, which are major contributors to pathogenicity, resulting in decreased wilt disease. The VOCs significantly up-regulated the expression of genes related to wilt resistance and pathogen defense. Over-expression of EDS1 and NPR1 suggest the involvement of SA pathway in induction of systemic resistance. Our findings provide new insights regarding the potential of antibacterial VOCs as a biocontrol tool against bacterial wilt diseases.
The phytopathogen Ralstonia solanacearum is a Gram-negative, soil-borne, aerobic, non-sporing, motile bacterium, which can cause bacterial wilt in a wide variety of potential host plants (approximately 450 crop species), especially in the Solanaceae family1,2,3,4. Rsc enters through the roots and produces an excessive amount of extracellular polysaccharides (EPS) within the vascular system, blocking the vascular tissue, causing wilt and ultimately leading to the death of infected plants in severe infections5. Bacterial wilt is a devastating disease of crops that occurs widely in tropical and subtropical regions of the world, resulting in severe economic loss6. Several studies have explored the control of soil-born plant diseases through plant growth promoting rhizobacteria (PGPRs), which have the ability to maintain plant pathogenic microbes below the threshold level in the soil7,8,9. A large number of biocontrol agents have been identified that manage the bacterial wilt pathogen, including Bacillus spp., Pseudomonas spp., Trichoderma spp., Streptomyces spp. and Paenibacillus spp3,10,11,12. Bacillus species are considered the most efficient species because they have the ability to produce spores that can survive in adverse environments13. Most of the processes involved in controlling phytopathogens through antagonistic microorganisms require physical contact among interacting partners as well as very close vicinity. However, many bacteria and fungi can inhibit other pathogenic fungi from a distance, which raises the possibility that these microorganisms must produce invisible volatile compounds that inhibit the growth of antagonistic organisms. Several recent studies have revealed that volatile organic compounds (VOCs) emitted by some antagonistic bacteria and fungi have antifungal or nematicidal properties14,15. Effmert (2012) reported 300 bacteria and fungi as VOC producers, and nearly 800 VOCs were recorded in the database of volatiles emitted by microorganisms (DOVE-MO). Among these, 671 VOCs belong to 212 bacterial species, and 335 belong to 96 species of fungi16.
Bacterial VOCs play a beneficial role in three ways: promoting plant growth, inhibiting the growth of plant pathogens and inducing systemic resistance17,18. Many bacterial VOCs have been reported to be plant growth promoters, such as 2,3-butanediol and acetoin by Bacillus subtilis GB03 and B. amyloliquefaciens IN937a, 2-pentylfuran by B. megaterium XTBG34, 13-tetradecadien-1-ol, 2-butanone and 2-methyl-n-1-tridecene by Pseudomonas fluorescens SS10119,20. In addition to their growth-promoting activity, the VOCs trigger plant tolerance to biotic17 and a-biotic factors3 by inducing systemic resistance. Ryu et al.17 first identified 2,3-butanediol from B. subtilis GB03 and B. amyloliquefaciens IN937a, which significantly induced resistance in the Arabidopsis plant against the pathogen Pectobacterium carotovorum sub sp. carotovorum. Similarly, Paenibacillus polymyxa E681 produced tridecane, which also induced systemic resistance in plants21. A large number of investigations, which were initiated in 1966, have reported that VOCs affect the growth and development of fungi22,23,24,25,26,27. However, studies examining the interactions between PGPR and phytopathogenic bacteria are in preliminary stages. The bacterial VOCs, 3-pentanol and 2-butanone, have been reported to induce resistance against the bacterial angular leaf spot pathogen, Pseudomonas syringae pv. lachrymans15.
A few investigations have been reported regarding the mode of action of antimicrobial VOCs. However, there is evidence that VOCs damage the DNA of the pathogenic microorganism and induce a change in the expression level of ontogenesis-related enzymes, which may alter the growth of targeted organism28. Various studies have collectively revealed that the type three secretion systems (T3SSs), encoded by the hypersensitive response and pathogenicity (hrp) genes, T4SS pili, EPS and chemotaxis, are the major contributors to pathogenicity in Rsc. The type three secretion system is essential for the pathogenicity of Rsc because it facilitates the injection of type three effector proteins (TTE) into plant cells29,30,31,32. The transcriptional expression of virulence and a-virulence factors is controlled by a complex regulatory network, and PhcA, a global virulence regulator, is the center of this network5,33. The homolog of bacterial wilt resistance gene RRS1 from Arabdiopsis was detected in tobacco, acting as major resistant gene against Rsc34. Similarly RE-bw, another homologue of bacterial wilt resistance gene RRS1 was observed as resistant gene against bacterial wilt caused by Rsc in egg plant. Over-expression of RE-bw enhanced endogenous salicylic acid (SA) content and up-regulated the transcriptional expression of EDS1, PAD4, NPR1 and SGT1 which are the main components of SA signaling pathway35. In this context, we used six Bacillus spp. that have been shown to have effective antagonistic activity against plant pathogens. The main objective of this study was to evaluate the toxicity of VOCs produced by selected Bacillus spp. against Rsc and the effect on the activation of induced systemic resistance in tobacco against bacterial wilt. The toxicity of VOCs in this study was evaluated in terms of the effect on physiological, ultra-structural and virulence-related characteristics and also on the transcriptional expression of related genes (PhcA, T3SS, T4SS pili, EPS and chemotaxis). Similarly induction of systemic resistance was observed, investigating the transcriptional expression of disease resistance and defense related genes in tobacco.
Experimental procedure
Microorganisms and growth conditions
Bacillus amyloliquefaciens FZB42 was kindly provided by R. Borriss (ABiTEP GmbH, Berlin, Germany). All other strains of Bacillus and Ralstonia used in this study (see Supplementary material Table S1) were obtained from the Laboratory of Biocontrol and Bacterial Molecular Biology, Nanjing Agriculture University, Nanjing, China. Bacillus strains were grown in Luria-Bertani (LB) medium at 37 °C overnight and maintained in LB broth supplemented with 30% glycerol at −20 °C as working stocks. Ralstonia solanacearum TBBS1 was grown in tetrazolium chloride (TZC) agar medium (supplemented with 0.05% TTC)36 for 48 h at 28 °C and maintained in sterilized distilled water at room temperature. Single colonies with a pink center were transferred to Casamino Acid Peptone Glucose (CPG) agar medium for routine use37.
Antibacterial activity evaluation of Bacillus spp
The agar diffusion method38 was used to test the antibacterial activity on CPG medium. A five-milliliter suspension of bacterial pathogen was mixed with150 ml of molten CPG medium (≤50 °C) after 18–24 h of shaking (200 rpm) at 28 °C, poured into plates and allowed to solidify. Three sterile filter papers (4 mm) were placed on the agar surface at equal distances, 2.5 cm apart from the center. Five μl cultures of Bacillus strains grown overnight were dropped on two of the filter papers, and the remainder was impregnated with 5 μl of sterile water as a control. The plates were sealed with Parafilm® M (Pechiney, Neenah, WI, USA) and incubated at 28 °C for 48 hours, and the antibacterial effect was determined by measuring the diameter of the growth inhibition zones around the filter papers inoculated with Bacillus strains.
Evaluation of the antibacterial activity of VOCs produced by FZB42 and LSSC2 against Rsc
Bacillus amyloliquefaciens FZB42 and B. artrophaeus LSSC22 were selected and further evaluated for volatile-mediated antibacterial activity against Rsc using the I-plate system17, which consisted of centrally partitioned plastic Petri dishes (85 × 15 mm) with no physical contact between the two microorganisms grown on either side. Ten μl of (18–24 h) Rsc culture (1 × 107 CFU ml−1) grown in CPG broth was dropped in one partition with CPG agar medium, and the plates were sealed and incubated for 24 hours. After 24 hours, 10 μl of liquid overnight-grown culture of each Bacillus (1 × 108 CFU ml−1) strain were dropped in the other partition containing modified minimal salt medium (MS) (1.5% agar, 1.5% sucrose, and 0.4% TSA (w/v)). The plates were double sealed and incubated at 28 °C for five days. Only the pathogen was inoculated on control plates. The diameter of Rsc measured in cm and viable cells of the pathogen were counted by the 10-fold serial dilution technique. Each experiment was performed with five replicates, and the experiment was repeated three times.
Scanning and transmission electron microscopy (SEM & TEM)
External morphological changes of the pathogen cells were observed using Scanning Electron Microscopy (SEM), and ultra-structural changes in the pathogen cells were observed using Transmission Electron Microscopy (TEM) after exposure to the VOCs of FZB42 and LSSC22 for three days at 28 °C. For the preparation of pathogen cells for SEM and TEM, filter paper discs were removed from the plates containing pathogen colonies with and without exposure to the VOCs of biocontrol strains using flame-sterilized micro-tweezers. The cells were then collected directly into Eppendorf tubes, washed three times with sterile water and fixated with 2% glutaraldehyde (Solarbio, (Beijing) Co. Ltd.) for 30 min at 4 °C. Fixed cells were rinsed thrice for 10 minutes with 100 mM phosphate buffer, post-fixed for 3 h in 1% osmium tetroxide, followed by dehydration through an ethanol gradient39. Later, the samples were coated with gold and electron micrographs were obtained using the Hitachi Science System SEM (Hitachi S-3000N, Tokyo, Japan). Similarly, for TEM, cells were collected as mentioned above, and the samples were embedded in Epon 812 and sectioned with an ultramicrotome40. Ultra-structural changes in the cells were observed using a Hitachi transmission electron microscope (H-600, Hitachi, Tokyo, Japan).
Swarming, swimming and twitching motility assays
The motility activity of Rsc exposed to volatile compounds from FZB42 and LSSC22 was assessed using divided Petri plates. Rsc cultures were normalized to an OD600 of 1.0, and then 2 μl of the cultures were spotted onto one compartment of the divided plates containing different percentages of agar in CPG medium for twitching and swarming motility, while for swimming motility, a sterile tooth pick was dipped and then touched gently in the center of a CPG agar medium compartment. Another compartment containing modified MS agar medium was spot inoculated with FZB42 and LSSC22 cell culture. The swarming and swimming motility halo were examined after incubation at 28 °C for 72 h, and twitching motility was observed after 48 hours. The experiment was repeated three times with five replicates each time. For swarming motility, 0.7% agar was used, for twitching motility, 1.6% agar was used, and for swimming motility, 0.2% agar was used.
Chemotaxis assay
The chemotaxis assay was performed in divided plates with modified MS medium on one side and chemotaxis buffer medium on the other side (10 mM phosphate buffer, 0.1 mM EDTA, 1 μM methionine, 10 mM lactic acid, 0.35% agar and pH 7.3). Ten mm of the agar was removed from the medium, and the holes were refilled with 100 μl of host plant (tobacco) root exudates. Rsc was inoculated at a distance of 15 mm from the holes in one compartment, while FZB42 and LSSC22 cultures were spot inoculated in the other compartment as described above. The plates were sealed with Parafilm and incubated at 28 °C. The Rsc cells, which had moved towards the root exudates, were removed after three days, and the CFU/ml was counted on the CPG agar plates as described earlier. For the collection of root exudates, the tobacco seeds were surface sterilized and then transferred to 1% water agar plates. These plates were incubated at 4 °C overnight and then at 28 °C in the dark for 4 to 6 days. For each extraction, 40 germinated seedlings were transferred into a sterile 50-ml conical tube containing 5 ml of sterile chemotaxis buffer and incubated at 28 °C for 24 h. They were then filtered using a Millipore membrane filter (0.22 μm) and stored at −80 °C. This experiment was performed with five replicates and was repeated three times.
Collection and analysis of VOCs by SPME/GC-MS
SPME Analysis
A 20-μl suspension of each Bacillus strain was inoculated into 30 ml of modified MS agar medium in a 100 ml-vial at 28 °C. To collect VOCs, 2 cm divinyl benzene/carboxen/PDMS (DCP, 50/30 μm) solid phase micro extraction (SPME) fiber (Supelco, Bellefonta, PA, USA) was used. Three days after incubation, the SPME fiber was inserted into the headspace of the vial containing bacteria and incubated at 50 °C for 30 min.
GC-MS analysis
GC-MS analysis was performed using a Bruker 450-GC gas chromatograph in combination with a Bruker 320-MS mass spectrometer. Helium gas was used as the carrier at a flow rate of 1 ml min−1. The SPME fibers were desorbed at 220 °C for 5 min, and GC–MS was run for 25 min. The starting temperature of the column was 35 °C for 3 min, which was increased to 180 °C at 10 °C/min and further increased to 240 °C at 4 °C/min, held for 5 min. The mass spectrometer was operated in the electron ionization mode at 70 eV with a source temperature of 220 °C, with continuous scanning from 50 m/z to 500 m/z. The mass spectra data for the volatile compounds were analyzed using the data in the NIST/EPA/NIH Mass Spectrum Library.
Evaluation of the antibacterial activity of synthetic VOCs identified through GCMS analysis
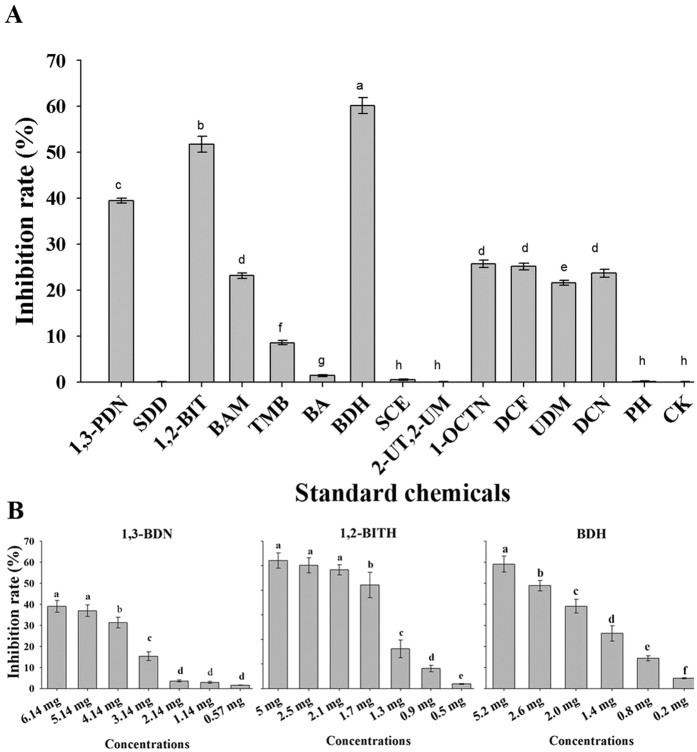
Standard chemicals were purchased from Sigma-Aldrich or Aladdin, previously collected and analyzed through GC-MS analysis. They were tested as pure chemicals for their antibacterial volatile activity against Rsc using the I-plate system. Solid-state VOCs were dissolved in dimethyl sulfoxide (DMSO) to obtain a final concentration of 1 mg ml−1. Ten μl (1 × 107 CFU/ml) of freshly grown Rsc (18–24 h) in CPG broth was dropped at the center of one half of the I-plate containing CPG agar, while on the other half, 100 μl of each of the tested VOCs or DMSO only was dropped. DMSO served as a control because it does not inhibit microorganisms. The plates were sealed and incubated for five days, and the results were measured as described earlier. Each experiment included three replicates and was repeated three times. Based on the results of the initial screening of individual VOCs, the most effective VOCs, BDH, 1,2-BIT and 1,3-BDN, were chosen for further detailed analysis regarding their antibacterial activity against Rsc at different concentrations to find minimum inhibitory content (MIC).
Effect of the degree of concentration of VOCs
To test the effect of the concentration of VOCs on the growth inhibition of the pathogen, an experiment was designed using three partition plates with completely sealed portions without any air movement except holes in two walls of the partitions. This arrangement was established so that the VOCs moved from the first partition, encompassing either Bacillus or a synthetic chemical, to the second partition and then from the second partition to the third partition. The second and third partition both were inoculated with 10 μl of Rsc (18–24 h) culture. The entire experiment was repeated three times with five replicates per experiment.
Effect of VOCs on the virulence and pathogenicity of Ralstonia solanacearum TBBS1
The pathogenicity assay of Rsc was performed after three days of exposure to FZB42 and LSSC22 VOCs, and then the Rsc cells were harvested in sterile distilled water to an OD of 0.1 at 600 nm. Five tobacco plants were inoculated at the three to four leaf stage by puncturing the stem at the third fully expanded leaf from the apex with the help of an inoculum-dipped needle, and then 100 μl of the VOC-treated Rsc suspension (1 × 107 CFU ml−1) was injected into each plant stem41. Similarly, 5 plants were inoculated with sterile water, and 5 plants were inoculated with a suspension of Rsc (1 × 107 CFU ml−1) cells that were not exposed to VOCs. The experiment was performed using a completely randomized design and repeated three times with five replicates in each experiment. The data for wilt was collected 21 days after inoculation using the following formula: Disease index (%) = [Σ (ni × vi) ÷ (V × N)] × 100, where ni indicates the number of plants with the respective disease rating; vi = disease rating; V = the highest disease rating (5) and N = the number of plants observed. The disease rating was calculated using the scale: 1 = no symptoms, 2 = one leaf wilted, 3 = two to three leaves wilted, 4 = four or more leaves wilted and 5 = whole plant wilted.
Pathogen inoculation and effect of VOCs on wilt disease development
The experiment was divided into two parts, i.e., the determination of the effect of VOCs on wilt disease in Petri plates and in plastic pots fitted on jars. I-plates prepared with one-half-strength Murashige and Skoog solid media and 5–6-day-old emerging tobacco seedlings (seven seedlings/plate) were dipped in the suspension of Rsc (1 × 107 CFU/ml) cells and then transplanted into one compartment. The non-inoculated control roots were dipped in sterile water. Bacterial strains, FZB42 and LSSC22, were cultured on LB broth one day before as described above. A 20-μl (108 CFU/ml) suspension of each strain or synthetic chemical was dropped into the other compartment. For individual chemical assay, each chemical with two different concentrations i.e. 10 mM 1,3-BDN and 1 mM 1,3-BDN, 1 mM 1,2-BITH, 0.1 mM 1,2-BITH, 0.1 mM BDH and 0.01 mM BDH were used in one compartment, while in other compartment tobacco seedlings as described earlier. Wilt symptoms were observed after one week of inoculation, and the data were recorded as described earlier. In the other experiment, plastic pots were fixed on glass jars (60 × 120 mm) and sealed with Parafilm to avoid the escape of VOCs from the jar, and a Petri dish (35 × 12 mm) was inoculated with FZB42, LSSC22 or sterilized water (control) and placed at the bottom of the jar. Filter paper was used at the bottom of the plastic pot containing tobacco seedlings to ensure that only VOCs produced by FZB 42 and LSSC22 could come into contact with the tobacco plants under soil conditions. Eight small holes (4 mm) were created at the bottom of the plastic pot for VOCs exposure. All pots were inoculated with a suspension of Rsc (at an OD of 0.1 at 600 nm) by dipping the roots in the suspension, except the non-inoculated control, and then the seedlings were re-planted in the pots. The inoculated seedlings were kept in a growth chamber at 28/22 °C day/night temperature for a 16/8-h light/dark photoperiod at 85% relative humidity. The experiment was performed using a completely randomized design with five replicates, and the whole experiment was repeated three times. The data for wilt was collected 21 days after inoculation using the formula mentioned above.
Real-time PCR
The Rsc cells were harvested after exposure to VOCs from FZB42 and LSSC22 for 72 hours. Total RNA was extracted using the Bacterial RNA Kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. First-strand cDNA was obtained using Reverse Transcriptase (TaKaRa Bio Inc., Tokyo, Japan) with random hexamer primers. The 16srRNA was used as an internal reference, and the transcriptional levels of PhcA, TSSS, Type IV pili, EPS and chemotaxis related genes were detected. For tobacco plants, leaves were collected at 3rd, 6th and 9th days after inoculation with Rsc. Total RNA was isolated using the TRIzol reagent (Invitrogen Biotechnology Co., Carlsbad, CA, USA) according to the manufacturer’s instructions. The EF-1α42 was used as an internal reference, and the transcriptional expression levels of RRS1, NPR1 and EDS1 genes were detected. Real-time PCR was performed using a SYBR Green/Fluorescent qPCR master mix (Takara) on a Roche-480 system (Roche). The qRT-PCR program consisted of denaturation at 95 °C for 1 min, followed by 40 amplification cycles at 95 °C for 5 sec, 57 °C for 30 sec, and 72 °C for 30 sec. The specific primers used in this study are listed in Table S2. Each sample was replicated thrice for qPCR, and 2−ΔΔCt method was used to analyze gene expression level43.
Statistical analysis
To evaluate the significance of the treatments, the data from each experiment was analyzed using analysis of variance (ANOVA) and Duncan’s multiple-range test was employed to assess differences among treatments at P = 0.05 using SPSS ver. 17.0 statistical software (SPSS, Chicago, IL). Graphs and figures were plotted using sigma plot version 10.0.
Results
Evaluation of antibacterial activity
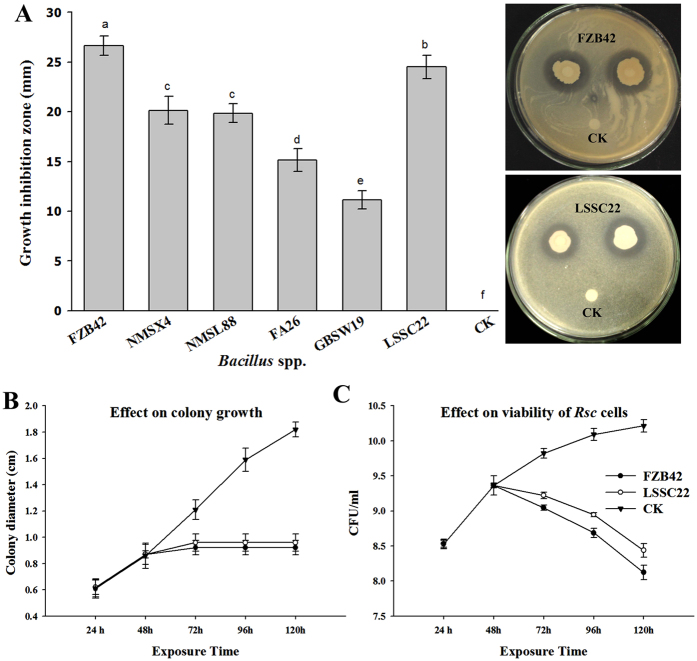
All antagonistic strains inhibited the growth of Rsc at varying levels, but Bacillus amyloliquefacieans FZB42 produced the largest zone of inhibition (26.65 mm ± 1.01 SD), followed by Bacillus artrophaeus LSSC22 (24.51 mm ± 1.55 SD). Bacillus amyloliquefacians NMSX4 and B. cereus NMSL88 inhibited the pathogen at the same level (20.15 mm ± 1.39 SD and 19.85 mm ± 0.95 SD) and were not significantly different. Bacillus pumulis GBSW 19 produced the smallest zone of inhibition of 11.15 mm ± 0.94 SD (Fig. 1A). FZB42 and LSSC22, which was shown to be most effective against Rsc in the GIZ experiment, were further evaluated in vitro for the production and activity of antibacterial VOCs against the pathogen Rsc. FZB 42 inhibited the colony growth of Rsc, restricting to 0.92 cm ± 0.055 SD and LSSC22 to 0.96 ± 0.065 SD cm as compared to control 1.82 cm ± 0.057 SD (Fig. 1B). Inhibition in colony diameter was 49.39% by FZB42 and 47.25% by LSSC22. The number of viable cells of Rsc also decreased when exposed to VOCs produced by FZB42 and LSSC22 and this decrease was maximum at 120 h (Fig. 1C).
Figure 1. The effect of strains of Bacillus on the growth of Rsc.
(A) The mean growth inhibition zones (mm) produced by antagonistic Bacillus spp. (B) Antibacterial volatile activity of B. amyloliquefacians FZB42 and B. artrophaeus LSSC22 against Ralstonia solanacearum. Error bars indicate standard deviations of the three replicates. Different letters above error bars represent significant differences according to Duncan’s multiple-range test (P = 0.05) using SPSS software (SPSS, Chicago, IL).
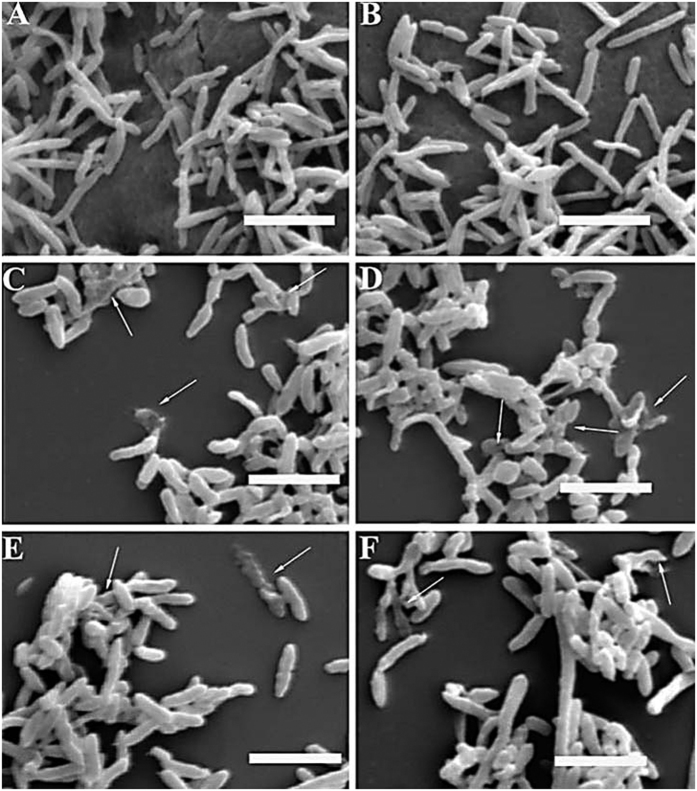
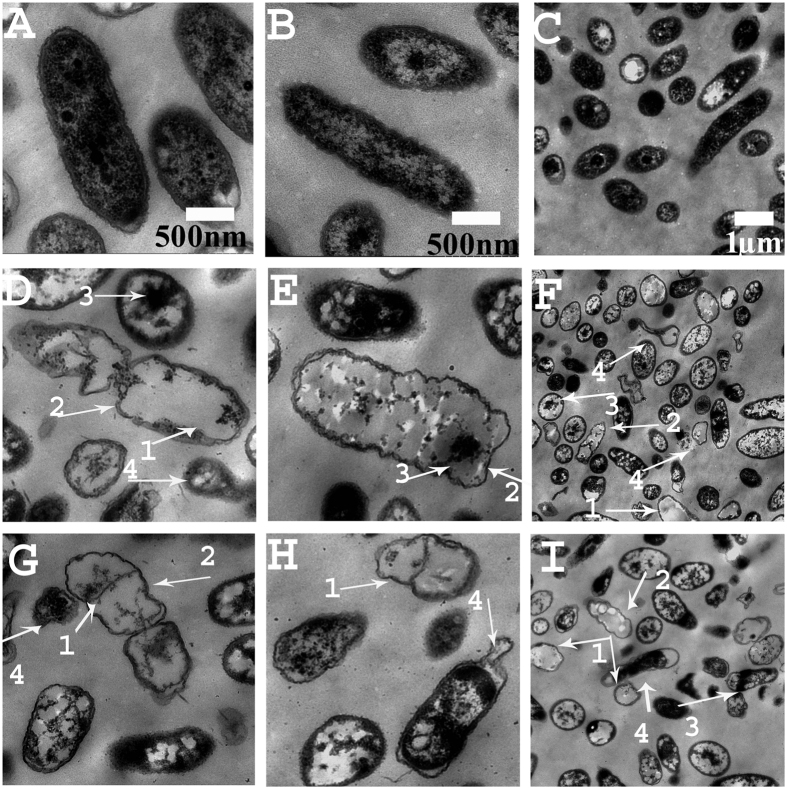
Bacillus VOCs caused morphological and ultra-structural abnormalities in Rsc cells observed by SEM and TEM
TEM and SEM analysis revealed a wide range of abnormalities in pathogenic cells that were affected by FZB 42 and LSSC22 when compared to the untreated control. SEM of the non-treated samples showed normal growth (Fig. 2A,B), whereas the colonies of pathogenic cells were severely disrupted following treatment with VOCs, as shown in Fig. 2C–F. The ultra-structural changes due to antibacterial VOCs of these Bacilli on the pathogenic cells of Rsc were further observed using TEM. Transmission electron micrographs of the Rsc cells obtained from the non-inoculated control showed apparently intact envelops with evenly distributed electron-dense cytoplasmic contents (Fig. 3A–C). The pathogenic cells exposed to VOCs of FZB42 and LSSC22 exhibited a large number of abnormalities. These abnormalities included loosening, and even at some places rupturing, of the cell wall, movement of the cytoplasmic content towards these ruptured wall areas, in some cells a lack of differentiated materials in the cytoplasm and also misshapen bacterial cells (Fig. 3D–I).
Figure 2. Bacillus VOCs caused a wide range of abnormalities in the morphology of the Ralstonia solanacearumcells observed by SEM.
Pathogenic cells were grown on CPG at 28 °C, and after three days of exposure to VOCs of FZB 42 and LSSC22, samples were collected from the I-plates. Electron micrographs were obtained using a Hitachi Science System SEM (Hitachi S-3000N, Tokyo, Japan). In the control (water), Rsc was grown on CPG without exposure to VOCs. The colony morphology of the pathogen was altered in the presence of volatiles produced by FZB 42 (C,D) and LSSC22 (E,F) and revealed several deformed pathogenic cells within the colony of the pathogen compared with the control (A,B). Arrow heads indicating distortion of Rsc cells. Bars; 5 μm.
Figure 3. Bacillus VOCs caused severe ultra-structural abnormalities in the Ralstonia solanacearum cells observed by TEM.
Ralstonia solanacearum cells were collected as previously mentioned and were prepared for TEM. Ultra-structural changes in the cells were observed using a Hitachi transmission electron microscope (H-600, Hitachi, Tokyo, Japan). Compared with the undamaged cells in the control (A–C), pathogenic cells showed a wide range of abnormalities after exposure to the VOCs of FZB42 (D–F) and LSSC22 (G–I). These changes included 1. Movement of cytoplasmic contents, 2. Loosening of the cell wall, 3. Formation of inclusion bodies and 4. Misshapen bacteria.
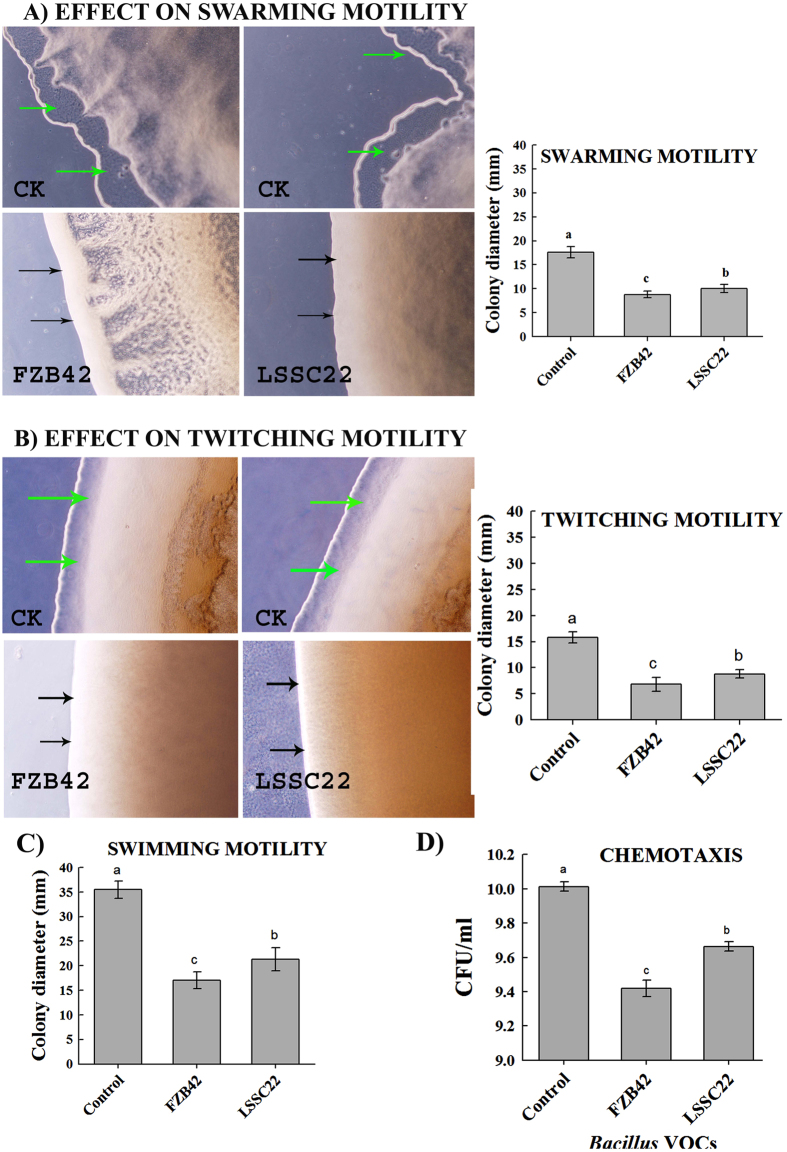
Bacillus VOCs inhibited motility (swarming, swimming and twitching) and chemotaxis of Rsc
The volatile compounds produced by FZB42 and LSSC22 had a negative effect on the motility of Rsc. VOCs produced by FZB 42 significantly reduced all three motility types. Swarming motility was restricted to 8.80 mm by FZB42 and 10.00 mm by LSSC22 as compared to control 17.60 mm, while swimming motility was restricted to 17.00 mm by FZB42 and 21.30 mm by LSSC22 as compared to control 35.00 mm. Similarly twitching motility was limited to 6.80 mm by FZB42 and 8.80 mm by LSSC22 as compared to control 15.80 mm (Fig. 4A–C). The inhibition in motility by FZB42 was 50%, 52.11% and 56.96% in swarming, swimming and twitching, respectively, compared with the non-exposed control. Swarming motility was reduced by 43.18%, swimming by 40% and twitching by 44.30%, compared with the control following exposure to VOCs produced by LSSC22. In the chemotaxis assay, Rsc showed significantly greater motility toward the hole containing root exudates in the control. FZB 42 and LSSC22 inhibited the chemotaxis of Rsc by 74.50% and 56.13%, respectively (Fig. 4D). In the negative control, there was no directed motility in any direction, but slight motility of Rsc cells in all directions was observed because sterile water was used instead of root exudates. The negative control justified that chemotaxis was present in Rsc along with motility.
Figure 4. Effect of VOCs produced by Bacillus amyloliquefacians FZB42 and Bacillus artrophaeus LSSC22 on motility of Ralstonia solanacearum.
(A) Swarming motility (B) twitching motility (C) swimming motility (D) chemotaxis Error bars indicate standard deviations of the means. The data were analyzed using analysis of variance (ANOVA) followed by Duncan’s multiple-range test (P = 0.05) using SPSS software (SPSS, Chicago, IL). Error bars indicate standard deviations of three replicates and different letters describe significant differences at P = 0.05 within the same data group.
GC-MS analysis of VOCs produced by FZB42 and LSSC22
Volatiles produced by FZB42 and LSSC22 were collected by a combination of HS-SPME and GC-MS. The volatile fractions of the antagonistic bacteria were compared with the volatiles retrieved from the control (non-inoculated medium). Mass spectra data of the volatile compounds were analyzed using the data in the NIST/EPA/NIH Mass Spectrum Library. Thirteen compounds were identified from FZB42 and 10 compounds were identified from LSSC22 (see supplementary material, Table S3), which had relatively high peak areas, e.g. ≥1%, and were not similar to the control. 1,2-Benzisothiazol-3(2 H)-one (1,2-BIT), (1 R)-2,6,6-Trimetyhlbicyclo[3.1.1]-hept-2-ene (TMB), Benzoic acid (BA), Dodecane, 1-fluoro (DCF), Dodecane (DCN) and Phenol, 2-(1,1-dimethylethyl)-6-methyl (PH) were common both in FZB42 and LSSC22. Seven other VOCs: Silanediol, dimethyl (SDD), Benzeneacetamide (BAM), Oxime, methoxy-phenyl (OMP), Benzaldehyde(BDH), Sulfurous acid, cyclohexyl-methyl isobutyl ester (SCE), 6-Tridecen, 2,2,4,10,12,12-hexamethyl-7-(3,5,5-trimrthylhexyl)-(6THT) and 2-Undecanethiol, 2-methyl (2-UT,2-M) were found in FZB42 while four VOCs: 1,3–Butadiene (1.3BDN), 1-octyn-3ol, 4-ethyl-(1OCTN), Undecanal, 2-methyl (UDM) and Cyclohexene, 3-(1,5-dimethyl-4-hexenyl)-6-methylene-(CHN) were found in LSSC22.
BDH, 1,2-BITH and 1,3-BDN had strong antibacterial activity against Rsc
The results showed that three chemicals, BDH, 1,2-BITH and 1,3-BDN, reduced the viability of the Rsc cells by 60.113%, 51.65% and 39.89%, respectively, compared with the control. These chemicals were selected for further study at different concentrations to find minimum inhibitory content (MIC). SDD, SCE, 2-UT2-M and PH did not inhibit the growth of Rsc, while BAM, 1-OCTN, DCF, UDM and DCN had a moderate effect, resulting in a 23.11%, 25.73%, 25.15%, 21.54% and 23.66% inhibition rate, respectively. TMB and BA had the least inhibition (Fig. 5A). BDH, 1,2-BITH and 1,3-BDNwere further evaluated at different concentrations, which showed that BDH, 1,2-BITH and 1,3-BDN reduced the viable cells even at 0.20 mg, 0.50 mg and 0.57 mg, respectively (Fig. 5B).
Figure 5. BDH, 1,2-BITH and 1,3-BDN showed strong antibacterial activity against Ralstonia solanacearum.
The data were analyzed using analysis of variance (ANOVA) followed by the Duncan’s multiple-range test (P ≤ 0.05) using SPSS software (SPSS, Chicago, IL). Error bars indicate the mean ± SD and different letters describe significant differences at P = 0.05 within the same data group. The compounds are ordered according to their retention time on a capillary GC column (Supelcowax®10). 1,3-BDN = 1,3–Butadiene, SDD = Silanediol, dimethyl, 1,2-BIT = 1,2-Benz isothiazol-3(2 H)-one, BAM = Benzeneacetamide, OMP = Oxime, methoxy-phenyl, TMB = (1 R)-2,6,6-Trimetyhlbicyclo[3.1.1]hept-2-ene, BA = Benzoic acid, 2-formyl-4,6-dimethoxy-,8,8-dimethoxyoct-2-yl, BDH = Benzaldehyde, SCE = Sulfurous acid, cyclohexylmethyl isobutyl ester, 2-UT,2-M = 2-Undecanethiol, 2-methyl, 1,OTN = 1-octyn-3ol, 4-ethyl-, DCF = Dodecane, 1-fluoro, UDM = Undecanal, 2-methyl DCN = Dodecane, PH = Phenol, 2-(1,1-dimethyl)-5-methyl.
Antibacterial activity of VOCs was dependent on the concentration of VOCs
The results showed that after five days of incubation, the 2nd partition had more growth inhibition in the form of less viable cells of Rsc than the 3rd partition following inoculation with FZB42. Similar results were obtained when synthetic VOC 1,2-BITH was used. However, the most effective VOC, BDH (undiluted), inhibited the growth of Rsc completely in both partitions, while the control did not inhibit growth (see Supplementary material Fig. S1).
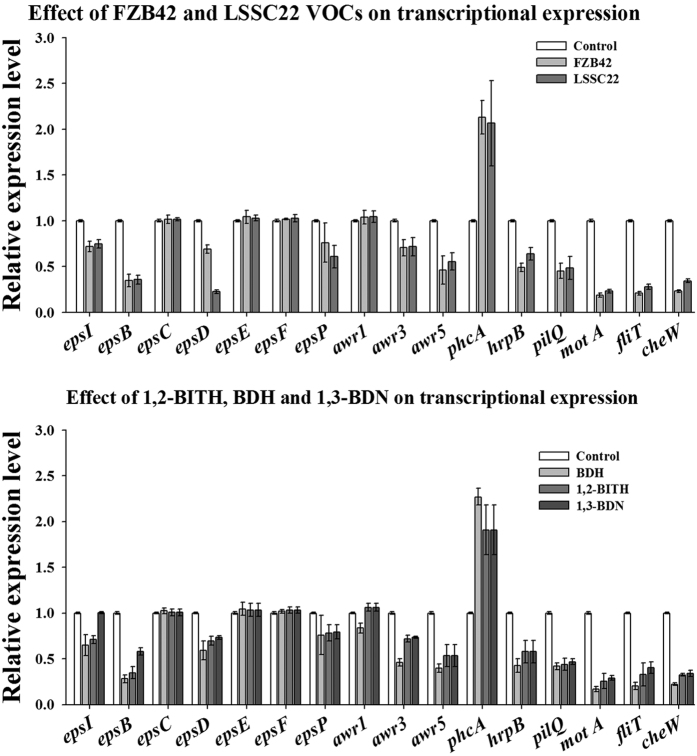
Bacillus VOCs altered the transcriptional expression of T3SS, T4SS pili, EPS and chemotaxis-related genes in Rsc
The transcriptional expression of genes related to extracellular polysaccharides (epsI, epsB, epsC, epsD, epsE, epsF, epsP), motility (motA, fliT), T3SS (hrpB), TTE (awr1, awr3, awr5), T4SS pili (pilQ), chemotaxis (cheW), and the global virulence regulator, PhcA, were significantly altered after exposure to VOCs of FZB42, LSSC22 and synthetic chemicals. Real-time PCR analysis elucidated that, of the 16 genes examined, 11 (epsI, epsB, epsD, epsP, motA, fliT, hrpB, awr3, awr5, pilQ, and cheW) were down-regulated at various levels, one (phcA) was up-regulated, and 4 (epsC, epsE, epsF and awr1) were unaffected after Rsc cells were exposed to VOCs of FZB 42 and LSSC22 for three days at 28 °C. BDH down-regulated 12 genes, including awr1, which was unaffected by FZB42 and LSSC22. Similarly, 1,2-BITH and 1,3-BDN altered the expression levels of 12 genes (epsI, epsB, epsD, epsP, awr3, awr5, phcA, hrpB, pilQ, motA, fliT, and cheW). However, 1,3-BDN had no effect on epsI along with four other unaffected genes (Fig. 6).
Figure 6. Transcriptional expression of virulence-related genes in Ralstonia solanacearum.
The Rsc cells were harvested after exposure to VOCs from B. amyloliquefacians FZB 42 and B. artrophaeus LSSC22, benzaldehyde, 1,2-benz isothiazol-3(2 H)-one and 1,3-butadiene (1,3-BDN) for 72 hours. Real-time PCR was performed using a SYBR Green/Fluorescent qPCR master mix (Takara) on a Roche-480 system (Roche). The 16srRNA was used as an internal reference, and the transcriptional levels of PhcA, T3SS, T4SS pili, EPS and chemotaxis-related genes were detected. Each sample was replicated thrice for qPCR, and 2−ΔΔCt method was used to analyze gene expression level. Error bars indicate standard deviations of the means of three replicates.
Bacillus VOCs reduced the virulence and pathogenicity of Rsc and induced systemic resistance
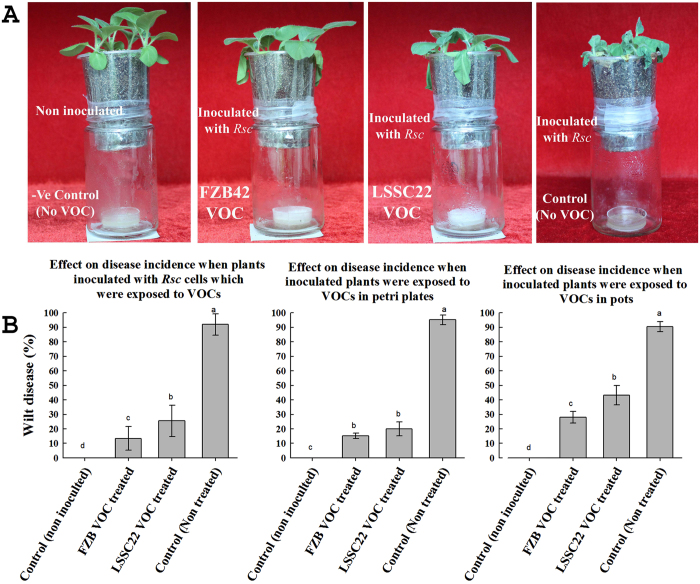
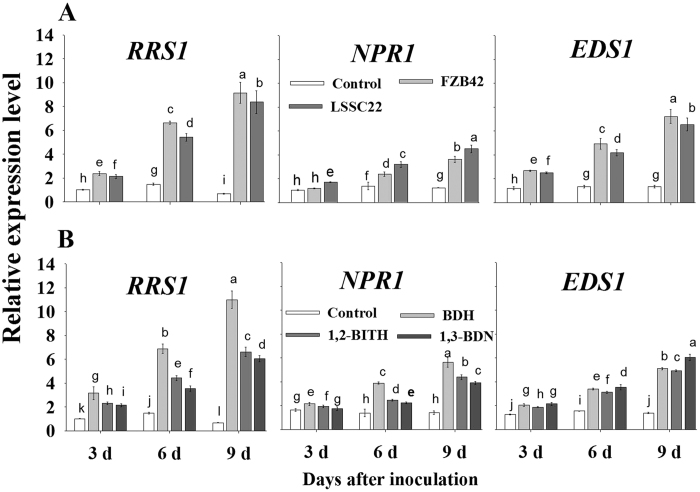
The pathogenicity assay of Rsc was performed after three days of exposure to VOCs of FZB42 and LSSC22, which resulted in a decrease in the virulence of Rsc. A 13.33% wilt disease index was observed when Rsc was exposed to VOCs from FZB42, and a 25.45% index was observed for LSSC22, while the index was 92.00% when Rsc cells were not exposed to any bacterial VOCs. FZB42- and LSSC22-derived VOCs reduced disease development significantly (by 15.2% and 16%, respectively) when tobacco seedlings were exposed to their VOCs, compared with the control (95%), but there was no significant difference between the treatments. Similarly BDH, 1,2-BITH and 1,3-BDN also reduced the disease development. A 14.4% and 15.2% disease index was observed by BDH (0.1 mM and 0.01 mM), 18.4 and 20.8% by 1,2-BITH (1 mM and 0.1 mM) while 24.8 and 28. % by 1,3-BDN (10 mM and 1 mM) as compared to (97.6%) water control. Reduction in disease development in the I plate system confirmed the induced systemic resistance in tobacco against Rsc by VOCs of FZB42 and LSSC22 as well as by synthetic chemicals (see Supplementary material Fig. S2). To demonstrate that the VOCs produced by FZB42 and LSSC22 induced systemic resistance at a broader level (in-vivo), we created the in planta system. The results showed that VOCs produced by FZB42 and LSSC22 caused a significant reduction in the wilt index, confirming the induced systemic resistance in plants against Rsc. FZB42 significantly reduced the disease and showed only a 28% wilt index, while LSSC22 had a 43.20% wilt index, as compared with a 92% wilt index in the non-exposed control (Fig. 7). To verify the resistance in tobacco against Rsc, due to the exposure of VOCs in pot experiment, we analyzed the genes of tobacco plant relating to resistance and defense by real time PCR. The transcriptional expression of R gene RRS1 which is related to resistance in tobacco against Rsc was induced by FZB42, LSSC22 and also by individual chemicals; BDH, 1,2-BITH and 1,3-BDN as compared to untreated control. However over-expression was more by FZB42 and BDH as compared to others and increased with the time, showing its maximum at 9th days after inoculation. An up-regulation in defense related genes NPR1 and EDS1 was observed after exposure to Bacillus VOCs. However, there was a difference in the relative expression levels of NPR1 and EDS1 among the VOCs produced by FZB42 and LSSC22. Increased expression of NPR1 was noticed when treated with VOCs of LSSC22 compared with FZB42, which reached the highest level at the 9th day. Expression of EDS1 was also induced by FZB42 and LSSC22-VOCs (Fig. 8A). Similarly, synthetic VOCs BDH, 1,2-BITH and 1,3-BDN induced the transcriptional expression of genes relating to defense at varying level as compared to control (Fig. 8B). These results indicate that VOCs stimulated the resistance and defense related genes which activated the induction of systemic resistance in tobacco against Rsc.
Figure 7. Volatiles produced by B. amyloliquefacians FZB42 and B. artrophaeus LSSC22 reduced the virulence of Ralstonia solanacearumand and induced systemic resistance.
%Wilted plants was calculated after 21 days of inoculation according to the Winstead formula. Five tobacco plants were inoculated at the three to four leaf stage by puncturing the stem at the third fully expanded leaf from the apex with the help of an inoculum-dipped needle. Then, 100 μl of the VOC-treated Rsc suspension was injected into each plant stem. In the control, plants were inoculated with sterile water, and in the non-treated group, plants were inoculated with a suspension of Rsc cells that were not exposed to VOCs. For the Petri plate experiment (B2), 5-6-day-old emerging tobacco seedlings inoculated with Rsc were transplanted into one compartment, bacterial strains were cultured in other compartment of I-plate. In the non-inoculated control, the roots were dipped in sterile water. Wilt symptoms were observed after 7 days of inoculation, and the data were recorded as described earlier. In B3, plastic pots, fixed on glass jars, having a Petri dish inoculated with FZB42, LSSC22 or sterilized water were used. All three pots were inoculated with a suspension of Rsc by the root dip method, except the non-inoculated control, and were kept in a growth chamber at 28/22 °C day/night temperature for a 16-h light/8-h dark photoperiod at 85% relative humidity. Error bars indicate standard deviations of the means. Different letters above error bars represent significant differences according to Duncan’s multiple-range test (P = 0.05) using SPSS software (SPSS, Chicago, IL).
Figure 8. Transcriptional expression of virulence-related genes in Ralstonia solanacearum.
Relative expression of the resistance gene RRS1 and defense related genes NPR1 and EDS1 in tobacco were observed after exposure to VOCs produced by FZB42 and LSSC22 (A) and synthetic chemicals (B) in pot experiment. Error bars indicate standard deviations of three replicates and 2−ΔΔCt method was used to analyze gene expression level.
Discussion
Microbial volatiles have been increasingly studied during the last two decades for their beneficial and environmentally friendly roles, such as induced systemic resistance against plant pathogens and abiotic factors21,44, plant growth enhancement15,20 and antagonistic action against plant pathogenic fungi and nematodes8,14. However, most investigations have been conducted using plant pathogenic fungi15. Recently, antibacterial activity of VOCs produced by Bacillus amyloliquefaciens SQR-9 and Pseudomonas fluorescens WR-1 was demonstrated against the tomato wilt pathogen, although this study did not further investigate the induction of systemic resistance against Rsc45,46. In our study, we demonstrated that bacterial VOCs not only control bacterial pathogens but can also induce systemic resistance. Bacillus amyloliquefaciens FZB42 is well known for plant growth regulation and the synthesis of complex bioactive molecules such as microcin B17, streptolysin S and amylocyclicin that inhibit the growth of plant pathogenic fungi and bacteria47,48,49. Rsc requires swimming motility and twitching motility for virulence31,50, as non-motile mutants of Rsc have been demonstrated to be significantly less virulent. There is a strong relationship between swimming motility and virulence, as swimming motility is a virulence trait of Rsc and is required for the wilt pathogen to properly invade and colonize the host plant50. Rsc not only requires motility but also chemotaxis toward root exudates to efficiently colonize and enter into plant roots for disease development51. Our results revealed a reduction in both motility and chemotaxis of Rsc following exposure to VOCs of FZB42 and LSSC22 (Fig. 4). This phenomenon was confirmed in the pathogenicity assay and the transcriptional expression of motility-related genes. Twitching motility is a form of bacterial translocation over firm surfaces that requires T4SS pili. Our results revealed down-regulation of pilQ, fliT, motA and the chemotaxis-related gene cheW (Fig. 7). T3SS, T4SS pili, EPS and chemotaxis are major pathogenicity determinants of Rsc. In Rsc, the expression of Hrp-T3SS-related genes is regulated by an AraC-type transcriptional activator, HrpB, which is down-regulated by VOCs from FZB42 and LSSC22. Thirty HrpB-regulated hpx (hrpB-dependent expression) genes and three hrpB-regulated genes, popA, popB and popC, have been identified in Rsc32. HrpB acts as a master regulatory gene, and the down-regulation of hrpB might ultimately affect these genes. T3SS encoded by hypersensitive response and pathogenicity (hrp) genes delivers bacterial effector proteins called type three effector proteins (TTE) directly into the host cell cytosol to promote disease30,52. AWR (alanine-tryptophan-arginine tryad) effectors are involved in virulence or avirulence in Rsc upon interaction with the host plant. Our results showed down-regulation of awr3 and awr5. Transcriptional expression of awr (type III effecter proteins) requires HrpB53 which was down-regulated in our study, justifying the down-regulation of awr3 and awr5. A complex regulatory network is required for the transcriptional expression of virulence or avirulence factors, and PhcA is the control center of this system33. PhcA is directly or indirectly involved in the regulation of genes, resulting in the expression of several virulence factors such as EPS, plant cell wall-degrading enzymes, TTSS and bacterial motility54. Results showed that transcriptional expression of PhcA was up-regulated up to 3-fold following treatment with VOCs of FZB42 and LSSC22. The global virulence regulator, PhcA, negatively regulates the expression of T3SS genes, supporting our results. Similarly, Hrp pili in Rsc were also repressed by phcA in cells grown in rich medium54. VOCs from FZB42 and LSSC22 repressed epsI, epsB, epsD and epsP, while epsC, epsE and epsF were not affected. EPS is the one of the most important virulence factors because EPS I-deficient mutants are nearly avirulent and do not colonize plant xylem vessels as efficiently as wild-type pathogens5. Our results are in conformity with previously reported results in which repressed transcriptional expression of virulence-associated genes in Xanthomonas oryzae pv. oryzae were noted55. Similarly VOCs produced by Bacillus amyloliquefaciens SQR-9 and Pseudomonas fluorescens WR-1 down regulated the expression of RSc proteins related to virulence46,47. At high concentrations, VOCs might damage the cell membrane, causing the movement of intracellular material out of the cell and resulting in cell death. Electron micrographs of the untreated Rsc samples revealed normal growth of the cells, while the Rsc colonies were disturbed, in the presence of VOCs of FZB42 and LSSC22. The transmission electron micrograph showed that VOCs caused damage to the pathogenic cells at high concentrations, while the non-inoculated control displayed normal cell growth. These abnormalities included loosening of the cell wall and, in some cells, rupturing of the cell wall and movement of the cytoplasmic contents towards the ruptured walls, resulting in the formation of misshapen cells and sometimes cell death (Figs 2 and 3). These results are consistent with previously reported studies in which altered morphology of fungal hyphae and conidiophores of Botrytis cinerea and Penicillium italicum were observed after exposure to microbial VOCs56. Similarly SEM and TEM analysis showed abnormal surface morphology in cells of Xanthomonas oryzae pv. oryzae after exposure to VOCs produced by Bacillus cereus D1355. One prominent finding of our study was that VOCs from FZB42 and LSSC22 negatively affected disease development and induced systemic resistance in tobacco plants. However, disease development appeared to be greater in the Rsc inoculated plants that were treated directly with VOCs. This observation might be due to the hindered contact of VOCs with Rsc cells (Fig. 7B1 and B3). The tobacco seedlings grown in divided Petri plates displayed a greater level of induced systemic resistance compared with the potted experiment, as in Petri plates, higher levels of VOCs were available (Fig. 7B1 and B2). The decreased disease development in plants inoculated with VOCs-treated Rsc cells might be a consequence of the VOCs not only reducing pathogen growth but also restricting the movement of the pathogen into tobacco plant roots. VOCs from FZB42 and LSSC22 might be influenced in two positive ways: promoting plant growth and inhibiting pathogen growth, and thus inducing systemic resistance. Our results suggested that FZB42 and LSSC22 emit VOCs that can elicit plant defense mechanisms. The results showed the up-regulation of transcriptional expression of R gene RRS1 along with the overexpression of NPR1 and EDS1 when exposed to VOCs after inoculation with Rsc. The over-expression of RRS1 confirmed the induction of systemic resistance in tobacco as the RRS1 is specifically the bacterial wilt resistance gene isolated from Arabdiopsis and detected in tobacco34. The R gene (RRS1-R) isolated from Nd-1 showed a significant enhancement of resistance when overexpressed in Col plants57,58. Overexpression of R gene, RE-bw in eggplant confirmed that the RE-bw was an important R gene against bacterial wilt. Furthermore, results showed the involvement of RE-bw in enrichment of SA content, ROS-scavenging enzymatic activities, cell wall, and lignin to minimize the Rsc colonization to roots. The up-regulation of transcriptional expression of NPR1 and EDS1 suggested that the induction of systemic resistance might be motivated by SA signaling path way as EDS1 and NPR1 are the main components of SA pathway35. The chemical compounds, BDH, 1,2-BITH and 1,3-BDN, are responsible for eliciting the defense response. Tobacco seedlings receive a sufficient amount of airborne chemical information to trigger ISR, as measured by the ability of the seedlings to resist infection. These results suggest that induced systemic resistance can be achieved without any contact between PGPRs and plants, indicating that VOCs might be involved normally in the process of induced systemic resistance59. These results are consistent with previous findings in which Bacillus amyloliqefaciens IN937a, B. subtilis GB03, Escherichia coli DH5a and Pseudomonas fluorescens 89B61 produced BDH, inducing systemic resistance in plants against pathogens and inhibited the growth of various pathogenic fungus mycelia and spore germination21,60,61. Similarly, benzothiazole inhibits mycelial growth14,62. BDH is an aromatic aldehyde with strong antimicrobial activity, but it is a generally regarded as safe (GRAS)-compound without any toxic effect in humans or on the environment63. In conclusion, our study showed the following novel results: (1) VOCs produced by FZB42 and LSSC22 inhibited colony growth, motility and negatively influenced the chemotaxis of Rsc, (2) caused morphological and ultra-structural abnormalities in Rsc cells, (3) decreased virulence levels both when Rsc cells were directly treated with VOCs and when plants inoculated with Rsc were exposed to VOCs, inducing systemic resistance, (4) altered the transcriptional expression of virulence related genes (T3SS, T4SS, EPS, motility and virulence regulator PhcA) and over-expressed genes related to wilt resistance and pathogen defense (5) BDH, 1,2-BIT, 1,3-BDN were not only the key inhibiting factors of Rsc, but also eliciting ISR. These results provide new insight into the inter-communication among the pathogenic bacteria Rsc and antagonistic bacteria, FZB42 and LSSC22 and tobacco plants in terms of the suppression of the wilt disease index and induction of systemic resistance in plants. This is the first report on the production and systemic resistance activity of VOCs derived from FZB42 and LSSC22 against bacterial wilt disease. VOCs produced by FZB42 and LSSC22 are good antibacterial compounds, providing an alternative to pesticides, and they can be used as an environmentally friendly biocontrol mechanism.
Additional Information
How to cite this article: Tahir, H. A. S. et al. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep. 7, 40481; doi: 10.1038/srep40481 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (31471811), the Agro-scientific Research in the Public Interest (20130315), and the Key research and development program of Jiangsu Province (BE2015354).
Footnotes
Author Contributions H.A.S.T., H.J.W., Q.G. and X.W.G. conceived and designed the experiments. H.A.S.T. conducted most of the experiments. Y.N. and R.H. performed the quantitative real time-PCR. H.A.S.T. analyzed the experimental data and wrote the manuscript. All authors reviewed the manuscript.
References
- Hayward A. C. Biology and epidemiology of bacterial wilt caused by Pseudomonas Solanacearum. Annu. Rev. Phytopathol. 29, 65–87 (1991). [DOI] [PubMed] [Google Scholar]
- Swanson J. K., Yao J., Tans-Kersten J. & Allen C. Behavior of Ralstonia solanacearum Race 3 Biovar 2 during latent and active infection of Geranium. Phytopathol. 95, 136–43 (2005). [DOI] [PubMed] [Google Scholar]
- Xue Q. Y. et al. Evaluation of the strains of Acinetobacter and Enterobacter as potential biocontrol agents against Ralstonia wilt of tomato. Biol. Cont. 48, 252–258 (2009).
- Genin S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187, 920–928 (2010). [DOI] [PubMed] [Google Scholar]
- Schell M. A. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38, 263–292 (2000). [DOI] [PubMed] [Google Scholar]
- Peeters N., Guidot A., Vailleau F. & Valls M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. plant pathol. 14, 651–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Wang H., Wang C., Tian B. & Hua Y. Global effect of an RNA polymerase β-subunit mutation on gene expression in the radiation-resistant bacterium Deinococcus radiodurans. Sci. China. Life Sci. 54, 854–62 (2011). [DOI] [PubMed] [Google Scholar]
- Guo Q. et al. Complete Genome Sequence of Bacillus subtilis BAB-1, a biocontrol agent for suppression of tomato gray mold. Genome Announc. 2(4), e00744–14, doi: 10.1128/genomeA.00744-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Molecular mechanism of plant growth promotion and induced systemic resistance to tobacco mosaic virus by Bacillus spp. J. Microbiol. Biotechnol, doi: 10.4014/jmb.0901.0008 (2016). [DOI] [PubMed] [Google Scholar]
- Ramesh R., Joshi a. a. & Ghanekar M. P. Pseudomonads: major antagonistic endophytic bacteria to suppress bacterial wilt pathogen, Ralstonia solanacearum in the eggplant (Solanum melongena L.). World J. Microbiol. Biotechnol. 25, 47–55 (2008). [Google Scholar]
- Maji S. & Chakrabartty P. K. Biocontrol of bacterial wilt of tomato caused by Ralstonia solanacearum by isolates of plant growth promoting rhizobacteria. Aust. J. Crop Sci. 8, 208–214 (2014). [Google Scholar]
- Gao S. et al. Efficient colonization and harpins mediated enhancement in growth and biocontrol of wilt disease in tomato by Bacillus subtilis. Lett. Appl. Microbiol. 57, 526–33 (2013). [DOI] [PubMed] [Google Scholar]
- Francis I., Holsters M. & Vereecke D. The Gram-positive side of plant-microbe interactions. Environ. Microbiol. 12, 1–12 (2010). [DOI] [PubMed] [Google Scholar]
- Weisskopf L. The potential of bacterial volatiles for crop protection against phytophathogenic fungi. Microbial Pathogens a Strategies for Combating Them: Sci., Technol. Education (ed. Méndez-Vilas A.) 1352–1363 (Badajoz, 2013). [Google Scholar]
- Song G. C. & Ryu C.-M. Two volatile organic compounds trigger plant self-defense against a bacterial pathogen and a sucking insect in cucumber under open field conditions. Intern. J. Mol. Sci. 14, 9803–9819 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effmert U., Kalderás J., Warnke R. & Piechulla B. Volatile mediated interactions between bacteria and fungi in the soil. J. Chem. Ecol. 38, 665–703 (2012). [DOI] [PubMed] [Google Scholar]
- Ryu C. M. et al. Bacterial volatiles promote growth in Arabidopsis. PNAS 100, 4927–4932 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J., Wu H., Huo R., Gao X. & Borriss R. Stimulation of plant growth and biocontrol by Bacillus amyloliquefaciens subsp. plantarum FZB42 engineered for improved action. Chem. Biol. Technol. in Agric. 1, 1–14 (2014). [Google Scholar]
- Zou C., Li Z. & Yu D. Bacillus megaterium strain XTBG34 promotes plant growth by producing 2-pentylfuran. J. Microbiol. 48, 460–466 (2010). [DOI] [PubMed] [Google Scholar]
- Park Y.-S. et al. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem. Biophys. Res. Commu. 461, 361–5 (2015). [DOI] [PubMed] [Google Scholar]
- Lee B. et al. Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PloS one 7, e48744 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7 (2005). [DOI] [PubMed] [Google Scholar]
- Ortíz-Castro R., Contreras-Cornejo H. A., Macías-Rodríguez L. & López-Bucio J. The role of microbial signals in plant growth and development. Plant Signal. Behav. 4, 701–712 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minerdi D., Bossi S., Gullino M. L. & Garibaldi A. Volatile organic compounds: a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35. Environ. Microbiol. 11, 844–854 (2009). [DOI] [PubMed] [Google Scholar]
- Xie X., Zhang H. & Pare P. Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus subtilis (GB03). Plant Signal. Behav. 4, 948–953 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M. et al. Serratia odorifera: analysis of volatile emission and biological impact of volatile compounds on Arabidopsis thaliana. Appl. Microbiol. Biotechnol. 88, 965–76 (2010). [DOI] [PubMed] [Google Scholar]
- Kim Y. C. et al. The multifactorial basis for plant health promotion by plant-associated bacteria. Appl. Environ. Microbiol. 77, 1548–55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A. M., Strobel G. a., Moore E., Robison R. & Sears J. Volatile antimicrobials from Muscodor crispans, a novel endophytic fungus. Microbiol. 156, 270–7 (2010). [DOI] [PubMed] [Google Scholar]
- Aldon D., Brito B., Boucher C. & Genin Â. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. Euop. Mol. Biol. Organ. J. 19, 2304–2314 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A., Cunnac S., Reymond N., Genin S. & Boucher C. Genome-wide analysis of gene expression in ralstonia solanacearum reveals that the hrpb gene acts as a regulatory switch controlling multiple virulence pathways. Molecul. Plant-Microb. Interac. 18, 938–949 (2005). [DOI] [PubMed] [Google Scholar]
- Schell M. A., Liu H., Kang Y. & Denny T. P. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiol. 147, 3215–3229 (2001). [DOI] [PubMed] [Google Scholar]
- Mukaihara T. & Tamura N. Identification of novel Ralstonia solanacearum type III effector proteins through translocation analysis of hrpB-regulated gene products. Microbiol. 155, 2235–2244 (2009). [DOI] [PubMed] [Google Scholar]
- Jacobs J. M., Lavanya B., Fanhong M., Annett M. & Caitilyn A. The In Planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L. et al. Detection on RRS1 gene of bacterial wilt resistance from Arabidopsis in tobacco for homolog. Chinese Agric. Sci. Bull. 28(31), 137–140 (2012). [Google Scholar]
- Xi’ou X. et al. Functional characterization of a putative bacterial wilt resistance gene (RE-bw) in Eggplant. Plant Molecul. Biol. Report. 33, 1058–1073 (2015). [Google Scholar]
- Kelman. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium chloride medium. Phytopathol. 44, 693–695 (1954). [Google Scholar]
- Hendrickt C. A. & Sequeira L. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48, 94–101 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Perry D. M. & Kirby W. M. M. Single disc antibiotic sensitivity testing of Staphylococci. A. M. A. Arch. Intern. Med. 104, 208–216 (1959). [DOI] [PubMed] [Google Scholar]
- Islam T., Hashidoko Y., Deora A., Ito T. & Tahara S. Suppression of damping-off disease in host plants by the rhizoplane bacterium Lysobacter sp. strain sb-k88 is linked to plant colonization and antibiosis against soilborne peronosporomycetes. Appl. Environ. Microbiol. 71, 3786–3796 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Xie S., Feng J., Wang F. & Xu Q. Protective effect of Polygonum orientale L. extracts against Clavibater michiganense subsp. sepedonicum, the causal agent of bacterial ring rot of potato. PloS one 8, e68480 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstead N. N. & K. A. Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathol. 42, 628–634 (1952). [Google Scholar]
- Berberich T., Sugawara K., Harada M. & Kusano T. Molecular cloning, characterization and expression of an elongation factor 1α gene in maize. Plant Mol. Biol. 29, 611–615 (1995). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25, 402–8 (2001). [DOI] [PubMed] [Google Scholar]
- Gu Y.-Q. et al. Evaluation and identification of potential organic nematicidal volatiles from soil bacteria. Soil Biol. Biochem. 39, 2567–2575 (2007). [Google Scholar]
- Raza W. et al. Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci. Rep. 6, 248–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza W. et al. Volatile organic compounds produced by Pseudomonas fluorescens WR-1 restrict the growth and virulence traits of Ralstonia solanacearum. Microbiol. Rese. 192, 103–113 (2016). [DOI] [PubMed] [Google Scholar]
- Scholz R. et al. Plantazolicin, a novel microcin B17/streptolysin S-like natural product from Bacillus amyloliquefaciens FZB42. J. bacteriol. 193, 215–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz R. et al. Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. bacteriol. 196, 1842–52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Wu H.-J., Qiao J., Gao X. & Borriss R. Novel routes for improving biocontrol activity of bacillus based bioinoculants. Front. Microbiol. 6, 1395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans-kersten J., Brown D. & Allen C. Swimming motility, a virulence trait of ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Molicul. Plant Microbe Interact. 17, 686–695 (2004). [DOI] [PubMed] [Google Scholar]
- Yao J. & Allen C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188, 3697–3708 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls M., Genin S. & Boucher C. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS pathogens 2, e82 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S., Christian B. & Stephane G. Characterization of the cis-Acting Regulatory Element Controlling HrpB-Mediated Activation of the type iii secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186, 2309–2318 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. et al. Complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J. Bacteriol. 177, 1259–1267 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Wu H., Rajjar F. & Gao X. Antibacterial activity of Bacillus cereus D13 volatile compounds against Xanthomonas oryzae pv. Oryzae. Mol. Plant Pathol, doi: 10.1111/mpp.12494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Fumigant activity of volatiles of Streptomyces globisporus JK-1 against Penicillium italicum on Citrus microcarpa. Postharvest Biol. Technol. 58, 157–165 (2010). [Google Scholar]
- Deslandes L. et al. Resistance to Ralstonia solanacearum In Arabidopsis thaliana is conferred by the recessive RRS1-R gene. PNAS 99(4), 2404–2409 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M. et al. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. The Plant J. cell mol. Biol. 60, 218–26 (2009). [DOI] [PubMed] [Google Scholar]
- Mortel R. C. et al. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 160, 2173–2188 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa R., Behrends V., Williams H. D., Bundy J. G. & Rojo F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ. Microbiol. 18, 807–18 (2016). [DOI] [PubMed] [Google Scholar]
- Farag M., Ryu C.-M., Sumner L. W. & Paré P. W. GC-MS SPME profiling of rhizobacterial volatiles reveals prospective inducers of growth promotion and induced systemic resistance in plants. Phytochem. 67, 2262–8 (2006). [DOI] [PubMed] [Google Scholar]
- Fernando W. G. D., Ramarathnam R., Krishnamoorthy A. S. & Savchuk S. C. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochemi. 37, 955–964 (2005). [Google Scholar]
- Andersen A. Final report on the safety assessment of benzaldehyde. Int. J. Toxicol. 25, 11–27 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.