The genomic revolution is having a tremendous impact on the study of natural variation. It is making it possible finally to discover the molecular basis of complex traits, a fundamental question in evolutionary biology, and a question of immense practical importance in many other fields. The availability of polymorphism data from genome-wide marker loci will also make various forms of evolutionary inference, e.g. questions concerning the history of selection at a locus, much more reliable. In this review, we discuss the impact of genomics on the study of natural variation, focusing both on technological and methodological advances.
Genomic approaches are revolutionizing biology. The study of naturally occurring genetic variation will be affected more strongly than most other fields for the simple reason that most questions in this field are naturally “genomic”—they either concern the whole genome, or they cannot be answered using a gene-by-gene approach. The purpose of this review is to describe how genomics is affecting our ability to answer questions related to natural variation, in particular for Arabidopsis.
Natural variation is at the core of evolutionary biology, of plant and animal breeding, and of human genetics. For very different reasons, these fields all seek to understand natural variation. However, it is becoming increasingly clear that natural variation should also be of interest to functional biology (this point is well made in the context of plant biology by Alonso-Blanco and Koornneef, 2000). Many genes may have functions that cannot easily be determined by mutagenesis or similar approaches (due to lethality or redundancy). Alleles may differ from each other in subtle or complex ways that would be very difficult to replicate experimentally using traditional loss-of-function genetics (consider for example epigenetic alleles or plant self-incompatibility alleles). Genetic studies involve natural variation whether we like it or not because there is always a genetic background. To take a simple example, most screens for mutations affecting flowering time in Arabidopsis were carried out in rapid-cycling accessions that already carry loss-of-function mutations in the vernalization response pathway (Simpson and Dean, 2002). Had this work been done in one of the many vernalization-dependent winter-annual accessions, other genes would no doubt have turned up. Crossing mutations into different genetic backgrounds may be a powerful method for detecting modifiers of the mutation and can yield important clues about function. A well-known example is the CAULIFLOWER gene, which was discovered in a cross between a standard lab accession carrying the apetala1 mutation (involved in flower development) and a “wild-type” accession from the Ukraine (Bowman et al., 1993).
We will consider two kinds of questions that can be asked using natural variation. The first concerns the genetic basis of complex traits. This is arguably one of the most important challenges facing modern biology, and several recent reviews exist (e.g. Glazier et al., 2002); we focus here on tools for the Arabidopsis community, and in particular on the impact of genomics. The second kind of question concerns evolutionary inference. We may, for example, wish to know more about the recent history of selection on a particular locus. The genomics revolution will greatly increase our power to answer such questions as well.
DISSECTING COMPLEX TRAITS
The basic method for identifying loci responsible for variation in complex traits (so-called quantitative trait loci [QTLs]) is genetic mapping (Glazier et al., 2002). Traditional linkage mapping is done using pedigrees or suitable crosses (Doerge, 2002). Recently, there has also been a great deal of excitement regarding the use of natural populations of unrelated individuals instead, in so-called linkage disequilibrium mapping (Ardlie et al., 2002; Nordborg and Tavaré, 2002).
With respect to mapping, there is no fundamental difference between Mendelian and complex traits; the distinction is often arbitrary. In practice, however, the difference can be enormous. Genes that contribute to variation in complex traits are much more difficult to identify for a number of reasons (see e.g. Glazier et al., 2002). In statistical terms, each allele is typically responsible for only a very small fraction of the total phenotypic variation. As result, very few QTLs have been molecularly identified (both in absolute terms and relative to the number of identified genes controlling Mendelian traits). There are currently probably more reviews about the genetics of complex traits than there are actual results. Despite what one might have predicted based on allocation of research resources, most successes have not been in humans. This is perhaps not so surprising considering the much greater power of controlled genetic crosses. More surprising is the number of successes in non-model organisms, like cattle (Bostaurus spp., Grisart et al., 2002), tomato (Lycopersicon esculentum; Frary et al., 2000; Fridman et al., 2000), maize (Zea mays; Doebley et al., 1997), and rice (Oryza sativa; Yano et al., 2000; Takahashi et al., 2001; Kojima et al., 2002). One possibility is that this reflects the traits more than the organism: The traits studied by plant and animal breeding typically have been subject to strong directional selection and may therefore may have simpler genetic architecture than, say, bristle number in fruitfly (Drosophila melanogaster; Lai et al., 1994; Long et al., 1998).
In many ways, Arabidopsis is an ideal organism for dissecting complex traits. It is highly suitable for linkage mapping because very large numbers of offspring can readily be raised under uniform conditions. This, in combination with a relatively high recombination rate, makes it possible to map to a finer scale than in many other organisms. The fact that it is naturally selfing makes it easy to construct and maintain recombinant inbred lines (RILs) and near-isogenic lines (NILs). As will be discussed further below, it also appears that Arabidopsis may be highly suitable for linkage disequilibrium mapping. Finally, the full power of Arabidopsis as a model system for molecular biology can be brought to bear on confirming a QTL.
A number of QTLs have been successfully identified in Arabidopsis. Two major QTLs controlling vernalization response, FRI (FRIGIDA; Johanson et al., 2000) and FLC (FLOWERING LOCUS C; Michaels and Amasino, 1999; Sheldon et al., 1999), were the first to be identified as important in flowering time variation (Simpson and Dean, 2002), although other important loci certainly remain to be identified. A novel allele of the CRYPTCHROME 2 photoreceptor was identified as the cause of the EDI (EARLY DAY LENGTH INSENSITIVE) flowering time QTL (Alonso-Blanco et al., 1998; El-Assal et al., 2001). Natural variation for hypocotyl length light response has also been investigated at the molecular level. Functional variation has been found for PHYTOCHROME A (Maloof et al., 2001) and PHYTOCHROME D (Aukerman et al., 1997), and QTL studies suggest that variation in PHYOCHROME B (Borevitz et al., 2002) and other loci may also be important. Finally, candidate genes have been cloned and biochemically characterized for several QTL that control the amount and type of glucosinolates, small molecules that assist in herbivory resistance (Kliebenstein et al., 2001; Kroymann et al., 2001; Lambrix et al., 2001).
GENOTYPING TECHNIQUES FOR LINKAGE MAPPING IN ARABIDOPSIS
The ideas behind linkage mapping are by now fairly standard and have been described many times (for recent review, see Doerge, 2002; for a more extensive treatment, see Lynch and Walsh, 1998). We note here that mapping is an inherently genomic approach. What is changing is our ability to type very large numbers of markers in very large samples: This is improving at a fantastic rate. The first mapping studies in Arabidopsis used RFLPs (Chang et al., 1988; Lister and Dean, 1993; Clarke et al., 1995). Amplified fragment length polymorphism technology (Vos et al., 1995) soon followed and has been effective (Alonso-Blanco et al., 1998b; Breyne et al., 1999; Miyashita et al., 1999; Sharbel et al., 2000; Peters et al., 2001). Single nucleotide polymorphisms (SNPs) are currently the most popular markers, and many techniques are available to type them in high-throughput fashion; for example, Taq-Man (Livak, 1999), pyrosequencing (Alderborn et al., 2000), or MassArray (Jurinke et al., 2001). Most methods involve PCR at individual loci followed by primer extension across the SNP. The amenity of SNPs to high-throughput genotyping is only one of their advantages. Equally important is their ubiquity. Levels of polymorphism in Arabidopsis seem to depend strongly on sampling (i.e. on what accessions are compared) but are generally higher than in humans (M. Nordborg, unpublished data). In global samples, two accessions differ at least every 500 bp, on average (see e.g. Hagenblad and Nordborg, 2002; Haubold et al., 2002; Kuittinen et al., 2002; Tian et al., 2002). Large collections of Arabidopsis SNPs are available (Jander et al., 2002), and more are currently being developed (see below).
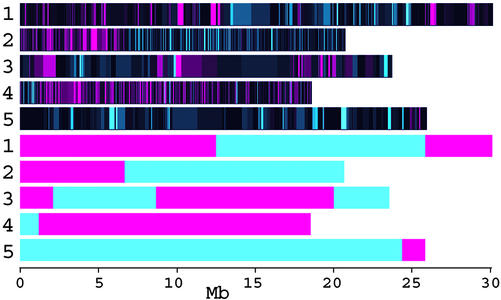
New methods based on oligonucleotide arrays (Winzeler et al., 1998; Cho et al., 1999; Nordborg et al., 2002; Borevitz et al., 2003) are attractive for several reasons. The highly parallel nature allows multiple markers to be assayed at once, dramatically reducing individual marker cost. Analysis of a single sample provides genome-wide coverage at high marker density such that recombination events are clearly defined (Fig. 1). Our recent work uses Affymetrix expression arrays as genotyping tools to genotype more than 8,000 SFPs between two Arabidopsis accessions for a cost of approximately $400 ($0.05 each). Total genomic DNA hybridization to arrays is both a marker discovery and genotyping platform. Bulk segregant mapping using array genotyping is a rapid and cost-effective way to map induced mutations or Mendelized QTL (Borevitz et al., 2003). The resolution in linkage mapping is now limited by the number of recombination events (i.e. the number of plants) rather than the number of markers that can be typed. It should be possible to localize large-effect QTLs very precisely (<100kb) in large intercross populations (http://naturalvariation.org) genotyped using arrays.
Figure 1.
A Columbia (Col) × Landsberg erecta RIL was hybridized to an Affymetrix expression array and genotyped at approximately 4,000 single feature polymorphisms (SFPs) as Col in green or Landsberg erecta in red (first five chromosomes). Recombination events are interpreted from these data (last five chromosomes). Adapted from Borevitz et al. (2003).
LINKAGE DISEQUILIBRIUM MAPPING IN ARABIDOPSIS
Although linkage mapping methods are well established and included in any advanced undergraduate textbooks on genetics, linkage disequilibrium mapping is a new and rapidly evolving field. With few exceptions, the only relevant source of information is the primary literature, in particular the human genetics literature. Because of this, we provide a brief introduction here.
Linkage disequilibrium mapping differs from standard linkage mapping methods in that marker-trait associations are sought in populations of unrelated individuals. For example, in an epidemiological study, we would look for markers that are over- or underrepresented in cases compared with controls. This idea is not new: Human geneticists started noticing associations between immunological markers and diseases a long time ago (Aird et al., 1953). However, for reasons that will be discussed shortly, these kinds of associations, known as linkage disequilibria, typically only exist between very closely linked loci. This means that linkage disequilibrium mapping is useful for fine mapping; it also means that it is not a practicable method unless a very dense marker map exists. Note that because the markers would be too closely linked to be ordered through standard methods (if you could order the markers using crosses, why would you need linkage disequilibrium mapping?), this means that linkage disequilibrium mapping will typically only be an alternative if the region (or genome) of interest has been sequenced (or at least has a good physical map). Exceptions exist, in particular for domesticated or selfing species where linkage disequilibria may be more extensive (see below).
Given a genome project and given modern genotyping technologies, marker density is no longer a problem. Linkage disequilibrium mapping is becoming a standard fine-mapping tool in human genetics: After a gene has been roughly localized using linkage mapping, linkage disequilibrium is used to further pinpoint the location. The canonical example of this approach is provided by Hästbacka et al. (1992), who narrowed a candidate region from 1 Mb to roughly 50 kb using linkage disequilibrium. The feasibility of genome-wide screens for linkage disequilibrium is less clear. It has been estimated that on the order of a million markers might be needed for genome-wide screens in humans, and even if genotyping costs could be reduced to a few cents per marker, this would be very expensive for an epidemiological study of reasonable size. Much of the current debate focuses on whether it may be possible to choose markers intelligently based on the observed pattern of linkage disequilibrium in smaller samples (so-called “haplotype blocks”; Daly et al., 2001; Patil et al., 2001; Gabriel et al., 2002).
Although the basic idea behind linkage disequilibrium mapping is straightforward, the statistics are decidedly less so. There are essentially two problems. The first problem concerns false positives. It is easy to see that marker-trait associations in natural populations can exist without the markers being linked to the trait loci. A very good example is that in the human population of San Francisco, skill with chop-sticks is strongly associated with the HLA-A1 allele (Lander and Schork, 1994). The reason is simply that this allele is more frequent among Chinese than among Europeans. Thus, in contrast to controlled crosses, P values calculated for the marker-trait association due to linkage disequilibrium have no absolute meaning. The only real solution to this problem is to use the genome as a form of control. Given genome-wide markers, we can do several things: (a) Attempt to infer the underlying population structure and then seek associations within appropriate subgroups (Pritchard et al., 2000). This approach has been utilized successfully in maize, which has considerable population structure (Thornsberry et al., 2001). (b) Try to estimate the true probability of false positives and adjust the P values accordingly (Devlin et al., 2001). (c) Accept a high rate of false positives. Decide how many candidate loci we are willing to test directly, rank the candidates in order of increasing probability, and start testing. False positives can be eliminated through standard F2 crosses.
The second problem concerns statistical power (i.e. false negatives). Although it is perfectly legitimate to test each marker for association with the trait, it is not very efficient. To understand why, we need to consider how linkage disequilibrium arises (for more details, see Nordborg and Tavaré, 2002). As we have seen, alleles at different loci may be statistically associated simply because of population structure. However, for mapping purposes, we are interested in those associations that are due to linkage, i.e. associations that exist because alleles at linked loci tend to be inherited together more often than alleles at unlinked loci. In an F2 population, the strength of the association between the alleles at two loci simply reflects the frequency of recombination between the two loci in the F1 generation. In natural populations, associations are much more complex and reflect the history of the chromosomal region. Consider a particular allele, and assume for the moment that it arose (through mutation) exactly once some time in the past (because mutation rates are low, this is usually a reasonable assumption). All currently existing copies of this allele can be traced back to a most recent common ancestor (MRCA) of the allele. Now, consider the chromosome that carries the locus in question. Every currently existing chromosome that carries the focal allele must have inherited a chromosomal segment containing the locus from the ancestral chromosome that carried the MRCA. The length of the segment shared with the ancestral chromosome depends on the rate of recombination and the age of the MRCA. However, in general, chromosomes that share a particular allele at a locus will also tend to share a short chromosomal segment surrounding this locus that they inherited along with the allele (see e.g. Nordborg and Tavare, 2002). Given sufficiently closely linked marker loci, this segment sharing will result in sharing of marker haplotypes. Testing for associations one marker at a time captures very little of this information: Statistical methods that utilize multilocus information typically have more power but pose difficult statistical problems because of the difficulty of taking shared ancestry into account (McPeek and Strahs, 1999; Morris et al., 2000, 2002; Liu et al., 2001).
Thinking about linkage disequilibrium this way also helps us understand its often confusing behavior. First of all, it is clear that it must be incredibly variable. The strength of association between alleles at two loci depends on a number of unknown factors: the ages of the alleles, the rate of recombination between them, the history of mutation at both loci, and the historical heterozygosity of the population, to name but a few (for more details, see Nordborg and Tavaré, 2002). Because of all these factors, it is common for more distantly linked markers to show stronger association than more closely linked ones. One important practical consequence is that although strong linkage disequilibrium may be evidence for linkage, the absence of associations is never evidence against it.
Second, although the average rate at which linkage disequilibrium decays does depend on the recombination rate, it also depends on population genetics parameters. For example, chromosomes in small populations tend to be more closely related to each other than chromosomes in large populations; thus, linkage disequilibrium will be more extensive in the former. Population structure will tend to increase linkage disequilibrium, whereas population expansions may reduce it. These factors contribute to explaining why linkage disequilibrium in humans can extend over 100 kb, whereas linkage disequilibrium in fruitfly rarely extends more than a few kilobase pairs, even though the average recombination rate per base pair differs only by a factor of four or five (Wall and Przeworski, 2000; Nordborg et al., 2002; Wall et al., 2002).
Population genetics also predicts that highly selfing species will harbor extensive linkage disequilibrium because recombination is only effective in breaking up associations between alleles in heterozygous individuals, which are much rarer in selfers. These predictions are clearly born out in Arabidopsis, in which linkage disequilibrium appears to decay on a scale roughly comparable with what is observed in humans (Nordborg et al., 2002). This and several other factors contribute to making Arabidopsis a very good candidate for genome-wide linkage disequilibrium mapping, certainly much better than humans: (a) The genome is small, which reduces costs. (b) There is much more polymorphism than in humans, which means that there will be many more informative markers. (c) The availability of highly inbred lines means that for the most part genotyping equals haplotyping. In outbred organisms, it is impossible to know the phase of the markers at two or more polymorphic loci. (d) False positives due to population structure can easily be eliminated by carrying out a simple cross.
A study to explore these possibilities is currently under way. This study, funded by the National Science Foundation 2010 Project, aims to sequence 2000 500-bp fragments in each of 96 accessions from around the world. The data will be made publicly available. As of writing, about one-half of the fragments have been sequenced, and the data are being processed (for more details, see http://walnut.usc.edu).
Although Arabidopsis may be an ideal candidate for linkage disequilibrium, it is arguably also an organism in which linkage disequilibrium mapping is not needed. Fine mapping in Arabidopsis can always be accomplished by testing enough offspring. However, the cost of genotyping accessions is incurred only once, whereas genotyping in crosses has to be done for each cross. Thus, the genotyped accessions will be a permanent mapping resource for Arabidopsis genetics. In the end, it seems likely that linkage and linkage disequilibrium mapping will complement each other, just as in human genetics. An additional benefit of the 2010 study just mentioned is that genome-wide markers for the 96 accessions, several of which are parents of RILs, will be generated.
Array genotyping can assist both linkage and linkage disequilibrium studies in several ways. One obvious way is in confirming potential associations. Candidate associations identified in a genome-wide linkage disequilibrium scan need to be confirmed in the F2 of specific crosses. Bulk segregant mapping with array genotyping performed on pools of extreme segregants is an efficient way to accomplish this. By analyzing several crosses, it should be possible to determine exactly which haplotypes are functionally different. It should be mentioned in this context that a general (and potentially very serious) problem with linkage disequilibrium mapping is genetic heterogeneity. Unrelated individuals with similar phenotypes may well be similar for different genetic reasons.
Array genotyping can also be used directly to construct a high-density (albeit lower quality than by sequencing) haplotype map. When several accessions are genotyped using arrays, we predict that at least one SFP will be identified in each gene. This marker density of approximately one per 5 kb (approximately 22,000 SFPs total) will provide a fine-scale haplotype map, which could be anchored with the high-quality sequence data from the 2010 project described above. Finally, array genotyping can be used to type other more extensive samples from populations that may show more extensive linkage disequilibrium (Nordborg et al., 2002).
TOOLS FOR CONFIRMING QTL
The mapping methods discussed above are used to predict genome regions containing functionally important naturally occurring genetic variation. State-of-the-art genotyping technologies and statistical mapping methods can provide very narrow candidate regions, on the order of hundreds of kilobase pairs. However, the gene(s) responsible and the functional change(s) ultimately must be identified, i.e. molecular “cloning” of QTLs. The fine mapping process often utilizes NILs or heterogeneous inbred families (HIFs). NILs contain a small chromosome segment from one parent containing the QTL introgressed into the background of the other parent, and HIFs take advantage of residual heterozygosity present in RILs (Alonso-Blanco and Koornneef, 2000). HIFs are chosen that are heterozygous only at a single QTL and, like NILs, are used for confirmation and fine mapping. At this stage, the QTL is said to be “Mendelized” because only a single gene is segregating and can be followed with a marker at a single locus. Once a narrow interval is defined, several tools are available in Arabidopsis for candidate gene identification and confirmation:
Genome-Wide Expression Arrays
If the QTL is the result of an alteration in the level of expression of a gene, it might be identified via transcriptional profiling. RNA is extracted from genotypes containing either QTL allele, typically the NIL and parental control. Differentially expressed genes in the QTL region are candidate genes, whereas differentially expressed genes that are unlinked to the QTL are part of the molecular phenotype and are a consequence of allelic variation at (or linked to) the QTL. Transcriptional profiling can also be done on pools of extreme RILs that are likely to be fixed for QTL in opposite directions. A further advancement involves sampling from different environments to identify genes differentially expressed only in the correct environment. To assign confidence to gene expression differences, independent biological replicates are used. Thresholds and false discovery rates are determined via comparison with a permutation distribution (Tusher et al., 2001). It must be noted that gene expression studies between different accessions will not discriminate between true gene expression differences and hybridization polymorphisms; however, both can suggest functional candidates.
Identification of candidate polymorphisms in coding regions through array hybridization (Borevitz et al., 2003) and direct sequencing can also suggest QTL candidate genes. It may be valuable to compare the identified polymorphisms to expression data and vice versa.
Knockout (KO) Collections
A nearly saturated collection of T-DNA KO lines is available (http://signal.salk.edu/). This is especially valuable for screening a collection of KO lines that cover most genes in a QTL interval for quantitative phenotypes. Once identified, the correct KO line can be used as a background for transgenic experiments or quantitative complementation. KO lines are available in both the Col (http://signal.salk.edu/) and Ws-2 backgrounds (Sussman et al., 2000). Natural alleles, whether change of function or null, are compared with KO lines for a similar phenotype.
Complementation Tests
Traditional complementation tests between recessive alleles can be used to test specific QTL candidate genes (Doebley et al., 1997; Maloof et al., 2001). Quantitative complementation, originally developed in fruitfly (Gurganus et al., 1999; Mackay, 2001), is an extension of the traditional test for partially dominant alleles. This test compares the QTL effect over a third allele or a null mutation in the candidate gene. The test directly compares the quantitative phenotype of four F1 lines (Parent1/test line, Parent1 QTL NIL/test line, Parent1/test line with null, and Parent1 QTL NIL/test line with null). If the candidate gene is responsible for the QTL, then it will quantitatively fail to complement the null mutation. This is detected as a significant interaction between the QTL genotype and the presence or absence of the null mutation in the test line. A significant interaction is evidence for the correct QTL gene; however, epitasis is an alternative explanation. Furthermore, lack of interaction could be caused by simple additivity of alleles; therefore, this test cannot rule out a candidate gene. It is especially useful that large collections of KO lines are available for Arabidopsis in two different backgrounds. The reciprocal hemizygosity test is related to quantitative complementation and makes use of null mutations in reciprocal backgrounds (Steinmetz et al., 2002).
Transgenics
Finally, the most direct way to confirm a QTL gene is to place the corresponding DNA fragment directly in the reciprocal background (El-Din El-Assal et al., 2001). This is readily done in Arabidopsis and other experimental systems. Position effect can be overcome by analysis of multiple independent transgenic lines. Alternative alleles can also be transformed into a KO background to avoid interaction with the endogenous allele. Furthermore, chimeric alleles or ones created with site-directed mutagenesis can be used to determine the precise functional change(s).
EVOLUTIONARY INFERENCE
We conclude by briefly discussing the impact of genomics on evolutionary inference from polymorphism data. This field is concerned with the past: migrations and demography (e.g. Innan and Stephan, 2000; Sharbel et al., 2000) or selection on particular alleles (e.g. Tian et al., 2002). Because evolution is a slow process, direct experimental evidence is never available. The general approach employed in this field has been to compare the data (or some particular feature of the data) with expectations under some theoretical model (explicit or implicit) and then decide whether the model fits the data. The decision making has sometimes involved sophisticated statistics and sometimes no statistics at all (for review, see Rosenberg and Nordborg, 2002). To make this more concrete, consider the following example. Hudson et al. (1994) sequenced 10 copies of Sod from fruitfly sampled in Barcelona. The 1,410-bp fragment sequenced contained a known SNP resulting in an amino acid polymorphism. Five of the sequences carried the allele known as “Fast-A” at this locus, and five carried the alternative allele. Interestingly, it was found that the five “Fast-A” sequences were completely identical, whereas the other five were all different and contained a total of 55 polymorphic sites. Computer simulations based on a standard population genetics model that includes no selection revealed that under the model, such a skewed sample would practically never be observed. An alternative explanation (favored by Hudson et al., 1994) is that selection has increased the frequency of “Fast-A” so rapidly that there has been no time for mutation (or recombination) to make the “Fast-A” haplotypes different from each other.
The problem with this approach is that there are often several alternative explanations. In particular, it is has long been known that demographic events can cause patterns of polymorphism that exactly mimic those expected under selection (Kreitman, 2000; Nordborg, 2001). For example, a population bottleneck followed by rapid growth can cause the kind of skew observed by Hudson et al. (1994). However, although selection acts on particular loci, demography affects the whole genome. This suggests that to detect selection, we should use the rest of the genome as control, analogously to what was described for linkage disequilibrium mapping above. We can either use the genomic data to develop more realistic null models or, more rigorously, simply compare our candidate locus with the rest of the genome. To continue the Sod example, if Hudson et al. (1994) had sequenced 1,000 other loci in the same sample and found that Sod showed the most extreme skew, then this would be strong evidence in favor of recent selection. Note that this approach requires prior beliefs about which loci have been subject to selection. Searching the genome blindly for loci that have been subject to recent selection is much more difficult because unlike the situation in linkage disequilibrium mapping (which can be confirmed in a new cross), it is usually impossible to verify independently that a locus has been under selection. Thus, simply ranking the top candidates is not a good option. It is necessary to use statistical models to determine what is due to selection. These models should be developed using the information in the data. Screening the genome for selected loci using standard models (e.g. Akey et al., 2002) is just the first step. Inference about historical demography will benefit from the availability of genomic polymorphism data for exactly the same reason (Rosenberg and Nordborg, 2002).
Arabidopsis has played an important role in helping population geneticists realize how difficult it is to disentangle the effects of selection and demography. The standard approach of detecting selection by rejecting the standard neutral model was largely developed in fruitfly, a species that was believed not to have strong population structure. Many years of polymorphism studies in fruitfly failed to find much evidence of selection (Hudson, 1996). In contrast, the first polymorphism survey in Arabidopsis was able to reject neutrality (Hanfstingl et al., 1994), and it quickly became evident that this was the rule rather than the exception (Aguadé, 2001; Hagenblad and Nordborg, 2002). Although it is possible that selection is more ubiquitous in Arabidopsis than in fruitfly, a simpler explanation is that population structure causes deviations from the standard neutral model on a genome-wide basis in the former but not the latter.
CONCLUSIONS
Our ability to identify the molecular basis for naturally occurring phenotypic variation is improving rapidly thanks to technological and methodological advances. This will be of great benefit to many areas of biology. Evolutionary biology, in particular, will be revolutionized because it will finally be possible to study genes that matter in populations.
Acknowledgments
We thank Dan Kliebenstein, Julin Maloof, and Rick Amasino for comments on the manuscript.
References
- Aguade M (2001) Nucleotide sequence variation at two genes of the phenylpropanoid pathway, the FAH1 and F3H genes, in Arabidopsis thaliana. Mol Biol Evol 18: 1–9 [DOI] [PubMed] [Google Scholar]
- Aird I, Bentall HH, Fraser Roberts JA (1953) A relationship between cancer of stomach and the ABO blood groups. Br Med J 1: 799–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12: 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderborn A, Kristofferson A, Hammerling U (2000) Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res 10: 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SE-D, Coupland G, Koornneef M (1998a) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Island ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, Van Den Bosch N, Pot J, Kuiper MTR (1998b) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M (2002) Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3: 299–309 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA (1997) A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Liang D, Plouffe D, Chang H-S, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large scale identification of single feature polymorphisms in complex genomes. Genome Res 13: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D et al. (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743 [Google Scholar]
- Breyne P, Rombaut D, Van Gysel A, Van Montagu M, Gerats T (1999) AFLP analysis of genetic diversity within and between Arabidopsis thaliana ecotypes. Mol Gen Genet 261: 627–634 [DOI] [PubMed] [Google Scholar]
- Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM (1988) Restriction fragment length polymorphism linkage map for Arabidopsis thaliana. Proc Natl Acad Sci USA 85: 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Mindrinos M, Richards DR, Sapolsky RJ, Anderson M, Drenkard E, Dewdney J, Reuber TL, Stammers M, Federspiel N et al. (1999) Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat Genet 23: 203–207 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Mithen R, Brown JK, Dean C (1995) QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29: 229–232 [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L (2001) Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 60: 155–166 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3: 43–52 [DOI] [PubMed] [Google Scholar]
- El-Assal SE-D, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Frary A, Nesbitt TC, Grandillo S, Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88 [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D (2000) A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA 97: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229 [DOI] [PubMed] [Google Scholar]
- Glazier AM, Nadeau JH, Aitman TJ (2002) Finding genes that underlie complex traits. Science 298: 2345–2349 [DOI] [PubMed] [Google Scholar]
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P et al. (2002) Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res 12: 222–231 [DOI] [PubMed] [Google Scholar]
- Gurganus MC, Nuzhdin SV, Leips JW, Mackay TF (1999) High-resolution mapping of quantitative trait loci for sternopleural bristle number in Drosophila melanogaster. Genetics 152: 1585–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad J, Nordborg M (2002) Sequence variation and haplotype structure surrounding the flowering time locus FRI in Arabidopsis thaliana. Genetics 161: 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanfstingl U, Berry A, Kellogg EA, Costa JT, 3rd, Rudiger W, Ausubel FM (1994) Haplotypic divergence coupled with lack of diversity at the Arabidopsis thaliana alcohol dehydrogenase locus: roles for both balancing and directional selection? Genetics 138: 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2: 204–211 [DOI] [PubMed] [Google Scholar]
- Haubold B, Kroymann J, Ratzka A, Mitchell-Olds T, Wiehe T (2002) Recombination and gene conversion in a 170-kb genomic region of Arabidopsis thaliana. Genetics 161: 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR (1996) Molecular population genetics of adaptation. In MR Rose, GV Lauder, eds, Adaptation, Academic Press, San Diego, pp 291–309
- Hudson RR, Bailey K, Skarecky D, Kwiatowski J, Ayala FJ (1994) Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136: 1329–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Stephan W (2000) The coalescent in an exponentially growing metapopulation and its application to Arabidopsis thaliana. Genetics 155: 2015–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jurinke C, van den Boom D, Cantor CR, Koster H (2001) Automated genotyping using the DNA MassArray technology. Methods Mol Biol 170: 103–116 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43: 1096–1105 [DOI] [PubMed] [Google Scholar]
- Kreitman M (2000) Methods to detect selection in populations with applications to the human. Annu Rev Genomics Hum Genet 1: 539–559 [DOI] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127: 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Kuittinen H, Salguero D, Aguade M (2002) Parallel patterns of sequence variation within and between populations at three loci of Arabidopsis thaliana. Mol Biol Evol 19: 2030–2034 [DOI] [PubMed] [Google Scholar]
- Lai C, Lyman RF, Long AD, Langley CH, Mackay TF (1994) Naturally occurring variation in bristle number and DNA polymorphisms at the scabrous locus of Drosophila melanogaster. Science 266: 1697–1702 [DOI] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia in herbivory. Plant Cell 13: 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265: 2037–2048 [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C (1993) Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J 4: 745–750 [DOI] [PubMed] [Google Scholar]
- Liu JS, Sabatti C, Teng J, Keats BJ, Risch N (2001) Bayesian analysis of haplotypes for linkage disequilibrium mapping. Genome Res 11: 1716–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14: 143–149 [DOI] [PubMed] [Google Scholar]
- Long AD, Lyman RF, Langley CH, Mackay TF (1998) Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland, MA
- Mackay TF (2001) The genetic architecture of quantitative traits. Annu Rev Genet 35: 303–339 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- McPeek MS, Strahs A (1999) Assessment of linkage disequilibrium by the decay of haplotype sharing, with application to fine-scale genetic mapping. Am J Hum Genet 65: 858–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita NT, Kawabe A, Innan H (1999) DNA variation in the wild plant Arabidopsis thaliana revealed by amplified fragment length polymorphism analysis. Genetics 152: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Whittaker JC, Balding DJ (2000) Bayesian fine-scale mapping of disease loci, by hidden Markov models. Am J Hum Genet 67: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP, Whittaker JC, Balding DJ (2002) Fine-scale mapping of disease loci via shattered coalescent modeling of genealogies. Am J Hum Genet 70: 686–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M (2001) Coalescent theory. In DJ Balding, MJ Bishop, C Cannings, eds, Handbook of Statistical Genetics, John Wiley & Sons, Inc, Chichester, UK, pp 179–212
- Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ et al. (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30: 190–193 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Innan H (2002) Molecular population genetics. Curr Opin Plant Biol 5: 69–73 [DOI] [PubMed] [Google Scholar]
- Nordborg M, Tavare S (2002) Linkage disequilibrium: what history has to tell us. Trends Genet 18: 83–90 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP et al. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719–1723 [DOI] [PubMed] [Google Scholar]
- Peters JL, Constandt H, Neyt P, Cnops G, Zethof J, Zabeau M, Gerats T (2001) A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol 127: 1579–1589 [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Nordborg M (2002) Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet 3: 380–390 [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T (2000) Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol 9: 2109–2118 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, Mc-Cusker JH, Davis RW (2002) Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330 [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124: 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M (2001) Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc Natl Acad Sci USA 98: 7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ESt (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28: 286–289 [DOI] [PubMed] [Google Scholar]
- Tian DC, Araki H, Stahl E, Bergelson J, Kreitman M (2002) Signature of balancing selection in Arabidopsis. Proc Natl Acad Sci USA 99: 11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Andolfatto P, Przeworski M (2002) Testing models of selection and demography in Drosophila simulans. Genetics 162: 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Przeworski M (2000) When did the human population size start increasing? Genetics 155: 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Richards DR, Conway AR, Goldstein AL, Kalman S, Mc-Cullough MJ, McCusker JH, Stevens DA, Wodicka L, Lockhart DJ et al. (1998) Direct allelic variation scanning of the yeast genome. Science 281: 1194–1197 [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y et al. (2000) Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]