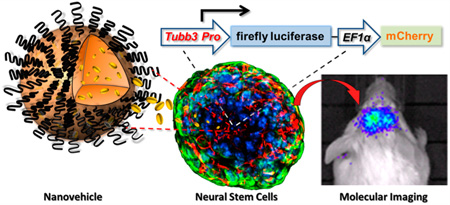
Abstract
Recent advances in neural stem cell (NSC) transplantation have led to an inspiring progress in alleviating central nervous system (CNS) damages and restoring brain functions from diseases or injuries. One challenge of NSC transplantation is directed differentiation of transplanted NSCs into desired neuronal subtypes, such as neurons, to compensate the adverse impact of brain injury; another challenge lies in the lack of tools to noninvasively monitor the dynamics of NSC differentiation after transplantation in vivo. In this study, we developed a polymer nanovehicle for morphogen sustained release to overcome the drawbacks of conventional methods to realize the long-term directed NSC differentiation in vivo. Moreover, we constructed a bicistronic vector with a unique neuron specific gene tubb3 promoter to drive reporter gene expression for real-time imaging of NSC differentiation and migration. The developed uniform nanovehicle showed efficient NSC uptake and achieved a controlled release of morphogen in cytosol to consistently stimulate NSC differentiation into neurons at a sustainably effective concentration. The spatiotemporal imaging results showed a multiplexed migration, proliferation, differentiation, and apoptosis orchestra of transplanted NSCs regulated by nanovehicles in TBI mice. The imaging results also uncovered the peak time of NSC differentiation in vivo. Although we observed only a handful of NSCs ultimately migrated to the TBI area and differentiated into neurons, those neurons were functional, ameliorating the detrimental impact of TBI. The imaging findings enabled by the nanovehicle and the neuron specific bicistronic vector provide additional understanding of the in vivo behaviors of transplanted NSCs in neuronal regenerative medicine.
Keywords: stem cells, imaging, traumatic brain injury, differentiation, nanoparticles
Graphical abstract
Stem cells remain potent self-renewal and differentiation capacities that have long been employed as an alternative therapeutic method,1,2 or applied in regenerative medicine for replacement of damaged or dysfunctional tissues/organs.3,4 The recent advance in neural stem cell (NSC) transplantation has led to an inspiring progress in alleviating central nervous system (CNS) damages and/or restoring the brain function from diseases5,6 or injuries.7 The successful implementation of NSC transplantation holds the potential of reverting the neurodegenerative diseases or CNS disorders, not just attenuating the severity of those disruptive insults.8 NSCs are also actively involved in regenerative medicine for ex vivo generation of functional cerebral organoid mimicking brain cortex or neural tissue engineering.9,10 The unique merits of NSCs in promoting CNS disorder recovery, pathology tropism and multipotent differentiation into neuron subtypes in tissue engineering have drawn much attention from basic investigations to clinical studies.
In brain trauma or ischemia where free radicals and inflammatory cells exponentially accumulate after insults, transplanted NSCs need to acclimate to the harsh environment to survive and self-renew. For brain function recovery, NSCs need to migrate to the lesion regions, differentiate into correct phenotypic cells, secrete numerous relevant cytokines and integrate into the local neural circuits.11 The capacities of transplanted NSCs in implementing above tasks have been documented from in vitro cell cultures to in vivo animal models of various brain disorders.12,13 In addition to transplanted NSCs for neuronal damage recovery, endogenous neurogenesis in adult brain collaboratively contributes to the recovery. The subventricular zone (SVZ) of lateral ventricles and the hippocampal dentate gyrus are two major stem cell niches in adult brain.14 After the onset of neuronal disorder, such as traumatic brain injury (TBI), endogenous NSCs accumulate at the penumbra of the lesions along with exogenous NSCs driven by an array of injury induced secreted chemokines to alleviate the injury consequence and contribute to the injury recovery.15
To promote neuronal damage recovery, NSCs should partially differentiate into regenerated neurons whose close interaction with surrounding environment through axons and dendrites help reconstruct the disrupted neural circuit network at the injury site.16 Although the ultimate NSC differentiation and function in neuronal disorders has been investigated,17,18 how NSCs differentiate spatially and temporally and how they interact with the stem cell niche in adult brain remain virtually unexplored. Understanding this dynamic and multiplexed interaction process is at the frontier of regenerative medicine and of significance to design proper NSC transplantation protocols in neural damage recovery. In addition to the challenge of understanding the dynamic behaviors of transplanted NSCs in living subjects, another critical issue is to reliably control the differentiation of transplanted NSCs into specific neural subtypes, such as neurons. Numerous experimental protocols have been reported on the directed differentiation of stem cells including NSCs into various neural subtype cells.19,20 However, most of these studies are based on in vitro cell manipulation and differentiation. Strategies of realizing spatiotemporal control of in vivo differentiation of transplanted NSCs in animal models of neural damage have not been fully explored.
In this report, we used a mouse model of TBI to noninvasively investigate the differentiation dynamics of transplanted NSCs. To promote the differentiation of NSCs into neurons, we established a robust nanoparticle system (nanovehicle) to enable sustained retinoic acid (RA) release and stimulate NSC differentiation into specific neuronal subtypes. This nanovehicle is composed of biodegradable polyesters as the core and phospholipids as the protecting shell, making it fairly biocompatible as an effective carrier to deliver RA and subsequently control NSC differentiation in vivo. We developed this nanovehicle to achieve a sustained RA release kinetics directly in the cytosol of cells and to maintain a constantly well-balanced concentration of RA to facilitate NSC differentiation in vivo. The differentiation of NSCs relies much on the extracellular cues from RA during NSC differentiation. However, the low efficiency and short half-life of RA in cell culture media usually lead to the failure of NSC differentiation unless a high concentration (1 µM) of RA is used with prolonged incubation time (2–3 days), which can be potentially toxic to NSCs.21 Our designed nanovehicle avoids such serious side effects and, more importantly, enables substantial NSC differentiation into mature neurons in living animals. To track the differentiation of transplanted NSCs in real-time, we cloned a neuron development relevant gene tubb3 (βIII tubulin) promoter to drive reporter gene firefly luciferase expression for bioluminescence imaging. The imaging results showed a complex migration, proliferation, differentiation, and apoptosis in the course of homing transplanted NSCs to brain injury sites. We also discovered a pivotal role of ipsilateral SVZ in reserving and promoting differentiating stem cells homing to a cerebral cortex injury lesion.
RESULTS AND DISCUSSION
The potential of NSC in regulating the pathological recovery and CNS repair has been well recognized.22,23 Recent studies of using NSCs for the treatment of neurological disorders or brain injuries have validated that NSC transplantation is an effective and safe approach.24–26 Despite extensive efforts on stem cell research in CNS repair, however, most studies concentrated on the ultimate therapeutic outcome after transplantation. Little is known about the dynamic process of transplanted NSCs in vivo. Knowledge about the fate of transplanted NSCs, especially their migration and differentiation behaviors, is critical to understand the impact of microenvironmental cues and stem cell niches on exogenous NSCs. To bridge this gap, we developed a molecular imaging system to track the real-time differentiation and migration dynamics of transplanted NSCs homing to the TBI insult in a preclinical mouse model. In addition to constructing an in vivo dynamic imaging system, a robust nanoparticle platform (nanovehicle) was successfully created and characterized. This nanovehicle can effectively regulate differentiation of NSCs in vivo over a long period without comprising the potency of NSCs.
Design of a Nanovehicle for Sustained Release of Retinoic Acid To Induce Neural Stem Cell Differentiation
Retinoic acid, or RA, is one of the most potent morphogens widely used to induce stem cell differentiation into neuron subtypes.27–30 It is, however, mostly used for the induction of stem cell differentiation in vitro at a high dose (1 µM) and long incubation time (over 48 h) leading to undesired side effects.21,31 The high dose and long incubation time collectively reflect the rather low efficiency of free RA to induce stem cell differentiation. In addition, free RA is not easily delivered to cells due to its hydrophobicity (LogP = 6.7), and highly polar organic solvents such as DMSO may lead to cytotoxicity.
To study the differentiation dynamics of transplanted NSCs in TBI mouse brain, a highly robust RA delivery platform to facilitate NSC differentiation while keeping the potency of RA is urgently required. In this study, we designed a nanovehicle to enable efficient NSC uptake and sustained intracellular release of RA for a long period to collectively enhance RA potency in stimulating transplanted NSC differentiation in vivo.
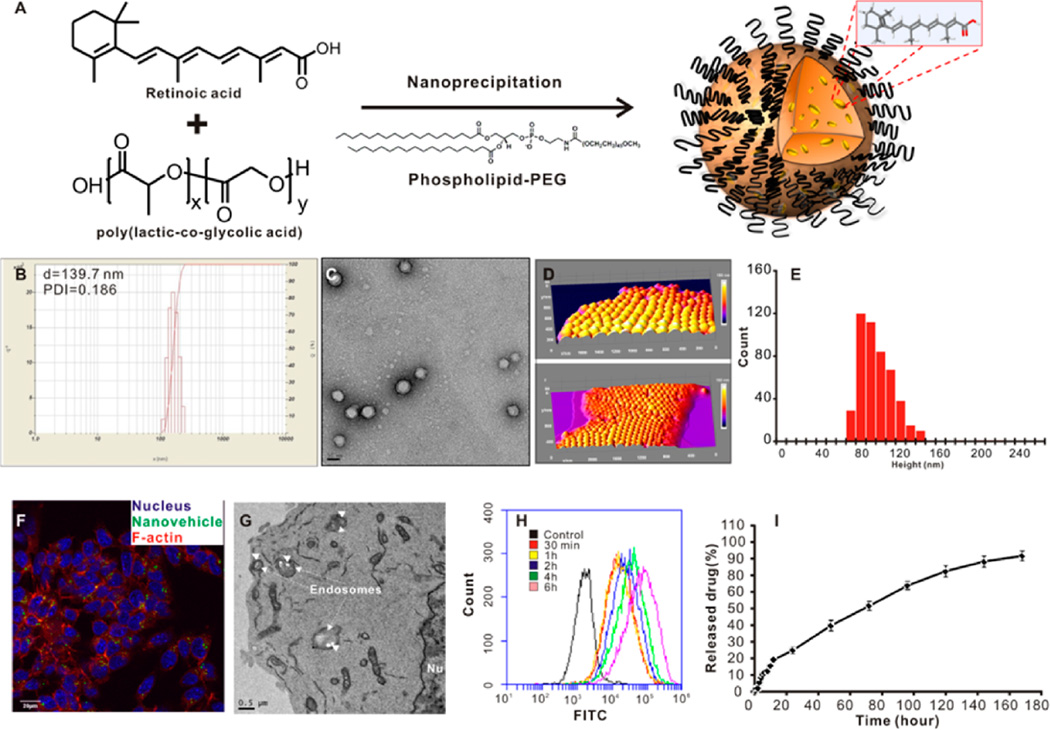
Biodegradable polyesters have been widely used to deliver a variety of drugs for disease management.32–34 In designing the RA delivery system, it is critical to employ a straightforward preparation method with few variables to ensure the high quality of the prepared nanovehicle. The excipients, which are usually used in nanovehicle preparation, should be free of any hazardous effects to intracellular organelles, while keeping the integrity of the nanovehicle for a reasonable period to realize sustained drug release. In our work, the matrix of this nanovehicle was composed of a biodegradable polyester PLGA.35 Another biocompatible material, phospholipid-PEG (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000 or DSPE-PEG), was used as the surfactant to facilitate the self-assembly of the nanovehicle.36 The hydrophobic molecule RA was sequestered in the polyester matrix during the self-assembly of nanovehicle via a nanoprecipitation method (Figure 1A). The average particle size is about 140 nm with fairly narrow size distribution (PDI = 0.186) (Figure 1B). The TEM (Figure 1C) and AFM (Figure 1D,E) images show that the nanovehicle is monodispersed with a core–shell structure. The loading efficiency (the mass of RA achieved in the nanovehicle/the mass of RA initially added in the preparation ×100%) of RA in the nanovehicle is 22.3% and the loading content (the mass of RA in the nanovehicle/the entire mass of nanovehicle loaded with RA × 100%) is 6.8% as estimated by HPLC. To study the NE-4C NSC uptake of this RA-carrying nanovehicle, we used confocal laser scanning microscopy (CLSM) (Figure 1F), TEM (Figure 1G), and flow cytometry (Figure 1H) to investigate the NSC uptake of fluorescently (6-courmrin) labeled nanovehicle. Results showed that NSC uptake of the nanovehicle was prominent into the cytosol after 2 h incubation in complete cell culture medium (Figure 1F,G). More nanovehicles were present in the cytosol of NSCs with longer incubation time (Figure 1H). This indicates the high efficiency of this nanovehicle to deliver RA into NSCs. The release profile of RA from this nanovehicle is another key factor determining its regulation potency in vivo. Figure 1I presents the sustained release profile of RA from the nanovehicle. The release kinetics of RA from this nanovehicle allows maintaining RA in an appropriate and constant local concentration in the cytosol to facilitate continuous NSC differentiation in vivo. In addition, the nanovehicle is highly biocompatible at up to 1 mg/mL concentration (Supporting Information Figure S1).
Figure 1.
Preparation and characterization of retinoic acid (RA) loaded nanovehicle. (A) Scheme of nanovehicle loaded with retinoic acid. (B) Dynamic light scattering analysis and (C) transmission electron microscopy image of RA-loaded nanovehicle. Scale bar = 50 nm. (D) Atomic force microscopy (AFM) image of RA nanovehicles at 1 nm per pixel resolution dried onto mica support in a 3D view (height scale bar, 225 nm). (E) The histogram of the height distribution of nanovehicles. In total, 480 nanovehicles were counted and analyzed by ImageJ with five thresholds for fwhm analysis (size = 0–15 000 pixel2, circularity = 0.50–1.00). The height frequency distribution is given by 91 ± 19 nm (mean ± SD). (F) NE-4C cellular uptake (2 h) of RA-loaded nanovehicle (100 ng/mL). Blue, DAPI; green, 6-coumarin; red, Alexa 568-conjugated phalloidin. Scale bar = 20 µm. (G) TEM image of NE-4C cell uptake of polymeric nanovehicles. Scale bar = 500 nm. (H) Flow cytometry analysis of nanovehicle uptake by NE-4C NSCs at different incubation time points. FITC channel was chosen. (I) Cumulative RA release profile from nanovehicles.
Efficient Induction of NSC Differentiation by RA-Loaded Nanovehicle
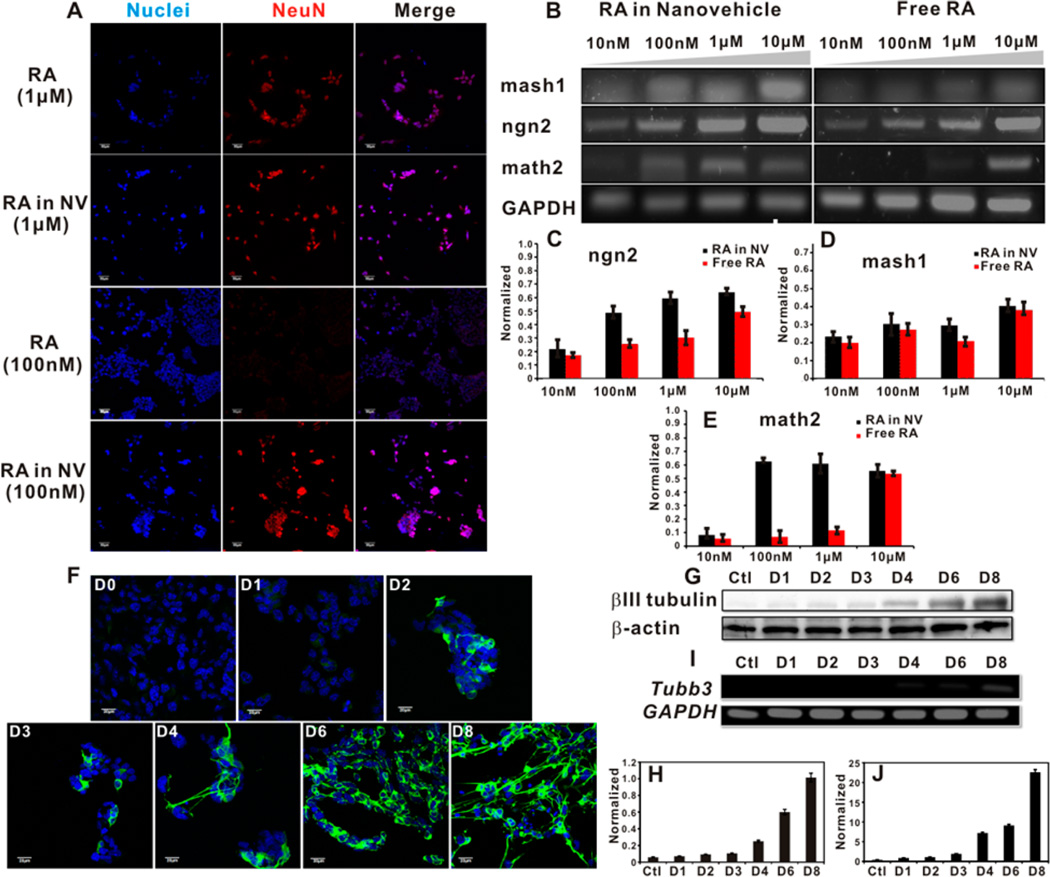
After constructing the RA-loaded nanovehicle, we studied the potency of this nanovehicle to induce NE-4C differentiation. In 8 days of continuous culture, both free RA and RA in nanovehicle at equivalent dose (1 µM) efficiently induced NE-4C differentiation into neurons, as marked by neuron marker, NeuN (Figure 2A). After incubation at a lower RA concentration (100 nM), RA-loaded nanovehicle managed to induce NE-4C differentiation, similar to that of 1 µM RA, while free RA at 100 nM failed to induce NeuN expression in NE-4C cells (Figure 2A). We next investigated the dose dependent gene expression of differentiating NSCs stimulated by free RA and RA-loaded nanovehicle (Figure 2B–E). The results showed that both free RA and RA-loaded nanovehicle at equivalently high doses (1 and 10 µM) were able to induce pro-neuron marker expression (mash1 and ngn2). For the mature neuron maker gene, math2, RA-loaded nanovehicle at 100 nM efficiently induced its expression, while free RA can only achieved such high expression level at the dose of 1 µM or higher. The dramatic dose difference in stimulating NSC differentiation confirms the advantages of RA-loaded nanovehicle over free RA in long-term neuron differentiation induction. The spatially positioned and locally high concentration of RA in NSCs over time significantly enhanced RA transportation into the nucleus to bind with transcription factors in initiating stem cell differentiation.37 This RA-loaded nanovehicle therefore is robust for long-term NSC differentiation studies at much lower but effective concentrations. All the following experiments used 100 nM RA-loaded nanovehicle to study the dynamics of NE-4C NSC differentiation.
Figure 2.
RA-loaded nanovehicle induces efficient neural stem cell differentiation at low concentration. (A) Confocal laser scanning microscopy imaging of NE-4C NSC differentiation by free RA and RA-loaded nanovehicle at 1 µM (two upper panels) or 100 nM (two lower panels) concentrations. Scale bar = 50 µm. NeuN: neuron specific marker stained by Cy3-conjugated secondary antibody. (B–E) PCR analysis of effect of free RA and RA-loaded nanovehicle on inducing NE-4C NSC differentiation. The pro-neuron markers mash1 and ngn2 and mature neuron marker math2 were measured. A 100 nM RA-loaded nanovehicle was able to efficiently induce NE-4C differentiation similar to the level of 1 µM free RA. GAPDH was used as the housekeeping gene. (F) Immunofluorescence staining of βIII tubulin expression in differentiating NE-4C NSCs stimulated by 100 nM RA-loaded nanovehicle. The βIII tubulin staining was illustrated by Alexa 488-conjugated secondary antibody. Scale bar = 20 µm. (G–J) Western blot (G and H) and PCR (I and J) analysis of βIII tubulin in differentiating NE-4C NSCs stimulated by 100 nM RA-loaded nanovehicle.
To track the in vitro expression of neuron specific marker, βIII tubulin, we incubated the nanovehicle (100 nm RA equivalent) with NSCs and stained the target protein by immunofluorescence staining at specific time points (Figure 2F and Supporting Information Figure S2). The imaging results show that the onset of sporadic βIII tubulin expression was on day 2, and its expression became prominent until the formation of mature neurons on day 8. Meanwhile, the βIII tubulin protein level (Figure 2G,H) and gene expression profile (Figure 2I,J) were tested using immunoblotting assay and PCR, respectively. Both results are consistent with the immunofluorescent staining data (Figure 2F and Supporting Information Figure S2) on the temporal expression of βIII tubulin of differentiating NSCs stimulated by the RA-loaded nanovehicle.
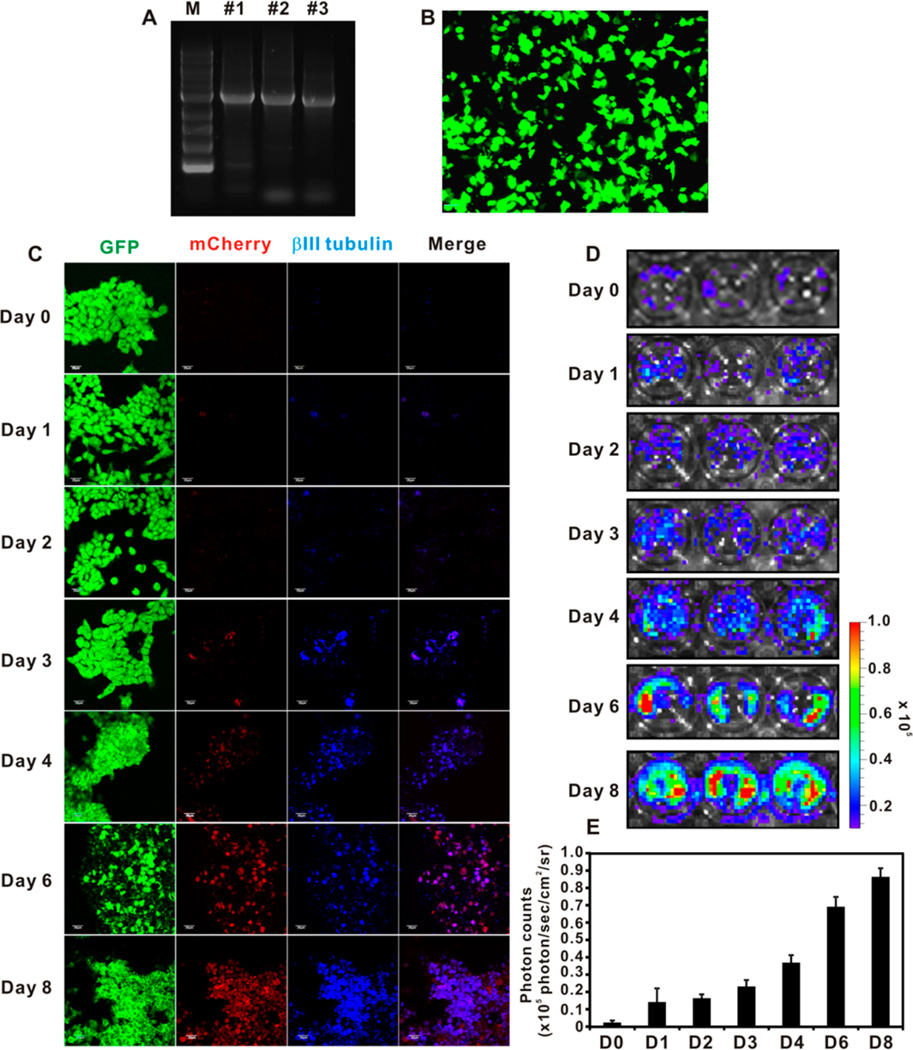
Cloning of tubb3 Promoter to Construct Bicistronic Vector
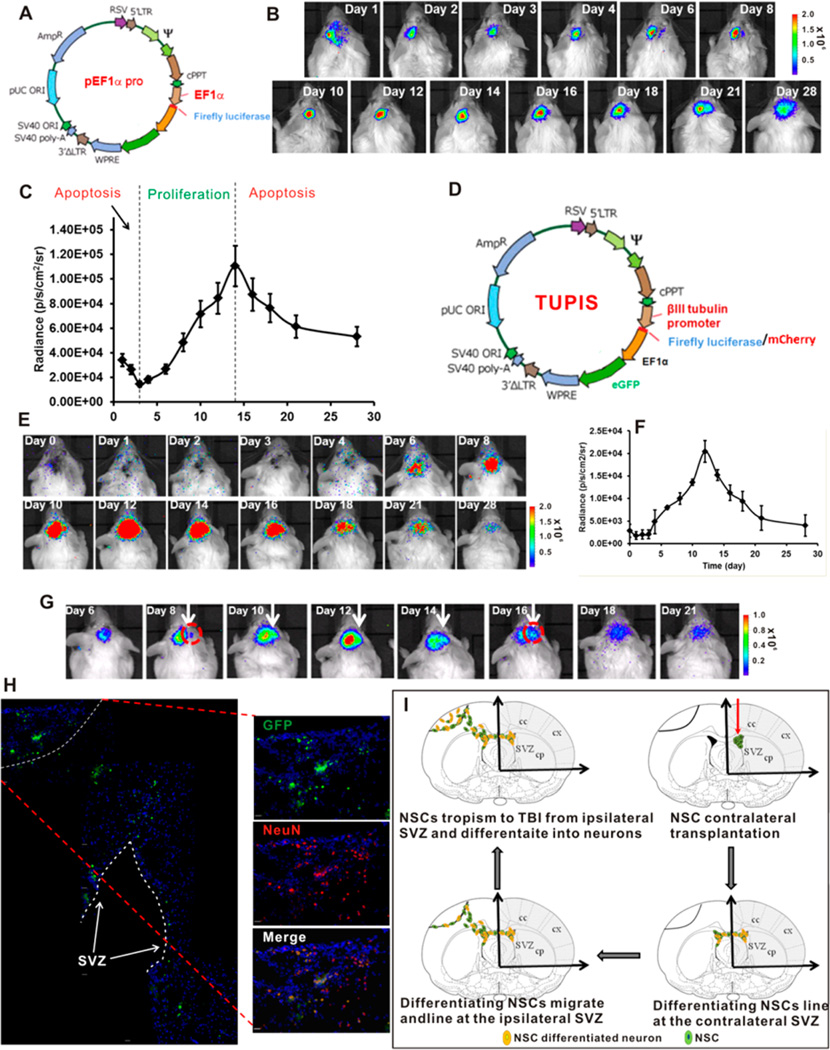
βIII tubulin is a neuron specific maker whose expression is proportional to the neuron differentiation status. It is absent in the mature NSCs but is expressed in both differentiating NSCs and differentiated mature neurons.38 Thus, the use of βIII tubulin promoter to drive reporter gene (fLuc) expression can facilitate imaging the dynamic differentiation status of transplanted NSCs in vivo. The tubb3 promoter was cloned directly from mouse genome DNA chromosome 8 covering the 2920 bp of the upstream region of the βIII tubulin starting codon without further optimization. Figure 3A shows the cloned promoter from three different brain tissue genome DNA collections. The #2 clone was used in the entire study. The bicistronic construct tubb3 promoter imaging system (TUPIS) in lentivirus was able to efficiently transduce NSCs as shown in Figure 3B. In this vector, the fLuc reporter gene was used mainly to track NSC differentiation, and the eGFP was independently expressed by a separate EF1α promoter at the downstream of the inserted reporter gene as an endogenous marker. To validate the correlation of the expression of TUPIS to the NSC differentiation condition, we used the RA-loaded nanovehicle (100 nM RA) to stimulate the TUPIS vector transduced NSCs with mCherry as the reporter gene (Figure 3C) for fluorescence microscopy imaging analysis. The results show consistent colocalization of βIII tubulin expression (blue) with mCherry (red) during the differentiation of NSCs. In another independent experiment, in which fLuc was used as the reporter gene for bioluminescence imaging, the imaging results confirmed the dynamics of TUPIS along with NSC differentiation (Figure 3D,E). Therefore, the TUPIS system was validated to represent the NSC dynamic differentiation progress under long-term, low-dose nanoformulated morphogen stimulation.
Figure 3.
Construction of bicistronic vector with tubb3 promoter driven reporter gene. (A) Gel electrophoresis of the cloned tubb3 promoter from three independent brain tissue extracts. The #2 clone was used in the entire study. (B) Fluorescence image of lentivirus transduced NE-4C NSCs with tubb3 promoter driven fLuc reporter gene and independent EF1α driven GFP reporter gene. Scale bar 50 µm. (C) Confocal laser scanning microscopy imaging of NE-4C NSC differentiation by 100 nM RA-loaded nanovehicle over 8 days. βIII tubulin was stained by Cy5.5-conjugated secondary antibody. The GFP and mCherry were endogenously expressed fluorescence proteins in transduced NSCs. Scale bar = 50 µm. (D) Bioluminescence imaging (BLI) of tubb3 promoter driven fLuc expression in transduced differentiating NSCs stimulated by 100 nM RA-loaded nanovehicle over 8 days. (E) Analysis of BLI results (n = 3).
Proliferation, Migration, and Differentiation of Contralaterally Transplanted NSC in TBI Mouse
We first investigated the proliferation and migration pattern of contralaterally transplanted NSCs in TBI mice as shown in Figure 4A–C. The results show that NSCs after transplantation experienced a tri-phase status during their self-renewal and migration to the TBI lesion. Immediately after transplantation, NSCs suffered from the first apoptotic condition in which NSCs acclimated to the new microenvironment; then, the NSCs underwent a steady proliferating period accompanied by the tropism to the injured semisphere starting at day 6 and afterward. After a proliferation peak (about 2 weeks after transplantation), the cellular apoptosis took the dominant role, leading to decreased activities until a balanced status was reached after 28 days (Figure 4B).
Figure 4.
Spatiotemporal imaging of the dynamics NE-4C NSC differentiation. (A) Map of vector used for tracking the migration, proliferation and apoptosis patterns of contralaterally transplanted NSCs in TBI mice. (B) BLI results of contralaterally transplanted NSCs in TBI mice over 28 days. (C) Analysis of differentiation stages (proliferation and apoptosis) of contralaterally transplanted NSCs in TBI mice. (D) Map of bicistronic vector with tubb3 promoter driven reporter gene and independent EF1α driven GFP. Drawing is not in scale. (E) BLI of the dynamic differentiation of NE-4C NSCs stimulated by 100 nM RA-loaded nanovehicle over 28 days. (F) Analysis of the differentiation pattern of BLI results. (G) BLI of the TBI tropism pattern of differentiating NSCs stimulated by 100 nM RA-loaded nanovehicle. (H) Montage of immunofluorescence staining of brain sections illustrating the migration pattern of differentiating NSCs at day 21. Scale bar = 50 µm. (I) Proposed NSC migration, proliferation, apoptosis and differentiation patterns of 100 nM RA-loaded nanovehicle stimulated NSCs in TBI mice. Drawing is not in scale.
To track the differentiation of NSCs, lentivirus transduced NSCs were transplanted in the lateral ventricle of the contralateral semisphere after incubation with RA-loaded nanovehicle for 24 h. Figure 4D–F shows the vector map and imaging results of differentiation pattern of NSC in vivo. In contrast to the initial stage of cell proliferation imaging results showing decreased signal due to apoptosis, no BLI signal was observed in the first 4 days indicating the relative quiescence of NSCs before starting differentiation. This observation is consistent with the in vitro study in which significant βIII tubulin protein expression was observed on days 6 and 8 of cell culture (Figure 2F and Supporting Information Figure S3). After experiencing the initial gene transcription activation period, dramatic increase of BLI signal driven by tubb3 promoter was observed. The differentiation peaked at day 12 after transplantation and returned to the background level after 28 days (Figure 4E,F). Figure 4G shows a series of high magnitude images of NSCs at different stages of differentiation, which demonstrates that NSC differentiation is accompanied by the progressive migration to the TBI lesion semisphere amid of cellular apoptosis as shown in Figure 4B,C. Immunohistochemical staining of the brain slide (Figure 4H and Supporting Information Figure S3) indicates that contralaterally transplanted NSCs are not directly attracted to the TBI insult, but are mainly deposited at the ipsilateral lateral ventricle after migrating through the corpus callosum channel. A portion of migrating cells finally reside at the periphery of TBI injury site and evolve into neurons as indicated by the neuron specific marker NeuN expression (Figure 4H). The entire NSC migration and differentiation process is outlined in Figure 4I.
By using the TUPIS vector in conjunction with the optimized RA-loaded nanovehicle, we were able to track the entire differentiation process of transplanted NSCs; identify the differentiation peak time (12 days after transplantation); and reveal the migration, differentiation, and apoptosis conditions (Figure 4). Instead of directly migrating to the TBI site, the contralaterally transplanted NSCs mainly accumulate in the ipsilateral lateral ventricle, followed by lining up on the subventricle zone (SVZ) before they gradually migrate to the penumbra, as well as the interior of the TBI site.
Improved Neuron Circuit Recovery after NSC Transplantation
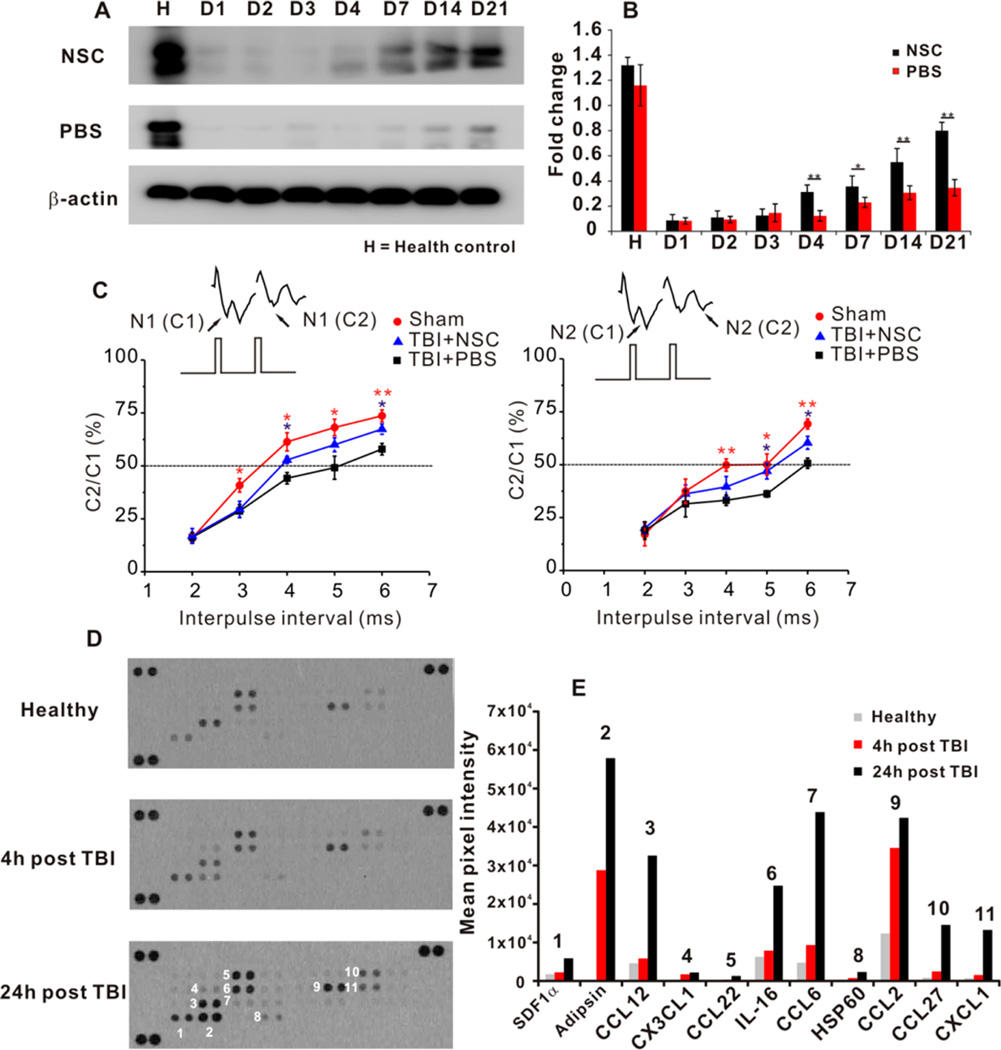
After we have observed significant NSC self-renewal, migration, and differentiation in the presence of optimized RA-loaded nanovehicles in the TBI area, we then explored the neuron circuit recovery capacity of migrated and differentiated NSCs at the TBI site. Figure 5A shows the dramatically enhanced expression of synapsin, a key protein regulating neurotransmitter release at synapses, in the NSC transplanted group, while samples from PBS treated group have limited expression over a long time. The electrophysiology on the whole brain slices (Figure 5B) consistently confirmed that the NSC transplantation significantly improved the neuron circuit connectivity with enhanced signal conductivity in both N1 and N2. A prior report of 20% improvement of synapsin I on the ipsilateral cortex of brain trauma by brain-derived neurotropic factor (BDNF) therapy is consistent with our results.39 The synapsin expression is indicative of neuron circuit recovery during the brain injury recovery. Electrophysiology analysis showed divergent effects on two components (N1 and N2), in which N1 presents signal from myelinated axons, and N2 indicates signal transduced from unmyelinated axons. Contralaterally transplanted NSCs ultimately helped the neuron conductivity recovery from both myelinated and unmyelinated axons. It was reported that NSC transplantation is beneficial to neuron myelination to enhance neuron disorder recovery.24,25 Furthermore, the highly differentiated NSCs into mature neurons at the TBI site could help re-establish the disrupted neuronal connection,16 leading to N1 signal enhancement.
Figure 5.
Transplanted NSCs in improving neuron circuit recovery and chemokines released at TBI for NSC tropism. (A and B) Western blot analysis of transplanted NSCs in improving synapsin expression at the cerebral cortex of the TBI lesion hemisphere in 3 weeks (n = 3). *P < 0.05, **P < 0.01. (C) Electrophysiology analysis of axon connectivity on brain slices from TBI mice at day 21. N1 indicates signals conducted from myelinated axons and N2 represents signals obtained from unmyelinated axons (n = 5–6/group, *P < 0.05 vs PBS, **P < 0.01 vs PBS). (D and E) Mouse chemokine array analysis of TBI hemisphere at 4 and 24 h after the onset of lesion. The film exposure was 2 min and all procedures were identical for the three membranes. The pixel intensity from the obtained membranes was analyzed by ImagJ software in arbitrary unit.
Chemokines Released after TBI for NSC Tropism
To study the chemokines released after brain injury, we used mouse chemokine assay kit as shown in Figure 5C. The released chemokines in both expression levels and types significantly increased at 24 h after TBI, when we transplanted NSCs. In addition to the well-known SDF1α, most released chemokines are highly related to recruiting T-cells (CCL2, CCL27, IL-16) and neutrophils (CXCL1, CCL6). Additionally, the extremely high expression of complement factor D (Adipsin) indicates the remarkable proteolytic events at the TBI site.
At 24 h after the onset of TBI, most chemokines are released to induce inflammatory reactions to recruit various types of immunocytes, such as T cells or neutrophils. We also observed the high expression of SDF1α, which was reported as the major driving force to attract NSC migration.40,41 The overexpression of proteolysis marker, Adipsin, implies the harsh microenvironment of the TBI site. This may partially explain the fact that only limited number of transplanted NSCs that could ultimately migrate, survive and highly differentiate into neurons in assisting brain injury recovery.
RNA Transcription Sequencing Analysis of TBI with Transplanted NSCs
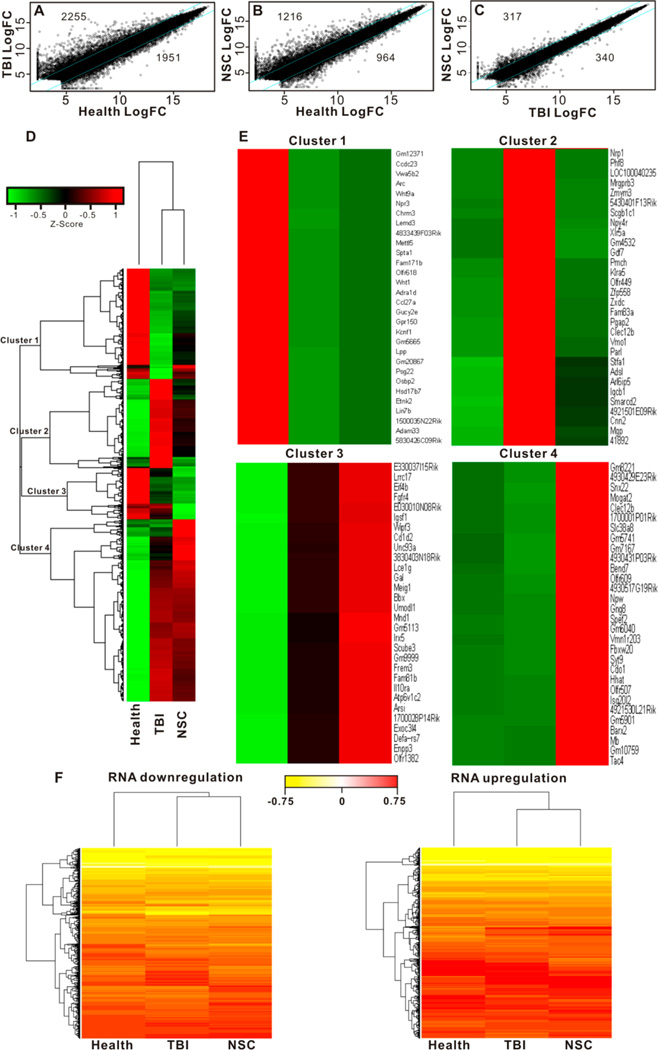
We have limited knowledge about the overall transcription profile in mouse TBI and the intervention effect of transplanted NSCs. Figure 6 provides the gene alteration plots and heat maps of transcription change in the brain area of TBI insult with or without NSC intervention. At the 2.5-fold significant change threshold, TBI resulted in 2255 transcript increase and 1951 transcript decrease in comparison to healthy control (Figure 6A), and the polymeric nanovehicle regulated NSC intervention led to 1216 transcript increase and 964 transcript decrease (Figure 6B). By comparing the TBI mice with and without NSC intervention, 317 gene transcriptions were upregulated, while 340 transcriptions were downregulated (Figure 6C). Hierarchical cluster analysis grouped a total of 4259 genes into 4 different clusters based on the similarity of expression patterns (Figure 6D). Genes in clusters 1 and 2 showed significant difference between TBI and NSC transplantation, indicating that the persistence of NSCs after TBI helped partial gene recovery. In contrast, gene expression in clusters 3 and 4 presented no statistical difference between TBI and NSC transplantation, implying that those genes in these two clusters can hardly be restored by NSC transplantation after TBI insult (Figure 6E). Figure 6F presents the overview of genes downregulated (left panel) and upregulated (right panel) in different experimental groups. Supporting Information associated with this manuscript provides elaborated information about specific gene transcript rankings and genetic codes. Overall, the transplanted polymeric nanovehicle regulated NSCs ameliorated the detrimental impact of TBI by partially rescuing genetic transcripts contributing to the genetic network of the damaged brain functions.
Figure 6.
RNA sequencing analysis of TBI and NSC intervention. (A) Scatter plots of fold changes (normalized log2 value) were used to assess the change of mRNA in TBI group and NSC treatment group. The values above the top cyan line and those below the bottom cyan line indicate >2.5-fold change in expression. Totals of 2255 genes increased, and 1951 genes decreased in TBI insult. (B) Comparison between NSC treatment and health control: 1216 gene transcripts increased, and 964 gene transcripts decreased in the NSC treatment group. (C) Comparison between NSC treatment and TBI insult: 317 gene transcripts were higher, and 340 ones were lower in the NSC treatment group. (D) The overall heat map of 4259 genes that displayed >2.5-fold changes between health control and TBI insult or NSC treatment. Hierarchical cluster analysis grouped a total of 4259 genes into 4 different clusters based on the similarity of expression patterns. (E) Top 30 expression genes in 4 clusters. Hierarchical clustering was conducted based on the complete linkage Euclidean distance of the Spearman correlation of the Z-score-transformed value of fold of changes. Green indicates lower expression and red indicates higher expression. (F) Heat maps of the overview of RNA transcription downregulation (left panel) and upregulation (right panel) associated with TBI and NSC intervention.
CONCLUSION
In this study, we developed a nanovehicle to encapsulate RA, a potent morphogen, to effectively induce NSC differentiation at 100 nM RA concentration in vivo. To enable real-time imaging of NSC differentiation, we cloned a neuron specific gene, tubb3, promoter to drive reporter gene expression to follow the entire differentiation process. We discovered at 2 weeks after NSC transplantation abundant NSC differentiation with increased apoptosis afterward. NSCs transplanted in the contralateral hemisphere underwent an indirect pathway to the TBI, viz., they preferentially accumulated in the ipsilateral SVZ before migrating to the injury site. NSC differentiation is a continuous process mixed with cell migration and apoptosis. Although a limited number of transplanted NSCs can finally accumulate at the TBI site. The transplanted NSCs partially reconstructed the damaged neuron circuit and improved the axon signal conductivity. The transplanted NSCs helped ameliorate the negative impact of TBI and partially rescued gene transcripts regulating brain functions. We envision that the discoveries in this report are beneficial for improving NSC therapeutic effects in TBI recovery.
MATERIALS AND METHODS
Preparation of Controlled Release Formulation for Retinoic Acid (RA)
The sustained RA release formulation was prepared by a nanoprecipitation method following the previously published protocol.42–44 Briefly, 1 mg of RA (Sigma-Aldrich) and 10 mg of poly(lactic-co-glycolic acid) (PLGA) (Sigma-Aldrich) were dissolved in 2 mL of acetonitrile (HPLC grade, Sigma-Aldrich). A solution of 4% 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(methoxy(polyethylene glycol)-2000) (DSPE-mPEG) (Avanti Polar Lipid, Inc.) in 1 mL of ethanol (Sigma-Aldrich) was diluted in 9 mL of hot water (65 °C) to form a uniform aqueous phase. With moderate magnetic stirring, the acetonitrile solution was added dropwise (1 mL/min) into the aqueous solution, followed by rapid magnetic stirring for 4 h. The resulting nanovehicle was washed three times with PBS using ultracentrifugation (100 K MWCO, 4000 rpm, 20 min). The final RA formula was dispersed in sterile PBS as a stock solution. The loaded RA was analyzed by HPLC: gradient mobile phase with water (0.1% TFA)/acetonitrile (0.1% TFA) 20%/80%–10%/90% over 12 min at 360 nm detection wavelength. The concentration of RA was determined using a RA calibration curve.
Construction of Vectors
Mouse βIII tubulin promoter was cloned from the genomic DNA (Chromosome 8:119906841–124345722) of the Balb/C mouse brain tissue. QIAamp DNA Mini Kit (Qiagen, Germany) was used to isolate the genomic DNA from brain tissue lysates according to the manufacturer's instruction. The promoter region extending 2920 upstream region of the βIII tubulin starting codon ATG was cloned from the extracted genomic DNA using the following primers: forward, 5′-GAACACTAGTAGCAAGGGTCAGACCACAG-3′; reverse, 5′-GTAAGAATTCGCTGACTTCACGCGGCTAGAG-3′. The underlined letters indicate the restriction enzyme sites, SpeI and EcoRI, respectively. For the PCR amplification, Expand High Fidelity PCR System (Cat# 11732641001, Roche) was used. The PCR conditions were set as the following: 30 cycles, 94 °C denature for 30 s, 62 °C annealing for 45 s, 72 °C extension for 180 s, and final 72 °C extension for 15 min. The cloned promoter was inserted into a lentivirus dual-promoter vector (Cal#: CD711B-1, System Biosciences) to replace the murine stem cell virus (MSCV) promoter on the backbone. The firefly luciferase reporter gene (fLuc) was cloned using the following primers: forward, 5′-GACCGAATTCGCCACCATGGAAGACGCCAAAAAC-3′; reverse, 5′-GGCGCGGATCCTTACACGGCGATCTTTCC-3′. The underlined letters indicate the restriction enzyme sites, EcoRI and BamHI, respectively. The following PCR conditions were used for amplification: 30 cycles, 94 °C denature for 30 s, 60 °C annealing for 45 s, 72 °C extension for 60 s, and final 72 °C extension for 15 min. The cloned fLuc DNA segment was inserted into the pCDH dual promoter backbone following closely to the βIII tubulin promoter sequence to yield the final pTubb3pro-fLuc vector. In this promoter, GFP expression was independently driven by EF1α promoter in the downstream of fLuc segment within the same single vector. The entire subclone sequence was confirmed by DNA sequencing. To prepare lentivirus for neural stem cell transduction, the packing plasmid pPAX2 and envelope plasmid pVSV-G were used in conjunction with pTubb3profLuc shuttle vector for lentivirus preparation. The HEK293T/17 cell line was used in the preparation procedure. The final copy number of prepared lentivirus was titrated by Lenti-X p24 Rapid Titer Kit (Clonetech) to be 107–108.
Neural Stem Cell Transduction
The NE-4C neural stem cell line established from cerebral vesicles of p53−/− mouse E9 embryos was obtained from the American Type Culture Collection (ATCC). The NE-4C cells were routinely cultured in Minimal Essential Medium Eagle (MEM Media) (Sigma-Aldrich) supplemented with 5% fetal bovine serum (FBS) (Gibco), 4 mM glutamine (Life Technologies), 100 unit/mL antibiotics (Life Technologies) at 37 °C with 5% CO2. For lentivirus transduction, no antibiotics were added in the culture medium. NSCs were transduced using above prepared lentivirus with polybrene (10 µg/mL) for 6 h. Then, fresh culture medium was added and cells were further incubated overnight. The next day, the sample transduction procedure was repeated. After culturing in the fresh medium for 48 h, the NSCs were washed twice with ice-cold PBS and harvested for injection.
Characterization of Nanovehicles
The average particle size and distribution of the nanovehicle were measured by dynamic light scattering (DLS) zetasizer (Marvin NanoZS90). The transmission electron microscope (TEM) images were obtained from Philips/FEI CM200. The atomic force microscope (AFM) imaging of the nanovehicle samples was carried out under a range of deposition conditions on freshly peeled mica surface under fluid. The excess fluid was blown away by an inert gas gush, followed by a complete drying of the sample surface under a gentle gas flow. The dried sample was imaged via standard optimizations and a range of imaging settings, e.g., using gentle tapping-mode and mostly with a Multimode AFM (Bruker, CA) consisting of a Nanoscope V controller, a type E scanner head, and a sharpened TESP-SS (Bruker, CA) or similar AFM cantilever. To enhance reliability for fuller comparison, the sample surfaces were often rinsed up to 2 times each with ca. 100 µL of deionized water to remove salt deposits, redried under gentle air flow, and reimaged similarly. AFM images were evaluated within the Nanoscope software (version 7.3–8.15, Bruker, CA), and exported to ImageJ (version 1.4x) for size analyses and 3D display.
For the TEM imaging of cellular uptake of nanovehicles, cells grown on thermonax coverslips were fixed in a mixture of 2.5% paraformaldehyde and 2.0% glutaraldehyde in 0.1 M sodium cacodylate buffer (SCB; pH = 7.4) for 1 h followed by extensive wash in SCB. Then samples were postfixed in 1.0% osmium tetroxide plus 0.8% potassium ferricyanide in the above buffer for 60 min. After several rinses in the SCB, the samples were dehydrated in a series of ethanol solutions (30%, 50%, 75%, and 95% for 5 min and 100% for 20 min with 3 changes) and infiltrated with Epon-Aradite (50% for 1 h and 100% for 1 day with 2 changes) (Ted Pella, Inc.). Samples were polymerized at 60 °C for 1 day.
Ultrathin sections (about 80 nm) were cut on Leica EM UC6 Ultramicrotome and collected on copper slot grids. Sections were counter-stained with uranyl acetate and lead citrate, and examined under a FEI Tecnai12 TEM (FEI) operated at beam energy of 120 keV. Images were acquired by using a Gatan 2k × 2k cooled CCD camera.
Cellular Uptake of Nanovehicle in NSCs
NSCs were cultured on polylysine precoated 8-well Lab-Tek chamber at the density of 2 × 104 cells/well overnight. Cells were incubated with 100 nM RA-loaded nanovehicle (1% 6-courmarin was added in the nanovehicle preparation as the marker for fluorescence imaging) for a predetermined time. Then, cell monolayer was washed 3 times with ice-cold PBS, followed by 3.7% neutralized formalin solution fixation for 10 min and 0.1%Triton X-100 penetration for 5 min. Alexa 568 conjugated phalloidin prediluted solution in 0.5% BSA PBS was incubated with cells for 40 min, followed by washing in 0.5% Tween-20 PBS (TPBS) solution 3 times, and cells were mounted with Vectashield mounting medium with DAPI (Vector Laboratory). The fluorescence images were taken from an Olympus FV10i confocal laser scanning microscopy (CLSM) using a 60× objective lens.
Immunoblot Assay
Cultured cells were lysed in RIPA buffer (Thermo Fisher) with proteinase inhibitor cocktail (Roche). For brain tissues, mouse was perfused from heart with 20 mL of saline before dissection of the injured brain issue. The dissected brain tissue was lysed in T-Per buffer (Thermo Fisher) with proteinase inhibitor cocktail (Roche). The lysate was gently sonicated using probe sonicator (Sonic Ultracell) on ice, followed by centrifugation at 10 000 rpm (5 min at 4 °C). The supernatant was retained and protein concentration was measured by a BCA Protein Assay Kit (Pierce) using BSA as the standard. The precasted 4–12% Bis-Tris minigel (Invitrogen) was used. The following primary antibodies were used to blot the transferred protein on PVDF membranes: βIII tubulin (1:1000, cat# ab18207, Abcam); Synapsin (1:1000, cat# 2312, Cell Signaling); β-actin (1:5000, cat# 8456, Cell Signaling). The blotting signals were captured from an Amersham Imager 600 (GE Healthcare) device, and semiquantified using ImageJ (NIH).
PCR Assay
mRNAs of cell culture or dissected brain issue were extracted by TRIzol reagent (Life Technologies, MD) following the manufacturer's protocol. The harvested total mRNAs were converted into cDNA by SuperScript III First-Strand Synthesis System (Invitrogen) according to the manufacture's instruction. The primers used in the PCS assay were as follows: Tubulin, β3 class III (Tubb3), forward 5′-CTTTTCGTCTCTAGCCGCGT-3′; reverse 5′-CTCATCGCTGATGACCTCCC-3′; mash1, forward 5′-TTAGTCCAGAGGAACAAGAGCTGC-3′; reverse 5′-AAGATGCAGGATCTGCTGCCATCC-3′; ngn2, forward 5′-AAGAGGACTATGGCGTGTGG-3′; reverse 5′-ATGAAGCAATCCTCCCTCCT--3′; math2, forward 5′-TGAGAATGGCTTGTCCAGAAGG-3′; reverse 5′-TGGTAGGGTGGGTAGAATGTGG-3′; GAPDH, forward 5′-ATCATCCCTGCATCCACT-3′; reverse 5′-ATCCACGACGGACACATT-3′.
Chemokine Array Assay
The proteins were analyzed for chemokine expression based on the instructions provided by the commercially available Mouse Chemokine Array kit (Proteome profiler Array) from R&D systems. Briefly, the proteins were isolated from different time points following treatment of the mice with NSCs. As a control, tissue from healthy mouse brain was used for the study. Protein concentration used for the study was calculated to be about 200 µg per membrane, calculated using the BCA Protein Assay kit. The experiment was performed in 4 well multidishes. Initially, the membranes containing the antibodies were blocked with blocking buffer for 1 h at room temperature. Concurrently, the detection antibody cocktail was reconstituted in buffer and added to the protein samples, incubating for 1 h at room temperature. Following the incubation, the membranes were washed 3 times with wash buffer and incubated with the secondary antibody (Streptavidin-HRP) for 30 min at room temperature. After the incubation time was complete, the secondary antibody was drained off and the membranes were washed again 3 times. The membranes are covered with the Supersignal West Pico Chemiluminescent substrate (Pierce) for detection and exposed to X-ray films for multiple exposure times. The membranes are compared for relative chemokine expression.
Traumatic Brain Injury (TBI) Mouse Model and NSC Contralateral Transplantation
Controlled cortical impact (CCI) model of TBI was conducted in adult male balb/c mice (6–7 weeks old). In short, mouse was anesthetized with 3% isoflurane at the beginning of the experiment. When the breath was smooth, 1.5% isoflurane was maintained throughout the entire surgery. The brain was fixed on a stereotaxic device. A 4 × 4 mm hole was drilled on the right skull, and a 2 mm diameter impactor tip was used to contuse the brain tissue. The impactor tip penetration depth was 2 mm with the velocity of 5 m/s. The mouse was kept on a warming pad at 37 °C during the surgery. The skull was discarded after CCI and the skin was sealed using 6− 0 suture. The mouse after surgery was kept under a warming light until awake. The mouse condition was closely monitored before returning to animal facility.
For the contralateral transplantation of NSCs after TBI, NSCs were transduced twice with lentivirus and incubated with equivalent amount of 100 nM RA-loaded nanovehicle for 24 h as described earlier. At 24 h after TBI onset, mouse was anesthetized initially with 3% isoflurane and maintained in 1.5% isoflurane during procedure. The transduced NSCs were harvested and prepared as 1 × 106 cell/mL in ice-cold PBS. Five microliters of cell solution (~5 × 103 cells) was gradually injected into the contralateral lateral ventricle (rostral, 0.5 mm; lateral, 0.75 mm; ventral, 2.5 mm) of TBI mouse using 10 µL volume Hamilton microinjection needle on a stereotaxic device. The needle was finally kept in place for 5 min before being gently withdrawn.
The animal studies were performed according to the principles and procedures outlined in the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Clinical Center, NIH.
Bioluminescence Imaging (BLI)
For imaging in cell culture, lentivirus transduced NSCs were initially cultured in 24-well plate at cell density of 0.4 × 105/well. The next day, RA-loaded nanovehicle was added into cell culture medium and incubated for 24 h, followed by changing fresh cell culture medium. At predetermined time points, 10 µL of d-luciferin (30 mg/mL) was added into each well. IVIS Lumina Series III (Caliper) was used for bioluminescence imaging. The following setting was used to obtain imaging data: exposure time = 1 min, bin = 1 × 1. Regions of interest (ROIs) were drawn and analyzed to quantify photon counts.
For in vivo bioluminescence imaging, mice were first subject to contralateral NSC transplantation as described in the above methods. At the predetermined time points, mice were anesthetized initially with 3% isoflurane and maintained in 1.5% isoflurane during the entire imaging procedure. A total of 150 µL of d-luciferin (30 mg/mL) was injected intraperitoneally in each mouse and we waited for 5 min for sufficient body circulation before imaging. Images were acquired at exposure time = 5 min, Bin = 1 × 1 for 25 min. The images with the highest captured signal were used for illustration.
Immunohistochemical (IHC) Staining
At designated time point, mice were perfused with 4% polyformaldehyde (PFA) (20–30 mL) before dissection of brain tissue. The brain tissue was dehydrated with 30% sucrose 24 h before being embedded in Tissue Tek OCT (Sakura Finetek), followed by snap freezing in liquid nitrogen. The OCT embedded tissue blocks were kept at −80 °C for cryosection. The obtained brain slices (15 µm-thick) were fixed in Z-fix solution (Anatech) for 10 min, and permeabilized with 0.1% Triton X-100 for 5 min. To stain the mature neuron cells at the periphery of TBI site, a mature neuron marker, NeuN, was chosen. Anti-NeuN primary antibody was obtained from Cell Signaling Technology (1:150, Cat# 12943). The Cy3-conjugated donkey anti-rabbit secondary antibody (1:200, Cat# 711165-152, Jackson ImmunoResearch) was used. The nuclei were counterstained with DAPI using Vectashield mounting medium (Vector Lab). The images were taken from Olympus X81 fluorescence microscope.
Brain Slice Electrophysiology
A brain slice of 400 µm thickness through craniotomy section was fixed in a chamber perfused with 2 mL/min artificial cerebrospinal fluid (ACSF) supplied with 95% O2/5% CO2 at 30 °C. Callosal recording was performed under the TBI area. The bipolar stimulating electrodes were positioned at the corpus callosum, and recording micropipette was placed in the contralateral hemisphere. The slices were stimulated by monophasic square wave pulse at 250 µA and 200 µs duration at one stimulating pulse every 10 s. All signals were recorded online and processed off-line using Clampex (version 10.1) program.
mRNA Sequencing and Analysis
Total RNA from each sample was quantified using the NanoDrop ND-1000 and the RNA integrity was assessed using standard denaturing agarose gel electrophoresis. For microarray analysis, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer's standard protocols. Briefly, total RNA from each sample was amplified and transcribed into fluorescent cRNA using the manufacturer's Agilent's Quick Amp Labeling protocol (version 5.7, Agilent Technologies). The labeled cRNAs were hybridized onto the Whole Mouse Genome Oligo Microarray (4 × 44 K, Agilent Technologies). After the slices were washed, the arrays were scanned by the Agilent Scanner G2505C. RStudio software was used to analyze the obtained results.
Supplementary Material
Acknowledgments
This study was supported, in part, by the Center for Neuroscience and Regenerative Medicine (CNRM) program at the Henry M. Jackson Foundation and the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH).
Footnotes
Conflict of Interest: The authors declare no competing financial interest.
Supporting Information Available: Additional figures showing MTT assay of nanovehicle to neural stem cells; chronological βIII tubulin protein expression of neural stem cells under different stimulation conditions; overview of TBI brain section; and sequencing ranking results. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.5b00690.
REFERENCES AND NOTES
- 1.Muller FJ, Snyder EY, Loring JF. Gene Therapy: Can Neural Stem Cells Deliver? Nat. Rev. Neurosci. 2006;7:75–84. doi: 10.1038/nrn1829. [DOI] [PubMed] [Google Scholar]
- 2.Martino G, Pluchino S. The Therapeutic Potential of Neural Stem Cells. Nat. Rev. Neurosci. 2006;7:395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- 3.Hsu YC, Li L, Fuchs E. Transit-Amplifying Cells Orchestrate Stem Cell Activity and Tissue Regeneration. Cell. 2014;157:935–949. doi: 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H, Loh KM, Nusse R. Stem Cell Signaling. An Integral Program for Tissue Renewal and Regeneration: Wnt Signaling and Stem Cell Control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 5.Beal MF, Hantraye P. Novel Therapies In the Search for a Cure for Huntington's Disease. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3–4. doi: 10.1073/pnas.98.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanberg PR. Neural Stem Cells for Parkinson's Disease: To Protect and Repair. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11869–11870. doi: 10.1073/pnas.0704704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindvall O, Kokaia Z, Martinez-Serrano A. Stem Cell Therapy for Human Neurodegenerative Disorders-How To Make It Work. Nat. Med. 2004;10(Suppl):S42–S50. doi: 10.1038/nm1064. [DOI] [PubMed] [Google Scholar]
- 8.Pluchino S, Zanotti L, Rossi B, Brambilla E, Ottoboni L, Salani G, Martinello M, Cattalini A, Bergami A, Furlan R, et al. Neurosphere-Derived Multipotent Precursors Promote Neuroprotection by an Immunomodulatory Mechanism. Nature. 2005;436:266–271. doi: 10.1038/nature03889. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brito C, Simao D, Costa I, Malpique R, Pereira CI, Fernandes P, Serra M, Schwarz SC, Schwarz J, Kremer EJ, et al. 3D Cultures of Human Neural Progenitor Cells: Dopaminergic Differentiation and Genetic Modification. Methods. 2012;56:452–460. doi: 10.1016/j.ymeth.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Abrous DN, Koehl M, Le Moal M. Adult Neurogenesis: From Precursors to Network and Physiology. Physiol. Rev. 2005;85:523–569. doi: 10.1152/physrev.00055.2003. [DOI] [PubMed] [Google Scholar]
- 12.Lie DC, Song H, Colamarino SA, Ming GL, Gage FH. Neurogenesis in the Adult Brain: New Strategies for Central Nervous System Diseases. Annu. Rev. Pharmacol. Toxicol. 2004;44:399–421. doi: 10.1146/annurev.pharmtox.44.101802.121631. [DOI] [PubMed] [Google Scholar]
- 13.Gross CG. Neurogenesis in the Adult Brain: Death of a Dogma. Nat. Rev. Neurosci. 2000;1:67–73. doi: 10.1038/35036235. [DOI] [PubMed] [Google Scholar]
- 14.Sanai N, Nguyen T, Ihrie RA, Mirzadeh Z, Tsai HH, Wong M, Gupta N, Berger MS, Huang E, Garcia-Verdugo JM, et al. Corridors of Migrating Neurons in the Human Brain and Their Decline during Infancy. Nature. 2011;478:382–386. doi: 10.1038/nature10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belmadani A, Tran PB, Ren D, Miller RJ. Chemokines Regulate the Migration of Neural Progenitors to Sites of Neuroinflammation. J. Neurosci. 2006;26:3182–3191. doi: 10.1523/JNEUROSCI.0156-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abematsu M, Tsujimura K, Yamano M, Saito M, Kohno K, Kohyama J, Namihira M, Komiya S, Nakashima K. Neurons Derived from Transplanted Neural Stem Cells Restore Disrupted Neuronal Circuitry in a Mouse Model of Spinal Cord Injury. J. Clin. Invest. 2010;120:3255–3266. doi: 10.1172/JCI42957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, et al. Dissecting Neural Differentiation Regulatory Networks through Epigenetic Footprinting. Nature. 2015;19:355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, Liu Y, Tu J, Wan J, Zhang J, Wu B, Chen S, Zhou J, Mu Y, Wang L. Activated Astrocytes Enhance the Dopaminergic Differentiation of Stem Cells and Promote Brain Repair Through Bfgf. Nat. Commun. 2014;5:5627. doi: 10.1038/ncomms6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis SA, Mak CK, Reynolds BA. Methods To Culture, Differentiate, and Characterize Neural Stem Cells from the Adult and Embryonic Mouse Central Nervous System. Methods Mol. Biol. 2013;946:479–506. doi: 10.1007/978-1-62703-128-8_30. [DOI] [PubMed] [Google Scholar]
- 20.Hu BY, Zhang SC. Directed Differentiation of Neural-Stem Cells and Subtype-Specific Neurons from hESCs. Methods Mol. Biol. 2010;636:123–137. doi: 10.1007/978-1-60761-691-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos T, Ferreira R, Maia J, Agasse F, Xapelli S, Cortes L, Braganca J, Malva JO, Ferreira L, Bernardino L. Polymeric Nanoparticles To Control the Differentiation of Neural Stem Cells in the Subventricular Zone of the Brain. ACS Nano. 2012;6:10463–10474. doi: 10.1021/nn304541h. [DOI] [PubMed] [Google Scholar]
- 22.Pollard SM, Conti L, Sun Y, Goffredo D, Smith A. Adherent Neural Stem (NS) Cells from Fetal and Adult Forebrain. Cereb. Cortex. 2006;16(Suppl. 1):I112–1120. doi: 10.1093/cercor/bhj167. [DOI] [PubMed] [Google Scholar]
- 23.Kokovay E, Shen Q, Temple S. The Incredible Elastic Brain: How Neural Stem Cells Expand Our Minds. Neuron. 2008;60:420–429. doi: 10.1016/j.neuron.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 24.Uchida N, Chen K, Dohse M, Hansen KD, Dean J, Buser JR, Riddle A, Beardsley DJ, Wan Y, Gong X, et al. Human Neural Stem Cells Induce Functional Myelination in Mice with Severe Dysmyelination. Sci. Transl. Med. 2012;4:155ra136. doi: 10.1126/scitranslmed.3004371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta N, Henry RG, Strober J, Kang SM, Lim DA, Bucci M, Caverzasi E, Gaetano L, Mandelli ML, Ryan T, et al. Neural Stem Cell Engraftment and Myelination in the Human Brain. Sci. Transl. Med. 2012;4:155ra137. doi: 10.1126/scitranslmed.3004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng YD, Benn SC, Kalkanis SN, Shefner JM, Onario RC, Cheng B, Lachyankar MB, Marconi M, Li J, Yu D, et al. Multimodal Actions of Neural Stem Cells in a Mouse Model of ALS: A Meta-Analysis. Sci. Transl. Med. 2012;4:165ra164. doi: 10.1126/scitranslmed.3004579. [DOI] [PubMed] [Google Scholar]
- 27.Tonge PD, Andrews PW. Retinoic Acid Directs Neuronal Differentiation of Human Pluripotent Stem Cell Lines in a Non-Cell-Autonomous Manner. Differentiation. 2010;80:20–30. doi: 10.1016/j.diff.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Okada Y, Shimazaki T, Sobue G, Okano H. Retinoic-Acid-Concentration-Dependent Acquisition of Neural Cell Identity During in Vitro Differentiation of Mouse Embryonic Stem Cells. Dev. Biol. 2004;275:124–142. doi: 10.1016/j.ydbio.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z, Zhang R, Wang HF, Wang Y, Zhao J, Wang F, Li W, Niu G, Kiesewetter DO, Chen X. Bioinspired Nanocomplex for Spatiotemporal Imaging of Sequential Mrna Expression in Differentiating Neural Stem Cells. ACS Nano. 2014;8:12386–12396. doi: 10.1021/nn505047n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic Acid Signaling Is Essential for Embryonic Hematopoietic Stem Cell Development. Cell. 2013;155:215–227. doi: 10.1016/j.cell.2013.08.055. [DOI] [PubMed] [Google Scholar]
- 31.Varga BV, Hadinger N, Gocza E, Dulberg V, Demeter K, Madarasz E, Herberth B. Generation of Diverse Neuronal Subtypes in Cloned Populations of Stem-like Cells. BMC Dev. Biol. 2008;8:89. doi: 10.1186/1471-213X-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. PLGA-Based Nanoparticles: An Overview of Biomedical Applications. J. Controlled Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Ho PC. A Nanocapsular Combinatorial Sequential Drug Delivery System for Antiangiogenesis and Anticancer Activities. Biomaterials. 2010;31:7115–7123. doi: 10.1016/j.biomaterials.2010.05.075. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Chui WK, Ho PC. Nanoparticulate Delivery System Targeted to Tumor Neovasculature for Combined Anticancer and Antiangiogenesis Therapy. Pharm. Res. 2011;28:585–596. doi: 10.1007/s11095-010-0308-2. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Niu G, Chen X. Polymeric Materials for Theranostic Applications. Pharm. Res. 2014;31:1358–1376. doi: 10.1007/s11095-013-1103-7. [DOI] [PubMed] [Google Scholar]
- 36.Gaudin A, Yemisci M, Eroglu H, Lepetre-Mouelhi S, Turkoglu OF, Donmez-Demir B, Caban S, Sargon MF, Garcia-Argote S, et al. Squalenoyl Adenosine Nanoparticles Provide Neuroprotection after Stroke and Spinal Cord Injury. Nat. Nanotechnol. 2014;9:1054–1062. doi: 10.1038/nnano.2014.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stavridis MP, Collins BJ, Storey KG. Retinoic Acid Orchestrates Fibroblast Growth Factor Signalling To Drive Embryonic Stem Cell Differentiation. Development. 2010;137:881–890. doi: 10.1242/dev.043117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Attardo A, Calegari F, Haubensak W, Wilsch-Brauninger M, Huttner WB. Live Imaging at the Onset of Cortical Neurogenesis Reveals Differential Appearance of the Neuronal Phenotype in Apical versus Basal Progenitor Progeny. PLoS One. 2008;3:E2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griesbach GS, Hovda DA, Molteni R, Gomez-Pinilla F. Alterations in BDNF and Synapsin I within the Occipital Cortex and Hippocampus after Mild Traumatic Brain Injury in the Developing Rat: Reflections of Injury-Induced Neuroplasticity. J. Neurotrauma. 2002;19:803–814. doi: 10.1089/08977150260190401. [DOI] [PubMed] [Google Scholar]
- 40.Carbajal KS, Schaumburg C, Strieter R, Kane J, Lane TE. Migration of Engrafted Neural Stem Cells Is Mediated by CXCL12 Signaling through CXCR4 in a Viral Model of Multiple Sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11068–11073. doi: 10.1073/pnas.1006375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Wang Y, Gutkind JS, Wang F, Lu J, Niu G, Teng G, Chen X. Engineered Mesenchymal Stem Cells with Enhanced Tropism and Paracrine Secretion of Cytokines and Growth Factors To Treat Traumatic Brain Injury. Stem Cells. 2015;33:456–467. doi: 10.1002/stem.1878. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Chui WK, Ho PC. Design of a Multifunctional PLGA Nanoparticulate Drug Delivery System: Evaluation of Its Physicochemical Properties and Anticancer Activity to Malignant Cancer Cells. Pharm. Res. 2009;26:1162–1171. doi: 10.1007/s11095-009-9837-y. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Ho PC. Self-Assembled Core-Shell Vascular-Targeted Nanocapsules for Temporal Antivasculature and Anticancer Activities. Small. 2010;6:2576–2583. doi: 10.1002/smll.201001122. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Zhang X, Huang P, Zhao W, Liu D, Nie L, Yue X, Wang S, Ma Y, Kiesewetter D, et al. Dual-Factor Triggered Fluorogenic Nanoprobe for Ultrahigh Contrast and Subdiffraction Fluorescence Imaging. Biomaterials. 2013;34:6194–6201. doi: 10.1016/j.biomaterials.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.