Abstract
Background
As more and more researchers are turning to big data for new opportunities of biomedical discoveries, machine learning models, as the backbone of big data analysis, are mentioned more often in biomedical journals. However, owing to the inherent complexity of machine learning methods, they are prone to misuse. Because of the flexibility in specifying machine learning models, the results are often insufficiently reported in research articles, hindering reliable assessment of model validity and consistent interpretation of model outputs.
Objective
To attain a set of guidelines on the use of machine learning predictive models within clinical settings to make sure the models are correctly applied and sufficiently reported so that true discoveries can be distinguished from random coincidence.
Methods
A multidisciplinary panel of machine learning experts, clinicians, and traditional statisticians were interviewed, using an iterative process in accordance with the Delphi method.
Results
The process produced a set of guidelines that consists of (1) a list of reporting items to be included in a research article and (2) a set of practical sequential steps for developing predictive models.
Conclusions
A set of guidelines was generated to enable correct application of machine learning models and consistent reporting of model specifications and results in biomedical research. We believe that such guidelines will accelerate the adoption of big data analysis, particularly with machine learning methods, in the biomedical research community.
Keywords: machine learning, clinical prediction rule, guideline
Introduction
Big data is changing every industry. Medicine is no exception. With rapidly growing volume and diversity of data in health care and biomedical research, traditional statistical methods often are inadequate. By looking into other industries where modern machine learning techniques play central roles in dealing with big data, many health and biomedical researchers have started applying machine learning to extract valuable insights from ever-growing biomedical databases, in particular with predictive models [1,2]. The flexibility and prowess of machine learning models also enable us to leverage novel but extremely valuable sources of information, such as wearable device data and electronic health record data [3].
Despite its popularity, it is difficult to find a universally agreed-upon definition for machine learning. Arguably, many machine learning methods can be traced back as far as 30 years ago [4]. However, machine learning started making a broad impact only in the last 10 years. The reviews by Jordan and Mitchell [5] and Ghahramani [6] provide accessible overviews for machine learning. In this paper, we focus on machine learning predictive methods and models. These include random forest, support vector machines, and other methods listed in Multimedia Appendix 1. They all share an important difference from the traditional statistical methods such as logistic regression or analysis of variance—the ability to make accurate predictions on unseen data. To optimize the prediction accuracy, often the methods do not attempt to produce interpretable models. This also allows them to handle a large number of variables common in most big data problems.
Accompanying the flexibility of emerging machine learning techniques, however, is uncertainty and inconsistency in the use of such techniques. Machine learning, owing to its intrinsic mathematical and algorithmic complexity, is often considered a “black magic” that requires a delicate balance of a large number of conflicting factors. This, together with inadequate reporting of data sources and modeling process, makes research results reported in many biomedical papers difficult to interpret. It is not rare to see potentially spurious conclusions drawn from methodologically inadequate studies [7-11], which in turn compromises the credibility of other valid studies and discourages many researchers who could benefit from adopting machine learning techniques.
Most pitfalls of applying machine learning techniques in biomedical research originate from a small number of common issues, including data leakage [12] and overfitting [13-15], which can be avoided by adopting a set of best practice standards. Recognizing the urgent need for such a standard, we created a minimum list of reporting items and a set of guidelines for optimal use of predictive models in biomedical research.
Methods
Panel of Experts
In 2015, a multidisciplinary panel was assembled to cover expertise in machine learning, traditional statistics, and biomedical applications of these methods. The candidate list was generated in two stages. The panel grew from a number of active machine learning researchers attending international conferences including the Asian Conference on Machine Learning, the Pacific Asia Conference on Knowledge Discovery and Data Mining, and the International Conference on Pattern Recognition. The responders were then asked to nominate additional researchers who apply machine learning in biomedical research. Effort was exercised to include researchers from different continents. Researchers from the list were approached through emails for joining the panel and/or recommending colleagues to be included. Two declined the invitation.
The final panel included 11 researchers from 3 institutions on 3 different continents. Each panelist had experience and expertise in machine learning projects in biomedical applications and has learned from common pitfalls. The areas of research expertise included machine learning, data mining, computational intelligence, signal processing, information management, bioinformatics, and psychiatry. On average, each panel member had 8.5 years’ experience in either developing or applying machine learning methods. The diversity of the panel was reflected by the members’ affiliation with 3 different institutions across 3 continents.
Development of Guidelines
Using an iterative process, the panel produced a set of guidelines that consists of (1) a list of reporting items to be included in a research article and (2) a set of practical sequential steps for developing predictive models. The Delphi method was used to generate the list of reporting items.
The panelists were interviewed with multiple iterations of emails. Email 1 asked panelists to list topics to be covered in the guidelines. An aggregated topic list was generated. Email 2 asked each panelist to review the scope of the list and state his or her recommendation for each topic in the aggregated list. Later iterations of email interviews were organized to evolve the list until all experts agreed on the list. Because of the logistic complexity of coordinating the large panel, we took a grow-shrink approach. In the growing phase, all suggested items were included, even an item suggested by only 1 panelist. In the shrinking phase, any item opposed by a panelist was excluded. As it turned out, most items were initially suggested by a panelist but seconded by other panelists, suggesting the importance of the group effort for covering most important topics.
The practical steps were developed by machine learning experts in their respective areas and finally approved by the panel. During the process, the panelists consulted extensively the broad literature on machine learning and predictive model in particular [16-18].
Results
A total of 4 iterations of emails resulted in the final form of the guidelines. Email 1 generated diverse responses in terms of topics. However the final scope was generally agreed upon. For email 2, most panelists commented on only a subset of topics (mostly the ones suggested by themselves). No recommendations generated significant disagreement except for minor wording decisions and quantifying conditions.
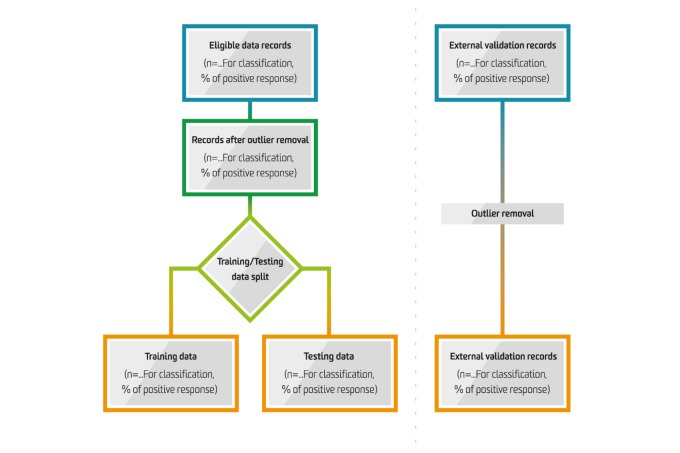
The final results included a list of reporting items (Tables 1-5,Textboxes 1-4, and Figure 1) and a template flowchart for reporting data used for training and testing predictive models, including both internal validation and external validation (Figure 2).
Table 1.
Items to include when reporting predictive models in biomedical research: title and abstract.
| Item number |
Section | Topic | Checklist item |
| 1 | Title | Nature of study | Identify the report as introducing a predictive model |
| 2 | Abstract | Structured summary | Background Objectives Data sources Performance metrics of the predictive model or models, in both point estimates and confidence intervals Conclusion including the practical value of the developed predictive model or models |
Table 5.
Items to include when reporting predictive models in biomedical research: discussion section.
| Item number |
Topic | Checklist item |
| 10 | Clinical implications | Report the clinical implications derived from the obtained predictive performance. For example, report the dollar amount that could be saved with better prediction. How many patients could benefit from a care model leveraging the model prediction? And to what extent? |
| 11 | Limitations of the model | Discuss the following potential limitations: • Assumed input and output data format • Potential pitfalls in interpreting the modela • Potential bias of the data used in modeling • Generalizability of the data |
| 12 | Unexpected results during the experiments | Report unexpected signs of coefficients, indicating collinearity or complex interaction between predictor variablesa |
aDesirable but not mandatory items.
Data leakage problem.
Leakage refers to the unintended use of known information as unknown. There are two kinds of leakage: outcome leakage and validation leakage. In outcome leakage, independent variables incorporate elements that can be used to easily infer outcomes. For example, a risk factor that spans into the future may be used to predict the future itself. In the validation leakage, ground truth from the training set may propagate to the validation set. For example, when the same patient is used in both training and validation, the future outcome in the training may overlap with the future outcome in the validation. In both leakage cases, the performance obtained is overoptimistic.
K-fold cross-validation.
K-fold validation refers to the practice of splitting the derivation data into K equal parts. The model is then trained on K−1 parts and validated on the remaining part. The process is repeated K times. The average results for K-folds are then reported. For small classes and rare categorical factors, stratified K-fold splitting should be used to ensure the equal presence of these classes and factors in each fold.
Figure 1.
Steps to identify the prediction problem.
Figure 2.
Information flow in the predictive modelling process.
Recognizing the broad meaning of the term “machine learning,” we distinguish essential items from desirable items (using appropriate footnotes in the tables). The essential items should be included in any report, unless there is a strong reason indicating otherwise; the desirable items should be reported whenever applicable.
Table 2.
Items to include when reporting predictive models in biomedical research: introduction section.
| Item number |
Topic | Checklist item |
| 3 | Rationale | Identify the clinical goal Review the current practice and prediction accuracy of any existing models |
| 4 | Objectives | State the nature of study being predictive modeling, defining the target of prediction Identify how the prediction problem may benefit the clinical goal |
Table 3.
Items to include when reporting predictive models in biomedical research: methods section.
| Item number |
Topic | Checklist item |
| 5 | Describe the setting | Identify the clinical setting for the target predictive model. Identify the modeling context in terms of facility type, size, volume, and duration of available data. |
| 6 | Define the prediction problem | Define a measurement for the prediction goal (per patient or per hospitalization or per type of outcome). Determine that the study is retrospective or prospective.a Identify the problem to be prognostic or diagnostic. Determine the form of the prediction model: (1) classification if the target variable is categorical, (2) regression if the target variable is continuous, (3) survival prediction if the target variable is the time to an event. Translate survival prediction into a regression problem, with the target measured over a temporal window following the time of prediction. Explain practical costs of prediction errors (eg, implications of underdiagnosis or overdiagnosis). Defining quality metrics for prediction models.b Define the success criteria for prediction (eg, based on metrics in internal validation or external validation in the context of the clinical problem). |
| 7 | Prepare data for model building | Identify relevant data sources and quote the ethics approval number for data access. State the inclusion and exclusion criteria for data. Describe the time span of data and the sample or cohort size. Define the observational units on which the response variable and predictor variables are defined. Define the predictor variables. Extra caution is needed to prevent information leakage from the response variable to predictor variables.c Describe the data preprocessing performed, including data cleaning and transformation. Remove outliers with impossible or extreme responses; state any criteria used for outlier removal. State how missing values were handled. Describe the basic statistics of the dataset, particularly of the response variable. These include the ratio of positive to negative classes for a classification problem and the distribution of the response variable for regression problem. Define the model validation strategies. Internal validation is the minimum requirement; external validation should also be performed whenever possible. Specify the internal validation strategy. Common methods include random split, time-based split, and patient-based split. Define the validation metrics. For regression problems, the normalized root-mean-square error should be used. For classification problems, the metrics should include sensitivity, specificity, positive predictive value, negative predictive value, area under the ROCd curve, and calibration plot [19].e For retrospective studies, split the data into a derivation set and a validation set. For prospective studies, define the starting time for validation data collection. |
| 8 | Build the predictive model | Identify independent variables that predominantly take a single value (eg, being zero 99% of the time). Identify and remove redundant independent variables. Identify the independent variables that may suffer from the perfect separation problem.f Report the number of independent variables, the number of positive examples, and the number of negative examples. Assess whether sufficient data are available for a good fit of the model. In particular, for classification, there should be a sufficient number of observations in both positive and negative classes. Determine a set of candidate modeling techniques (eg, logistic regression, random forest, or deep learning). If only one type of model was used, justify the decision for using that model.g Define the performance metrics to select the best model. Specify the model selection strategy. Common methods include K-fold validation or bootstrap to estimate the lost function on a grid of candidate parameter values. For K-fold validation, proper stratification by the response variable is needed.h For model selection, include discussion on (1) balance between model accuracy and model simplicity or interpretability, and (2) the familiarity with the modeling techniques of the end user.i |
aSee Figure 1.
bSee some examples in Multimedia Appendix 2.
cSee Textbox 1.
dROC: receiver operating characteristic.
eAlso see Textbox 2.
fSee Textbox 3.
gSee Multimedia Appendix 1 for some common methods and their strengths and limitations.
hSee Textbox 4.
iA desirable but not mandatory item.
Table 4.
Items to include when reporting predictive models in biomedical research: results section.
| Item number |
Topic | Checklist item |
| 9 | Report the final model and performance | Report the predictive performance of the final model in terms of the validation metrics specified in the methods section. If possible, report the parameter estimates in the model and their confidence intervals. When the direct calculation of confidence intervals is not possible, report nonparametric estimates from bootstrap samples. Comparison with other models in the literature should be based on confidence intervals. Interpretation of the final model. If possible, report what variables were shown to be predictive of the response variable. State which subpopulation has the best prediction and which subpopulation is most difficult to predict. |
Calibration.
Calibration of a prediction model refers to the agreement between the predictions made by the model and the observed outcomes. As an example, if the prediction model predicts 70% risk of mortality in the next 1 year for a patient with lung cancer, then the model is well calibrated if in our dataset approximately 70% of patients with lung cancer die within the next 1 year.
Often, the regularized prediction may create bias in a model. Therefore, it is advisable to check for the calibration. In the case of regression models, the calibration can be easily assessed graphically by marking prediction scores on the x-axis and the true outcomes on the y-axis. In the case of binary classification, the y-axis has only 0 and 1 values; however, smoothing techniques such as LOESS algorithm may be used to estimate the observed probabilities for the outcomes. In a more systematic way, one can perform the Hosmer-Lemeshow test to measure the goodness of fit of the model. The test assesses whether the observed event rates match the predicted event rates in subgroups of the model population.
Perfect separation problem.
When a categorical predictor variable can take an uncommon value, there may be only a small number of observations having that value. In a classification problem, these few observations by chance may have the same response value. Such “perfect” predictors may cause overfitting, especially when tree-based models are used. Therefore, special treatment is required.
One conservative approach is to remove all dummy variables corresponding to rare categories. We recommend a cutoff of 10 observations.
For modeling methods with feature selection built in, an alternative approach is to first fit a model with all independent variables. If the resulting model is only marginally influenced by the rare categories, then the model can be kept. Otherwise, the rare categories showing high “importance” score are removed and the model refitted.
Discussion
We have generated a set of guidelines that will enable correct application of machine learning models and consistent reporting of model specifications and results in biomedical research.
Because of the broad range of machine learning methods that can be used in biomedical applications, we involved a large number of stakeholders, as either developers of machine learning methods or users of these methods in biomedicine research.
The guidelines here cover most popular machine learning methods appearing in biomedical studies. We believe that such guidelines will accelerate the adoption of big data analysis, particularly with machine learning methods, in the biomedical research community.
Although the proposed guidelines result from a voluntary effort without dedicated funding support, we still managed to assemble a panel of researchers from multiple disciplines, multiple institutions, and multiple continents. We hope the guidelines can result in more people contributing their knowledge and experience in the discussion.
As machine learning is a rapidly developing research area, the guidelines are not expected to cover every aspect of the modeling process. The guidelines are expected to evolve as research in biomedicine and machine learning progresses.
Acknowledgments
This project is partially supported by the Telstra-Deakin COE in Big data and Machine Learning.
Multimedia Appendix 1
Guidelines for fitting some common predictive models.
Multimedia Appendix 2
Glossary.
Footnotes
Conflicts of Interest: None declared.
References
- 1.Ayaru L, Ypsilantis P, Nanapragasam A, Choi RC, Thillanathan A, Min-Ho L, Montana G. Prediction of outcome in acute lower gastrointestinal bleeding using gradient boosting. PLoS One. 2015;10(7):e0132485. doi: 10.1371/journal.pone.0132485. http://dx.plos.org/10.1371/journal.pone.0132485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogutu J, Schulz-Streeck T, Piepho HP. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions. BMC Proc. 2012;6(Suppl 2):S10. doi: 10.1186/1753-6561-6-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran T, Luo W, Phung D, Harvey R, Berk M, Kennedy RL, Venkatesh S. Risk stratification using data from electronic medical records better predicts suicide risks than clinician assessments. BMC Psychiatry. 2014 Mar 14;14:76. doi: 10.1186/1471-244X-14-76. https://bmcpsychiatry.biomedcentral.com/articles/10.1186/1471-244X-14-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiman L, Friedman J, Stone C, Olshen R. Classification and regression trees. New York: Chapman & Hall; 1984. [Google Scholar]
- 5.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015 Jul 17;349(6245):255–60. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- 6.Ghahramani Z. Probabilistic machine learning and artificial intelligence. Nature. 2015 May 28;521(7553):452–9. doi: 10.1038/nature14541. [DOI] [PubMed] [Google Scholar]
- 7.Bone D, Goodwin MS, Black MP, Lee C, Audhkhasi K, Narayanan S. Applying machine learning to facilitate autism diagnostics: pitfalls and promises. J Autism Dev Disord. 2015 May;45(5):1121–36. doi: 10.1007/s10803-014-2268-6. http://europepmc.org/abstract/MED/25294649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metaxas P, Mustafaraj E, Gayo-Avello D. How (not) to predict elections. 2011 IEEE Third International Conference on Privacy, Security, Risk and Trust and 2011 IEEE Third International Conference on Social Computing; 2011; Boston. 2011. [Google Scholar]
- 9.Jungherr A, Jurgens P, Schoen H. Why the pirate party won the German election of 2009 or the trouble with predictions: a response to Tumasjan A, Sprenger TO, Sander PG, & Welpe IM “Predicting elections with Twitter: what 140 characters reveal about political sentiment”. Social Science Computer Review. 2011 Apr 25;30(2):229–234. doi: 10.1177/0894439311404119. [DOI] [Google Scholar]
- 10.Lazer D, Kennedy R, King G, Vespignani A. Big data. The parable of Google flu: traps in big data analysis. Science. 2014 Mar 14;343(6176):1203–5. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 11.Foster KR, Koprowski R, Skufca JD. Machine learning, medical diagnosis, and biomedical engineering research - commentary. Biomed Eng Online. 2014 Jul 05;13:94. doi: 10.1186/1475-925X-13-94. https://biomedical-engineering-online.biomedcentral.com/articles/10.1186/1475-925X-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smialowski P, Frishman D, Kramer S. Pitfalls of supervised feature selection. Bioinformatics. 2010 Feb 1;26(3):440–3. doi: 10.1093/bioinformatics/btp621. http://bioinformatics.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=19880370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Babyak M. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med. 2004;66(3):411–21. doi: 10.1097/01.psy.0000127692.23278.a9. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins DM. The problem of overfitting. J Chem Inf Comput Sci. 2004;44(1):1–12. doi: 10.1021/ci0342472. [DOI] [PubMed] [Google Scholar]
- 15.Subramanian J, Simon R. Overfitting in prediction models - is it a problem only in high dimensions? Contemp Clin Trials. 2013 Nov;36(2):636–41. doi: 10.1016/j.cct.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference and prediction. New York, NY: Springer; 2009. [Google Scholar]
- 17.Kuhn M, Johnson K. Applied predictive modeling. Berlin: Springer; 2013. [Google Scholar]
- 18.Steyerberg E. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating (Statistics for Biology and Health) New York: Springer; 2009. [Google Scholar]
- 19.Cox D. Two further applications of a model for binary regression. Biometrika. 1958 Dec;45(3/4):562–565. doi: 10.2307/2333203. [DOI] [Google Scholar]
- 20.Quinlan J. Simplifying decision trees. Int J Man Mach Stud. 1987 Sep;27(3):221–234. doi: 10.1016/S0020-7373(87)80053-6. [DOI] [Google Scholar]
- 21.Quinlan J. Induction of decision trees. Mach Learn. 1986 Mar;1(1):81–106. doi: 10.1007/BF00116251. [DOI] [Google Scholar]
- 22.Podgorelec V, Kokol P, Stiglic B, Rozman I. Decision trees: an overview and their use in medicine. J Med Syst. 2002 Oct;26(5):445–63. doi: 10.1023/a:1016409317640. [DOI] [PubMed] [Google Scholar]
- 23.Kotsiantis S. Decision trees: a recent overview. Artif Intell Rev. 2011 Jun 29;39(4):261–283. doi: 10.1007/s10462-011-9272-4. [DOI] [Google Scholar]
- 24.Kingsford C, Salzberg SL. What are decision trees? Nat Biotechnol. 2008 Sep;26(9):1011–3. doi: 10.1038/nbt0908-1011. http://europepmc.org/abstract/MED/18779814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo W, Gallagher M. Unsupervised DRG upcoding detection in healthcare databases. IEEE International Conference on Data Mining Workshops (ICDMW); 2010; Sydney. 2010. pp. 600–605. [DOI] [Google Scholar]
- 26.Siddique J, Ruhnke GW, Flores A, Prochaska MT, Paesch E, Meltzer DO, Whelan CT. Applying classification trees to hospital administrative data to identify patients with lower gastrointestinal bleeding. PLoS One. 2015;10(9):e0138987. doi: 10.1371/journal.pone.0138987. http://dx.plos.org/10.1371/journal.pone.0138987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bae S, Lee SA, Lee SH. Prediction by data mining, of suicide attempts in Korean adolescents: a national study. Neuropsychiatr Dis Treat. 2015;11:2367–75. doi: 10.2147/NDT.S91111. https://dx.doi.org/10.2147/NDT.S91111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satomi J, Ghaibeh AA, Moriguchi H, Nagahiro s. Predictability of the future development of aggressive behavior of cranial dural arteriovenous fistulas based on decision tree analysis. J Neurosurg. 2015 Jul;123(1):86–90. doi: 10.3171/2014.10.JNS141429. [DOI] [PubMed] [Google Scholar]
- 29.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 30.Boulesteix A, Janitza S, Kruppa J, König Ir. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. WIREs Data Mining Knowl Discov. 2012 Oct 18;2(6):493–507. doi: 10.1002/widm.1072. [DOI] [Google Scholar]
- 31.Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods. 2009 Dec;14(4):323–48. doi: 10.1037/a0016973. http://europepmc.org/abstract/MED/19968396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Touw W, Bayjanov JR, Overmars L, Backus L, Boekhorst J, Wels M, van Hijum SA. Data mining in the Life Sciences with random forest: a walk in the park or lost in the jungle? Brief Bioinform. 2013 May;14(3):315–26. doi: 10.1093/bib/bbs034. http://bib.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=22786785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asaoka R, Iwase A, Tsutsumi T, Saito H, Otani S, Miyata K, Murata H, Mayama C, Araie M. Combining multiple HRT parameters using the 'Random Forests' method improves the diagnostic accuracy of glaucoma in emmetropic and highly myopic eyes. Invest Ophthalmol Vis Sci. 2014 Apr 17;55(4):2482–90. doi: 10.1167/iovs.14-14009. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida T, Iwase A, Hirasawa H, Murata H, Mayama C, Araie M, Asaoka R. Discriminating between glaucoma and normal eyes using optical coherence tomography and the 'Random Forests' classifier. PLoS One. 2014;9(8):e106117. doi: 10.1371/journal.pone.0106117. http://dx.plos.org/10.1371/journal.pone.0106117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol. 1996:267–288. [Google Scholar]
- 36.Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. Journal of the Royal Statistical Societyries B (Statistical Methodology) 2011;73(3):273–282. doi: 10.1111/j.1467-9868.2011.00771.x. [DOI] [Google Scholar]
- 37.Vidaurre D, Bielza C, Larrañaga P. A survey of L1 regression. Int Stat Rev. 2013 Oct 24;81(3):361–387. doi: 10.1111/insr.12023. [DOI] [Google Scholar]
- 38.Hesterberg T, Choi Nh, Meier L, Fraley C. Least angle and ℓ 1 penalized regression: A review. Statist Surv. 2008;2:61–93. doi: 10.1214/08-SS035. [DOI] [Google Scholar]
- 39.Fujino Y, Murata H, Mayama C, Asaoka R. Applying “lasso” regression to predict future visual field progression in glaucoma patients. Invest Ophthalmol Vis Sci. 2015 Apr;56(4):2334–9. doi: 10.1167/iovs.15-16445. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu Y, Yoshimoto J, Toki S, Takamura M, Yoshimura S, Okamoto Y, Yamawaki S, Doya K. Toward probabilistic diagnosis and understanding of depression based on functional MRI data analysis with logistic group LASSO. PLoS One. 2015;10(5):e0123524. doi: 10.1371/journal.pone.0123524. http://dx.plos.org/10.1371/journal.pone.0123524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee T, Chao P, Ting H, Chang L, Huang Y, Wu J, Wang H, Horng M, Chang C, Lan J, Huang Y, Fang F, Leung SW. Using multivariate regression model with least absolute shrinkage and selection operator (LASSO) to predict the incidence of Xerostomia after intensity-modulated radiotherapy for head and neck cancer. PLoS One. 2014;9(2):e89700. doi: 10.1371/journal.pone.0089700. http://dx.plos.org/10.1371/journal.pone.0089700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J. Stochastic gradient boosting. Comput Stat Data Anal. 2002 Feb;38(4):367–378. doi: 10.1016/S0167-9473(01)00065-2. [DOI] [Google Scholar]
- 43.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21. doi: 10.3389/fnbot.2013.00021. https://dx.doi.org/10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De'ath G. Boosted trees for ecological modeling and prediction. Ecology. 2007 Jan;88(1):243–51. doi: 10.1890/0012-9658(2007)88[243:btfema]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Mayr A, Binder H, Gefeller O, Schmid M. The evolution of boosting algorithms. From machine learning to statistical modelling. Methods Inf Med. 2014;53(6):419–27. doi: 10.3414/ME13-01-0122. [DOI] [PubMed] [Google Scholar]
- 46.González-Recio O, Jiménez-Montero JA, Alenda R. The gradient boosting algorithm and random boosting for genome-assisted evaluation in large data sets. J Dairy Sci. 2013 Jan;96(1):614–24. doi: 10.3168/jds.2012-5630. [DOI] [PubMed] [Google Scholar]
- 47.Cristianini N, Shawe-Taylor J. An introduction to support vector machines and other kernel-based learning methods. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 48.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. doi: 10.1023/A:1022627411411. [DOI] [Google Scholar]
- 49.Burges C. A tutorial on support vector machines for pattern recognition. Data Min Knowl Discov. 1998;2(2):121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 50.Smola A, Schölkopf B. A tutorial on support vector regression. Stat Comput. 2004 Aug;14(3):199–222. doi: 10.1023/B:STCO.0000035301.49549.88. [DOI] [Google Scholar]
- 51.Vapnik V, Mukherjee S. Support Vector Method for Multivariate Density Estimation. Neural Information Processing Systems; 1999; Denver. 2000. [DOI] [Google Scholar]
- 52.Manevitz L, Yousef M. One-class SVMs for document classification. J Mach Learn Res. 2001;2:139–154. [Google Scholar]
- 53.Tsochantaridis I, Hofmann T, Altun Y. Support vector machine learning for interdependent and structured output spaces. the twenty-first international conference on Machine learning; 2004; Banff. 2004. [DOI] [Google Scholar]
- 54.Shilton A, Lai DT, Palaniswami M. A division algebraic framework for multidimensional support vector regression. IEEE Trans Syst Man Cybern B Cybern. 2010 Apr;40(2):517–28. doi: 10.1109/TSMCB.2009.2028314. [DOI] [PubMed] [Google Scholar]
- 55.Shashua A, Levin A. Taxonomy of large margin principle algorithms for ordinal regression problems. Adv Neural Inf Process Syst. 2002;15:937–944. [Google Scholar]