Abstract
Lignin, one of the most abundant terrestrial biopolymers, is indispensable for plant structure and defense. With the availability of the full genome sequence, large collections of insertion mutants, and functional genomics tools, Arabidopsis constitutes an excellent model system to profoundly unravel the monolignol biosynthetic pathway. In a genome-wide bioinformatics survey of the Arabidopsis genome, 34 candidate genes were annotated that encode genes homologous to the 10 presently known enzymes of the monolignol biosynthesis pathway, nine of which have not been described before. By combining evolutionary analysis of these 10 gene families with in silico promoter analysis and expression data (from a reverse transcription-polymerase chain reaction analysis on an extensive tissue panel, mining of expressed sequence tags from publicly available resources, and assembling expression data from literature), 12 genes could be pinpointed as the most likely candidates for a role in vascular lignification. Furthermore, a possible novel link was detected between the presence of the AC regulatory promoter element and the biosynthesis of G lignin during vascular development. Together, these data describe the full complement of monolignol biosynthesis genes in Arabidopsis, provide a unified nomenclature, and serve as a basis for further functional studies.
Lignin is an aromatic heteropolymer that is mainly present in secondary thickened plant cells, where it provides rigidity and impermeability to the cell walls. In addition, lignin deposition may be induced upon wounding and infection to protect plant tissues against invading pathogens. Lignin is composed of different phenylpropanoids, predominantly the monolignols p-coumaryl, coniferyl, and sinapyl alcohols that differ in their degree of methoxylation (Fig. 1). When these monolignols are incorporated into lignin, they are called p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively. In addition to the three monolignols, other phenylpropanoids, such as hydroxycinnamyl aldehydes, hydroxycinnamyl acetates, hydroxycinnamyl p-hydroxybenzoates, hydroxycinnamyl p-coumarates, and hydroxycinnamate esters, are also present in the polymer (Ralph et al., 2001; Boerjan et al., 2003). Considerable variation exists in lignin composition between taxa, cell types, and developmental and environmental conditions.
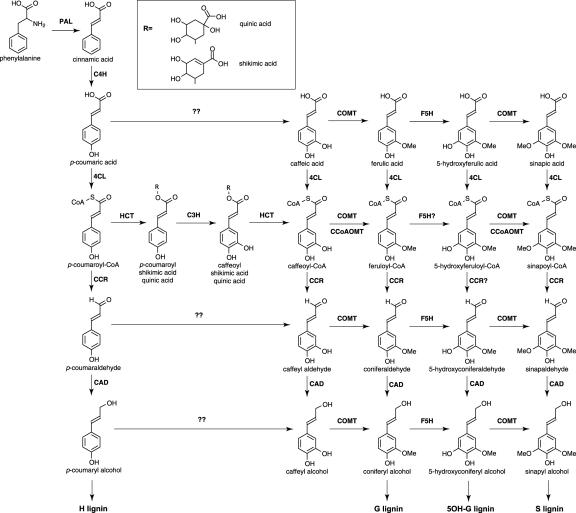
Figure 1.
The monolignol biosynthetic pathway. All the enzymatic reactions presented in the pathway have been demonstrated at least in vitro. Because of the variety in isoenzymes and kinetic properties, alternative routes through the metabolic pathway may exist. A question mark after an enzyme name means that the substrate has not been tested yet with this enzyme. For reactions with a single question mark, direct conversion has been detected, but the respective enzyme is unknown, whereas for those with a double question mark, no direct conversion has been detected.
Over the last decade, there has been a tremendous effort in cloning new genes involved in the monolignol biosynthetic pathway and in tackling the enzyme kinetics of the corresponding proteins and the role these enzymes play in controlling the amount and composition of lignin to be deposited in the cell wall (Anterola and Lewis, 2002; Humphreys and Chapple, 2002; Boerjan et al., 2003). As a consequence, the monolignol biosynthetic pathway has virtually been rewritten, although the exact route toward the monolignols is still a matter of debate (Fig. 1).
Although enzymatic assays and transgenic plants have contributed extensively to our understanding of the in vivo role of the enzymes, the role of individual gene family members has been more difficult to tackle, a limitation that can only be overcome in plant species such as Arabidopsis, for which the genome sequence and efficient reverse genetics tools are available (Arabidopsis Genome Initiative [AGI], 2000). Furthermore, the advent of genome-wide microarrays will make it possible to study the transcriptional differences that are the consequence of single gene perturbations and will allow the often pleiotropic phenotype of particular mutants to be explained at the molecular level.
As a first step toward studying the role of individual family members, we have undertaken a bioinformatics approach to identify, in Arabidopsis, all the gene family members of all monolignol biosynthesis genes known today. In many cases, only a subset of a given gene family, mostly obtained by homology-based gene isolation, has been characterized in the past. As a consequence, more distant family members might not have been discovered when, for example, primers were designed on only a few members of the family. This has led to an important bias in the range of sequence data available in public databases.
Here, we have used sensitive computational methods to delineate, in Arabidopsis, all members of the gene families currently known to be involved in monolignol biosynthesis. The integration of expression studies and promoter sequence analyses of the individual family members with phylogenetic analysis of the family has allowed us to select 12 genes as the most likely candidates to be involved in the developmental lignification in vascular tissues. Importantly, the promoter comparisons revealed a possible link between G lignin biosynthesis and the presence of the AC element that is correlated with a strong xylem expression. Together, these data describe the full complement of monolignol biosynthesis genes in Arabidopsis, introduce a unifying nomenclature for all genes of the pathway (Table I), and serve as a basis for further functional studies.
Table I.
Unifying nomenclature for gene families investigated in this study
Nomenclature was chosen to accommodate as much as possible previously published names. For explanation, see text.
| Gene Family | Gene | AGI No. | Other Names |
|---|---|---|---|
| Phe ammonia lyase (PAL) | Arath;PAL1 | At2g37040 | PAL1 |
| Arath;PAL2 | At3g53260 | PAL2 | |
| Arath;PAL3 | At5g04230 | PAL3 | |
| Arath;PAL4 | At3g10340 | ||
| Trans-cinnamate 4-hydroxylase (C4H) | |||
| Class I | Arath;C4H | At2g30490 | CYP73A5 |
| Class II | — | — | |
| 4-Coumarate:CoA ligase (4CL) | |||
| Class I | Arath;4CL1 | At1g51680 | 4CL1 |
| Arath;4CL2 | At3g21240 | 4CL2 | |
| Arath;4CL4 | At3g21230 | ||
| Class II | Arath;4CL3 | At1g65060 | 4CL3 |
| Class 4CL likes | Arath;4CL-like1 | At1g20510 | |
| Arath;4CL-like2 | At1g20500 | ||
| Arath;4CL-like3 | At1g20490 | ||
| Arath;4CL-like4 | At1g20480 | ||
| Arath;4CL-like5 | At1g62940 | ||
| Arath;4CL-like6 | At4g19010 | ||
| Arath;4CL-like7 | At4g05160 | ||
| Arath;4CL-like8 | At5g63380 | ||
| Arath;4CL-like9 | At5g38120 | ||
| Hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase (HCT) | Arath;HCT | At5g48930 | |
| p-Coumarate 3-hydroxylase (C3H) | |||
| Class I | Arath;C3H1 | At2g40890 | CYP98A3 |
| Class II | Arath;C3H2 | At1g74540 | CYP98A8 |
| Arath;C3H3 | At1g74550 | CYP98A9 | |
| Caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) | |||
| Class I | Arath;CCoAOMT1 | At4g34050 | CCoAOMT |
| Class II | Arath;CCoAOMT2 | At1g24735 | |
| Arath;CCoAOMT3 | At3g61990 | ||
| Arath;CCoAOMT4 | At3g62000 | ||
| Arath;CCoAOMT5 | At1g67990 | CCoAOMT | |
| Arath;CCoAOMT6 | At1g67980 | ||
| Arath;CCoAOMT7 | At4g26220 | ||
| Cinnamoyl-CoA reductase (CCR) | |||
| Class I | Arath;CCR1 | At1g15950 | |
| Arath;CCR2 | At1g80820 | ||
| Class CCR likes | Arath;CCR-like1 | At1g76470 | |
| Arath;CCR-like2 | At2g02400 | ||
| Arath;CCR-like3 | At2g33590 | ||
| Arath;CCR-like4 | At2g33600 | ||
| Arath;CCR-like5 | At5g58490 | ||
| Ferulate 5-hydroxylase (F5H) | Arath;F5H1 | At4g36220 | CYP84A1 |
| Arath;F5H2 | At5g04330 | CYP84A4 | |
| Caffeic acid O-methyltransferase (COMT) | |||
| Class I | Arath;COMT | At5g54160 | OMT1 |
| Class COMT likes | Arath;COMT-like1 | At1g21100 | |
| Arath;COMT-like2 | At1g21110 | ||
| Arath;COMT-like3 | At1g21120 | ||
| Arath;COMT-like4 | At1g21130 | ||
| Arath;COMT-like5 | At1g33030 | ||
| Arath;COMT-like6 | At1g51990 | ||
| Arath;COMT-like7 | At1g63140 | ||
| Arath;COMT-like8 | At1g76790 | ||
| Arath;COMT-like9 | At1g77520 | ||
| Arath;COMT-like10 | At1g77530 | ||
| Arath;COMT-like11 | At3g53140 | ||
| Arath;COMT-like12 | At5g37170 | ||
| Arath;COMT-like13 | At5g53810 | ||
| Cinnamyl alcohol dehydrogenase (CAD) | |||
| Class I | Arath;CAD2 | At3g19450 | LCAD-C |
| Arath;CAD6 | At4g34230 | LCAD-D | |
| Class II | Arath;CAD3 | At4g37970 | LCAD-A |
| Arath;CAD4 | At4g37980 | LCAD-B, ELI3-1 | |
| Arath;CAD5 | At4g37990 | ELI3-2, BAD | |
| Class III | Arath;CAD1 | At4g39330 | CAD1 |
| Arath;CAD7 | At2g21730 | LCAD-E | |
| Arath;CAD8 | At2g21890 | LCAD-F | |
| Not classified | Arath;CAD9 | At1g72680 |
RESULTS
A semiautomatic structural annotation and a phylogeny-based classification were performed using prediction results, experimental data, and information from homologous sequences (see “Materials and Methods”). A total of 34 candidate monolignol biosynthesis genes were annotated, of which nine had, to our knowledge, never been described before (Table I). In addition, 27 closely related superfamily members (“likes”) were identified in this process (Table I). To get a first insight into whether all these genes are indeed expressed and, more importantly, whether their expression pattern correlates with developmental lignification, their expression was analyzed in a set of tissues and for six developmental stages of inflorescence stem known to contain a high portion of lignifying cells. These data were compared with previous expression data from Arabidopsis and with information extracted from public databases of expressed sequence tag (EST). In addition, putative promoter elements, which drive expression during lignification, in pathogen and wound responses, and after induction by stress-related hormones, and potential subcellular localization signals were identified (due to size limitations, tables compiling all these data are available as supplemental data and at http://www.psb.ugent.be/bioinformatics/lignin/and are indexed by an “s” throughout the manuscript).
PAL
PAL (E.C. 4.3.1.5) is the first enzyme of the general phenylpropanoid pathway and catalyzes the nonoxidative deamination of Phe to trans-cinnamic acid and NH3 (Fig. 1). PAL mediates the influx from primary metabolism into the phenylpropanoid pathway and becomes rate limiting when its activity is reduced below a threshold of 20% to 25% in transgenic tobacco (Nicotiana tabacum; Bate et al., 1994; Sewalt et al., 1997).
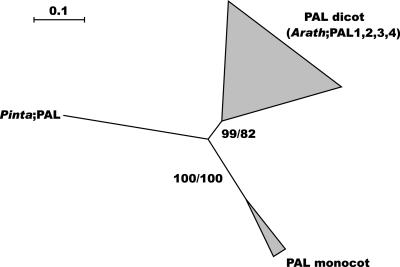
By using a thorough semiautomated annotation method, four genes encoding PAL proteins were detected in the Arabidopsis genome, three of which have been described previously (Ohl et al., 1990; Wanner et al., 1995). The phylogenetic analysis of PAL genes from various species provided no evidence for different classes in the PAL gene family (Fig. 2), although PAL1 is most closely related to PAL2, and PAL3 always clusters together with PAL4 (data not shown). The duplication that created the two PAL groups (PAL1 and PAL2 and PAL3 and PAL4) in Arabidopsis has been postulated to have predated the monocot-dicot split (Wanner et al., 1995), but the latter is not confirmed by our phylogenetic tree (Fig. 2).
Figure 2.
Neighbor-joining tree of the PAL family, inferred from Kimura corrected evolutionary distances. Bootstrap values (Neighbor-Joining/Maximum Likelihood; NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per nucleic acid. Clusters of sequences are represented as triangles with a height equal to the average distance separating the terminal nodes from the deepest branching point in the cluster and a base proportional to the number of sequences composing it. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: dicots, Populus (169453, 485808, and 1109640), Glycine (18376), Trifolium (437711), Citrus (4808125, 4808127, and 1276902), Rubus (7208613 and 7208615), Camellia (662270), Petroselinum (534892), Nicotiana (170349), Digitalis (2631994), and Lactuca (18001006); monocots, Oryza (20280 and 871493); and gymnosperms, Pinus (1143311). Arath, Arabidopsis; Pinta, pine (Pinus taeda).
PAL1 and PAL2 are not only structurally very similar, but they also share common promoter elements and a similar expression pattern (supplemental Table Is). mRNAs from both genes are most abundant in roots and stems, where the expression increases during the later stages of development (Fig. 3; supplemental Table Is; Wanner et al., 1995). Analysis of the fusion between the AtPAL1 promoter and β-glucuronidase (GUS) revealed that the expression is located in the vascular tissues (Ohl et al., 1990; Leyva et al., 1995). Besides PAL1 and PAL2, PAL4 also is highly expressed in stem tissue, as shown by our RT-PCR expression analysis (Fig. 3). In addition, PAL2 and PAL4 are abundantly expressed in the seeds, as judged from the EST data (Fig. 3; supplemental Table Is). Although all four genes are almost ubiquitously expressed in the tissues investigated in this study, PAL3 seems to be generally expressed at a lower level (supplemental Table Is; Wanner et al., 1995; Mizutani et al., 1997; Ruegger et al., 1999). PAL1 was one of the first plant defense genes identified, and its involvement in pathogen infection and abiotic stress has been studied. Among the ESTs derived from diverse stresses, PAL1 and PAL2 are clearly the most important stress-responsive family members, with 20 of 41 ESTs and 17 of 50 ESTs in total, respectively, even taking into account the relative database sizes (supplemental Table Is). In line with this, a number of regulatory elements, shown to be involved in promoter responsiveness to elicitors, wounding, and pathogen infection, were found in these genes using the stringent search method (see “Materials and Methods”; supplemental Table Is).
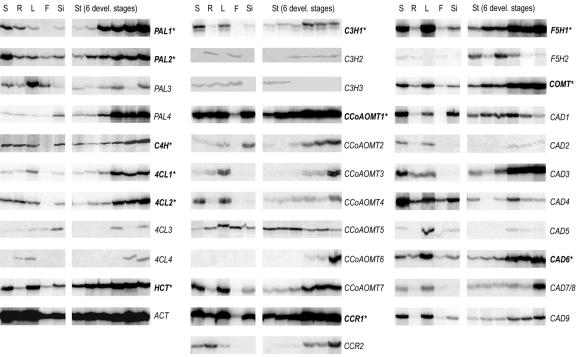
Figure 3.
Expression profiles of all 34 monolignol biosynthesis genes. Semiquantitative expression was determined using reverse transcription (RT)-PCR (see “Materials and Methods”). Due to different PCR dynamics of shorter or longer amplification products, only different tissues for a particular gene may be compared. It should be noted that CAD7 and CAD8 arose during a recent duplication event (described in detail by Tavares et al., 2000) and could not be distinguished in the RT-PCR analysis because of their high sequence similarity: 98% and 94% identity in the coding regions and putative 3′-untranslated regions, respectively. S, Seedling; R, root; L, leaf; F, flower; Si, green siliques; St, stem (at 1-, 3-, 5-, 10-, 15-, and 20-cm length).
In addition, and in accordance with the expression pattern, the promoters of PAL1 and PAL2 contain well-conserved AC elements that specify vascular expression of phenylpropanoid genes (supplemental Table Is; Ohl et al., 1990; Hauffe et al., 1993; Hatton et al., 1995; Wanner et al., 1995; Lacombe et al., 2000). An A box, proposed to work in conjunction with the AC elements in the parsley (Petroselinum crispum) PAL1 and PAL4 genes (Logemann et al., 1995), was not detected in the Arabidopsis PAL1 and PAL2 promoters (supplemental Table Is). PAL4 contains an A box but lacks an AC element. Interestingly, an H box and a G box were found in the PAL4 promoter. This combination of cis-elements was shown to be sufficient for the feed-forward induction of the chalcone synthase (CHS) promoter by p-coumaric acid in bean (Phaseolus vulgaris; Loake et al., 1992; Lindsay et al., 2002). This observation may indicate that PAL4 is regulated by the reaction product of C4H.
In conclusion, all PAL genes are expressed in the inflorescence stem, a tissue with a high portion of lignifying cells. However, the presence of an AC element qualifies PAL1 and PAL2 as the most likely candidates to be involved in monolignol biosynthesis in the vascular lignifying cells. In accordance, the corresponding mutants show defects in lignin formation (A. Rohde and W. Boerjan, unpublished data).
C4H
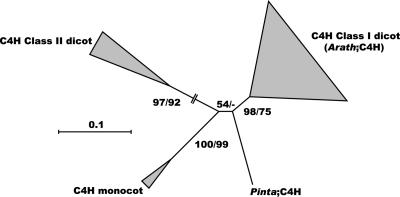
C4H (E.C. 1.14.13.11) controls the conversion of cinnamate into p-coumarate (Fig. 1). C4H (CYP73A5) belongs to the cytochrome P450-dependent monooxygenases, like the two other hydroxylases in the pathway (C3H, F5H). So far, only one C4H gene has been described in Arabidopsis (Bell-Lelong et al., 1997; Mizutani et al., 1997; Urban et al., 1997). Although multiple family members have been detected in other plants (Betz et al., 2001, and refs. therein), we could not find any evidence for additional CYP73 genes in Arabidopsis. Phylogenetic analysis shows that two classes of C4H genes exist in plants (Fig. 4; Nedelkina et al., 1999; Betz et al., 2001). Furthermore, the tree topology indicates that the origin of these two classes has predated the divergence of gymnosperms and angiosperms, suggesting that class II members must have existed at some time in the evolution for most plant lineages. The Arabidopsis C4H gene belongs to class I; a class II homolog was most probably lost during the evolution of this species.
Figure 4.
Neighbor-joining tree of the C4H family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank Identifier numbers of non-Arabidopsis sequences included in this tree are: Class I dicots, Populus (12276037, 3915089, and 3915096), Gossypium (9965899 and 9965897), Petroselinum (3915088), Ruta (13548653), Citrus (8572559 and 14210375), Catharanthus (1351206), Lithospermum (16555879 and 16555877), Capsicum (3603454 and 12003968), Zinnia (3915112), Helianthus (417863), Glycine (3915111), Phaseolum (586082), Glycyrrhiza (3915095), Cicer (14917048), Medicago (586081), and Pisum (3915077 and 9957081); Class II dicots, Mesembryanthemum (4206116), Citrus (7650489), Phaseolus (7430650), and Nicotiana (14423323 and 14423325); monocots, Triticum (10442761) and Sorghum (14192803); and gymnosperms, Pinus (4566493). Arath, Arabidopsis; Pinta, pine.
C4H is expressed in all tissues and upon exposure to light, wounding, and fungal infection (supplemental Table IIs; Bell-Lelong et al., 1997; Meyer et al., 1998; Nair et al., 2002). In our RT-PCR experiment, C4H expression increased during the later stages of stem development (Fig. 3). Activity of AtC4H::GUS coincides with vascular cells in the inflorescence stem and in leaves, but in roots the promoter is active in all cells, giving the strongest expression in this tissue (Bell-Lelong et al., 1997; Nair et al., 2002). A strong C4H expression is also found in siliques and seeds, where it could be involved in the production of sinapate esters (Chapple et al., 1994). In addition, the C4H promoter contains an H box, which might be responsible for induction of C4H expression after elicitation.
By TargetP (Emanuelsson et al., 2000), the C4H protein is predicted to contain an endoplasmic reticulum (ER)-targeting peptide. However, this peptide coincides with the membrane anchor region of P450 enzymes, whose features are a stretch of hydrophobic amino acids, followed by a small region rich in basic amino acids and a hinge region of the conserved (P/I)PGPx(G/P)xP sequence (Chapple, 1998). All class II C4H proteins included in the phylogenetic analysis (Fig. 4) show a divergent hinge and basic amino acid region. Although the function of these class II C4H proteins is unclear at the moment, the shared degeneration of this crucial region could be an important clue in discovering their function.
4CL
4CL (E.C. 6.2.1.12) catalyzes the formation of CoA esters of p-coumaric acid, caffeic acid, ferulic acid, 5-hydroxyferulic acid, and sinapic acid (Fig. 1; Lee et al., 1997; Hu et al., 1998). The plethora of additional potential substrates may explain why there are many 4CL isoenzymes in most plants. In addition to the different substrate specificities, the genes typically have a distinct spatio-temporal expression pattern (Lewis and Yamamoto, 1990; Hu et al., 1998; Harding et al., 2002).
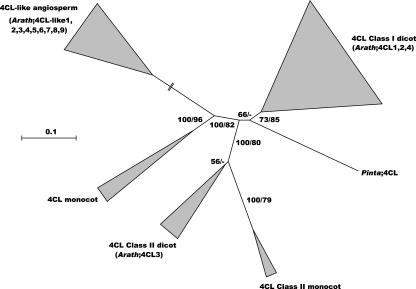
We detected four 4CL and nine 4CL-like genes in the Arabidopsis genome. Phylogenetic analysis of the predicted proteins, together with characterized 4CL proteins and luciferases, acetate, and fatty acid CoA-ligases (other adenylate-forming enzymes; data not shown), shows that 4CL proteins fall into two classes (Fig. 5; Ehlting et al., 1999; Cukovic et al., 2001). Three of the Arabidopsis proteins belong to class I (4CL1, 4CL2, and 4CL4) and 4CL3 to class II; the remaining nine are classified as 4CL like because they do not correspond to any of the 4CL or other enzyme classes mentioned above.
Figure 5.
Consensus of two neighbor-joining trees of the 4CL and 4CL-like proteins, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: Class I dicots, Solanum (398963, 398965, and 5163399), Capsicum (12003966), Nicotiana (12229631, 7428495, and 12229632), Lithospermum (1117778), Petroselinum (112800 and 112801), Rubus (9651915 and 9651917), Populus (7437854, 7437855, 14289344, 18032806, 7437852, and 15636677), and Amorpha (17063848); gymnosperms, Pinus 4CL (7437872); Class II monocots, Lolium (7188335) and Oryza (12229650); Class II dicots, Lithospermum (9988455), Glycine (18266852), and Populus (7437853 and 14289346); monocots, Oryza (112802), Lolium (7188337 and 7188339); and 4CL-like, Oryza (12039389). Arath, Arabidopsis; Pinta, pine.
Our expression analysis showed that 4CL genes are expressed in almost all investigated tissues, with 4CL4 having the most restricted expression (Fig. 3). The latter observation is supported by the smallest number of ESTs found for 4CL4 among the 4CL genes (supplemental Table IIIs). 4CL1 and 4CL2 are expressed throughout inflorescence stem development and expression increases during the later stages (supplemental Table IIIs; Lee et al., 1995; Mizutani et al., 1997; Ehlting et al., 1999). On the contrary, 4CL3 and 4CL4 are expressed only during the later stages of inflorescence stem development (Fig. 3; supplemental Table IIIs). The expression of 4CL3 is not affected by wounding and Peronospora parasitica infection, in clear difference to the class I 4CL genes (supplemental Table IIIs; Ehlting et al., 1999). In accordance with the expression analysis, the promoters of both 4CL1 and 4CL2 contain AC elements. Furthermore, the promoter analysis identified an AT-rich sequence motif in the 4CL4 promoter and an H box in the 4CL3 and 4CL4 promoters, hinting to a role in particular stress responses (Seki et al., 1996; Rushton et al., 2002).
In conclusion, 4CL1 and 4CL2 are the best candidates for a function in monolignol biosynthesis during developmental lignification, as suggested previously by Ehlting et al. (1999). Their expression correlates with tissues containing a high portion of lignifying cells, and AC elements are present in their promoters. To the contrary, 4CL3 (class II) was suggested to channel activated p-coumarate to CHS and subsequently to the flavonoid biosynthesis (Ehlting et al., 1999). 4CL4 (class I), although expressed more specifically or at a lower level, might have yet another substrate specificity. In soybean (Glycine max), a single amino acid deletion determines whether or not 4CL can use sinapic acid as a substrate (Lindermayr et al., 2003), a function lacking for 4CL1, 4CL2, and 4CL3 in Arabidopsis (Ehlting et al., 1999). Interestingly, 4CL4 shows a similar deletion in the region coding for the substrate–binding pocket, suggesting that this gene may have acquired an altered substrate specificity toward sinapic acid after duplication. A recent paper shows that 4CL4 is indeed able to convert sinapic acid (Schneider et al., 2003).
HCT
HCT belongs to a large family of acyltransferases that are involved in the biosynthesis of diverse secondary metabolites. Only recently, the first HCT has been purified from tobacco stems, and the corresponding gene was cloned (Hoffmann et al., 2003). In tobacco, HCT catalyzes the conversion of p-coumaroyl-CoA and caffeoyl-CoA to the corresponding shikimate or quinate esters (Fig. 1). These shikimate and quinate esters, themselves being important intermediates in the phenylpropanoid pathway, have been shown recently to be good substrates for C3H (Kühnl et al., 1987; Schoch et al., 2001; Franke et al., 2002a, 2002b; Nair et al., 2002). Moreover, HCT catalyzes also the reverse transesterification (Hoffmann et al., 2003). Therefore, HCT might play a critical role up- and downstream of C3H. For the Arabidopsis HCT homolog, a biochemical activity similar to that of the tobacco HCT has been shown (Hoffmann et al., 2003).
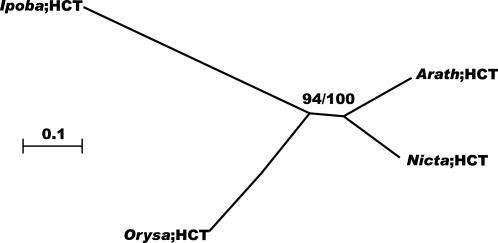
Here, one HCT gene was detected in the Arabidopsis genome (Fig. 6). Because only two homologs were characterized and the family is apparently well conserved (approximately 60% identity between monocot and dicot members; data not shown), no more distantly related genes were included.
Figure 6.
Neighbor-joining tree of the HCT family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: Ipomoea (6469032), Oryza (21740518), and Nicotiana (27475615). Arath, Arabidopsis; Ipoba, Ipomoea batatas; Nicta, tobacco; Orysa, rice (Oryza sativa).
The expression analysis shows that HCT is expressed in all tissues investigated but strongly in the inflorescence stem (Fig. 3; supplemental Table IVs). The promoter contains an AC element. The high and ubiquitous expression is confirmed by the second highest number of ESTs found for the 10 gene families analyzed (supplemental Table IVs). Interestingly, the combined presence of an H and a G box was observed, as for PAL4 and F5H2, suggesting transcriptional regulation by the pathway intermediate p-coumaric acid (Loake et al., 1992).
C3H
C3H was originally named after its suspected function in C3-hydroxylation of p-coumaric acid, but recently, CYP98A3 (C3H1) was shown to preferentially convert the shikimate and quinate esters of p-coumaric acid into the corresponding caffeic acid conjugates, whereas p-coumaric acid and p-coumaroyl-CoA were not substrates of this enzyme (Fig. 1; Schoch et al., 2001; Franke et al., 2002b; Nair et al., 2002).
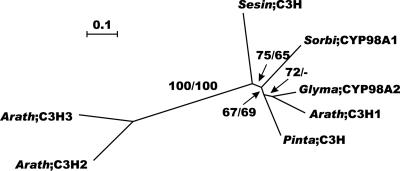
We detected three C3H genes in the Arabidopsis genome, which all belong to the CYP98 class of the P450 enzymes. Only a few proteins of this class could be found from other species for phylogenetic analysis (Fig. 7). Arabidopsis C3H1 clusters with all known C3Hs of other species, whereas C3H2 and C3H3 (CYP98A8 and CYP98A9, respectively) probably constitute a different class that diverged before the gymnosperm-angiosperm split (Fig. 7).
Figure 7.
Neighbor-joining tree of the C3H family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: Sesamum (17978831), Sorghum (5915857), Pinus (17978651), and Glycine (5915858). Arath, Arabidopsis; Glyma, soybean; Sesin, Sesamum indicum; Sorbi, Sorghum bicolor; Pinta, pine.
The expression analysis shows that C3H1 is expressed in all tissues, an observation that is supported by ESTs from various tissues (supplemental table Vs). Previous studies detected the highest expression in the vascular tissues of stem and root (supplemental table Vs; Schoch et al., 2001; Franke et al., 2002b; Nair et al., 2002). On the contrary, C3H2 and C3H3 are expressed only during particular stages of inflorescence stem development: C3H2 is expressed in older stems and C3H3 in young developing stems (Fig. 3). The fact that only one EST is found for C3H2 and none for C3H3 suggests that they are either conditionally regulated or expressed at low levels (supplemental table Vs). The promoter analysis reveals a well-conserved AC element in the promoter of C3H1, in agreement with its vascular expression detected by the GUS reporter system (Nair et al., 2002).
Analysis of the N terminus by TargetP predicts the C3H1 protein to contain an ER-targeting peptide, but it overlaps, as for C4H, with the membrane anchor region of P450 enzymes. The C3H1 protein has previously been localized in the membrane fraction in yeast (Franke et al., 2002b). In contrast to C3H1, the sequences of C3H2 and C3H3 are divergent in both the stretch of basic amino acids and the hinge region of the membrane anchor. Because these regions are necessary for the correct insertion of the enzyme in the membrane (Chapple, 1998), the degeneration of this region suggests they are not membrane-anchored proteins. It should be noted that C3H2 and C3H3 do not hydroxylate shikimate and quinate esters of p-coumaric acid (Schoch et al., 2001). In conclusion, C3H1 is involved in the monolignol pathway, as is functionally demonstrated with the ref8 (reduced epidermal fluorescence) mutant (Franke et al., 2002a, 2002b).
CCoAOMT
CCoAOMT (E.C. 2.1.1.104) catalyzes the methylation of caffeoyl-CoA to feruloyl-CoA (in vitro and in vivo) and 5-hydroxyferuloyl-CoA to sinapoyl-CoA (at least in vitro) and is, together with COMT, responsible for the methylation of the monolignol precursors (Fig. 1; Ye et al., 1994; Zhong et al., 1998; Pinçon et al., 2001).
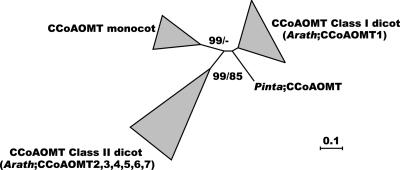
Seven putative members of the CCoAOMT gene family were detected in the Arabidopsis genome (Fig. 8). Plant CCoAOMT genes fall into two classes: Class I contains the Arabidopsis CCoAOMT1 gene together with the majority of experimentally characterized CCoAOMT genes (e.g. Zhong et al., 1998; Meyermans et al., 2000), whereas class II consists of six Arabidopsis genes and a few sequences from other species. The latter class does not closely resemble most of the certified CCoAOMT genes but contains an experimentally characterized chickweed (Stellaria longipes) CCoAOMT able to methylate caffeoyl-CoA (Zhang and Chinnappa, 1997).
Figure 8.
Neighbor-joining tree of the CCoAOMT family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per nucleic acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: Class I dicots, Populus (2960355, 857577, 13249170, and 2960357), Zinnia (533120), Petroselinum (169648), Nicotiana (2511736), Citrus (6561880), Vitis (1000518), and Eucalyptus (5739372 and 1934858); gymnosperms, Pinus CCoAOMT (4104458); Class II dicots, Stellaria (438896) and Populus (1785476); and monocots, Zea (5101869, 5101867) and Oryza (5091496 and 5257255 [three genes]). Arath, Arabidopsis; Pinta, pine.
CCoAOMT1 is expressed in all tissues investigated and has by far the highest number of ESTs (Fig. 3; supplemental Table VIs). Moreover, the CCoAOMT1 gene has two AC elements in its promoter. CCoAOMT1 is highly expressed in the basal portion of the inflorescence as compared with the apical portion (Goujon et al., 2003). Of the class II genes, CCoAOMT5 and CCoAOMT7 are expressed in all tissues, but only the expression of CCoAOMT7 increases during the later stages of inflorescence stem development. Furthermore, CCoAOMT4 and CCoAOMT5 are also expressed at all stages of inflorescence stem development. Others, such as CCoAOMT2, CCoAOMT3, and CCoAOMT6, are expressed toward the end of inflorescence stem development (Fig. 3). Few ESTs have been found for most genes of class II (supplemental Table VIs).
CCoAOMT genes of other species were shown to be responsive to pathogens or elicitors (e.g. Pakusch et al., 1991; Chen et al., 2000); corresponding promoter elements were identified in CCoAOMT1, CCoAOMT2 and CCoAOMT3 (supplemental Table VIs). CCoAOMT3 has an extended N-terminal sequence, not shared by any of the other CCoAOMTs, predicted to be an ER-targeting peptide.
Based on its clustering in class I, its expression characteristics and level, and the presence of two AC elements in its promoter, CCoAOMT1 is the main candidate gene to be involved in the monolignol pathway during developmental lignification.
CCR
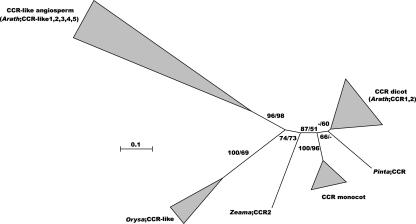
CCR (E.C.1.2.1.44) catalyzes the conversion of cinnamoyl-CoA esters to their respective cinnamaldehydes and is the first enzyme of the monolignol-specific part of the lignin biosynthetic pathway (Fig. 1). The two previously described CCR genes and five new CCR-like genes were found (Fig. 9; Jones et al., 2001; Lauvergeat et al., 2001).
Figure 9.
Neighbor-joining tree of the CCR family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank identification numbers of non-Arabidopsis sequences included in this tree are: CCR dicots, Eucalyptus (7431407, 7431408, and 10304406) and Populus (7239228, 2960364, and 9998901); CCR monocots, Lolium (9964087), Saccharum (3341511 and 17978549), and Zea (7431410 and 3242328); gymnosperms, Pinus CCR (17978649), Zea CCR2 (3668115), and Oryza CCR-like (13486725, 13486726, and 18307514); and CCR-like angiosperms, Oryza (15624051). Arath, Arabidopsis; Orysa, rice; Pinta, pine; Zeama, maize (Zea mays).
CCR1 is highly expressed in all tissues examined, whereas CCR2 is in all tissues but flowers, siliques, and the earliest stage of inflorescence stem development (Fig. 3). Although CCR2 was hardly detected in stem by RNA gel blots (Lauvergeat et al., 2001), the more sensitive RT-PCR clearly reveals CCR2 expression in the inflorescence stem (Fig. 3). For both genes, expression increases with age during inflorescence stem development (Fig. 3). Corresponding with the differences in expression levels of CCR1 and CCR2 (Lauvergeat et al., 2001), 10-fold more ESTs are found for CCR1 than for CCR2 (supplemental Table VIIs). Both genes are induced by Xanthomonas campestris infection and ESTs linked with stress and pathogen infection have been detected (Lauvergeat et al., 2001; supplemental Table VIIs). The promoter of CCR1 contains a well-conserved AC element and conforms with its function in lignification and the strong expression in stems (Lauvergeat et al., 2001; supplemental Table VIIs).
In conclusion, CCR1 and CCR2 are expressed during both developmental lignification and pathogen response, as documented by our expression analysis and ESTs (Fig. 3; supplemental Table VIIs). The role of CCR1 in lignification has clearly been established through the irx4 (irregular xylem) mutant characterization (Jones et al., 2001). Although CCR2 seems to be implicated in stress and elicitor response (Lauvergeat et al., 2001), the expression results do not exclude a (minor) role for CCR2 in developmental lignification.
F5H
F5H, also called coniferaldehyde 5-hydroxylase, is a cytochrome P450-dependent monooxygenase (CYP84) that is required for the production of syringyl lignin because it is responsible for the 5-hydroxylation of coniferaldehyde and/or coniferyl alcohol (Fig. 1; Humphreys et al., 1999; Li et al., 2000; Humphreys and Chapple, 2002).
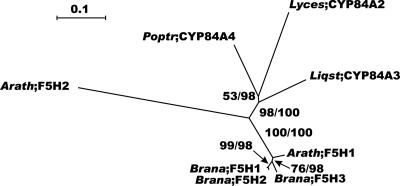
The Arabidopsis genome harbors two F5H homologs, both belonging to the CYP84 family of the P450 monooxygenases. F5H1 (CYP84A1) has been characterized in Arabidopsis, Liquidambar styraciflua, and Brassica napus (Meyer et al., 1996; Osakabe et al., 1999; Nair et al., 2000), whereas F5H2 (CYP84A4), a more divergent member of the CYP84 family, is described for the first time, to our knowledge, in this study. So far, no genes that closely resemble F5H2 have been detected in other plants, although the phylogeny indicates that the two proteins found in Arabidopsis diverged before the divergence of the different Rosidae subfamilies (Fig. 10).
Figure 10.
Neighbor-joining tree of the F5H family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. GenBank identification numbers of non-Arabidopsis sequences included in this tree are: Populus CYP84A4 (6688937), Lycopersicon CYP84A2 (5002354), Liquidambar CYP84A3 (5731998), and Brassica F5H1, F5H2, and F5H3 (10197650, 10197652, and 10197654). Arath, Arabidopsis; Brana, Brassica napus; Liqst, Liquidambar styraciflua; Lyces, tomato (Lycopersicon esculentum); Poptr, Populus trichocarpa.
Our expression analysis revealed F5H1 expression in all tissues and an increasing expression during inflorescence stem development (Fig. 3), in accordance with results of earlier studies (supplemental Table VIIIs; Meyer et al., 1998; Ruegger et al., 1999; Goujon et al., 2003). F5H1 was also expressed in several other tissues but mainly in young and senescent leaves and in roots (Meyer et al. 1996; Ruegger et al., 1999). In contrast to F5H1, F5H2 had the strongest expression in the early stages of inflorescence stem development (Fig. 3). Only two ESTs were found for F5H1 and none for F5H2 (supplemental Table VIIIs).
In the promoter analysis, for both genes an H box was found and for F5H2 a G box was also found, suggesting that both genes may be inducible and that F5H2 may be regulated by p-coumarate (Loake et al., 1992; Lindsay et al., 2002). Moreover, F5H1 and F5H2 contain a fully conserved membrane anchor region. In addition, F5H2 is predicted to contain an ER-targeting peptide that coincides with the region of the membrane anchor of P450 enzymes. Remarkably, no AC element was detected for either F5H gene, although F5H1 had been shown to be involved in lignification through the analysis of the fah1 mutant (Chapple et al., 1992).
COMT
COMT (E.C. 2.1.1.68) was originally postulated to be a bifunctional enzyme methylating caffeic acid and 5-hydroxyferulic acid. However, in vitro and transgenic studies revealed that the predominant role of COMT is the methylation of 5-hydroxyconiferaldehyde and/or 5-hydroxyconiferyl alcohol to sinapaldehyde and/or sinapyl alcohol, respectively (Fig. 1; Osakabe et al., 1999; Li et al., 2000; Chen et al., 2001; Guo et al., 2001; Parvathi et al., 2001; Goujon et al., 2003).
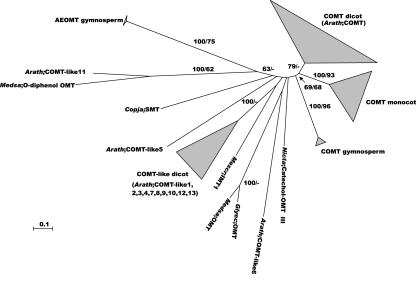
We detected only one COMT gene in the Arabidopsis genome. Furthermore, 13 proteins similar to COMT were detected that clustered in-between the functionally characterized COMT clade and the cluster containing the hydroxycinnamic acid/hydroxycinnamoyl-CoA ester O-methyltransferase protein (AEOMT; Li et al., 1997, 1999), i.e. among proteins that have been shown to use a wide variety of substrates (Fig. 11; Vernon and Bohnert, 1992; Maxwell et al., 1993; Pellegrini et al., 1993; Takeshita et al., 1995). Because the role of AEOMT in the monolignol pathway is still a matter of debate (Anterola et al., 2002), and other COMT candidate genes of conifers clustered much more closely to the known COMTs, it is unclear whether these 13 genes play any role in the monolignol pathway. Therefore, these genes were classified as COMT likes. As a consequence, only one class of COMTs exists in plants (Fig. 11; Maury et al., 1999).
Figure 11.
Neighbor-joining tree of the COMT family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank Identifier numbers of non-Arabidopsis sequences included in this tree are: COMT dicots, Populus (7528266, 762870, 231757, 444327, 7332271, 7447887, and 762872), Stylosanthes (1582580), Medicago (116908), Prunus (3913295), Fragaria (6760443), Liquidambar (5732000), Chrysosplenium (1184041 and 567077), Vitis (7271883), Capsicum (3421382, 7488967, and 12003964), Nicotiana (480082 and 480083), Eucalyptus (1169009 and 5739365), Clarkia (2832224 and 3913289), Mesembryanthemum (7447880), Thalictrum (4808522, 4808524, 4808526, 4808528, and 4808530), Catharanthus (18025321), Ocimum (5031492, 5031494), and Zinnia (642952); COMT monocots, Lolium (4104220, 4104222, 4104224, and 2388664), Sorghum (18033964), Saccharum (3341509), Zea (729135), and Festuca (14578611, 14578613, 14578615, and 14578617); COMT gymnosperms, Pinus (15524083), Picea (COMT-C7 and COMT-C16; M.H. Walter, personal communication); Nicotiana Catechol-OMT III (542050); Glycyrrhiza OMT (1669591), Medicago OMT (7447884), Mesembryanthemum IMT1 (1170555), Coptis sinapoyl-Glc:malate sinapoyltransferase (SMT; 758580), and Medicago O-diphenol OMT (6688808); and AEOMT gymnosperms, Pinus (7447883, 1777386, and 4574324). Arath, Arabidopsis; Copja, Coptis japonica; Glyec, Glycyrrhiza echinata; Medsa, alfalfa; Mescr, Mesembryanthemum crystallinum; Nicta, tobacco.
Our RT-PCR data show that COMT is expressed in all tissues investigated, and the numerous ESTs point toward a generally high and ubiquitous expression (Fig. 3; supplemental Table IXs). Ninety-nine COMT ESTs, with a fifth being stress related, is almost twice the number found for any other gene in this analysis (supplemental Table IXs). COMT expression is particularly high in the inflorescence stem, with an increase during the later stages of development (Fig. 3; supplemental Table IXs). Correspondingly, COMT::GUS expression occurs in xylem, differentiating fibers, and mature phloem (Goujon et al., 2003). Unlike many other monolignol biosynthesis genes, COMT has no AC elements in its promoter. In fact, to the best of our knowledge, AC elements have never been reported in COMT promoters of other plants either.
Interestingly, the COMT protein might be myristoylated. The N-terminal MGSTAETQLTPVQVTDDE sequence was identified as a “twilight zone” myristoylation signal, which corresponds both with truly myristoylated proteins and with false positives (Maurer-Stroh et al., 2002). Myristoylation is generally associated with cell membrane anchoring or, as recently shown for an Arabidopsis protein kinase, ER attachment (Lu and Hrabak, 2002). Pending the experimental verification of this observation, the putative localization of the COMT protein indicates a new research avenue in the field of monolignol channeling and export.
CAD
CAD (E.C. 1.1.1.195) catalyzes the last step in monolignol biosynthesis, i.e. the reduction of cinnamyl aldehydes into their corresponding alcohols (Fig. 1). CAD reduces various aldehydes, present in different cell types or during different stages of development. Besides the function in developmentally regulated lignification, a number of CAD genes have been characterized for their response to plant pathogens (Kiedrowski et al., 1992).
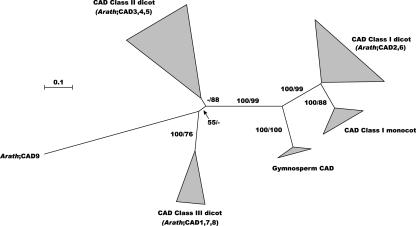
Here, nine putative CAD genes were detected in the Arabidopsis genome (Table I; Tavares et al., 2000; Sibout et al., 2003). Our phylogenetic analysis revealed that eight of the CAD proteins fall into three classes, whereas CAD9 is more divergent (Fig. 12). CAD2 and CAD6, belonging to the class I CADs, closely resemble CAD proteins that have been characterized for their involvement in lignification in other species. The topology of the tree indicates furthermore that the class I “true” CAD clade diverged from the other CADs before the angiosperm-gymnosperm split (Fig. 12).
Figure 12.
Neighbor-joining tree of the CAD family, inferred from Kimura corrected evolutionary distances. Bootstrap values (NJ/ML) above 50% are shown at the internodes. The scale measures evolutionary distance in substitutions per amino acid. Clusters of sequences are represented as described in Figure 2. Species and GenBank Identifier numbers of non-Arabidopsis sequences included in this tree are: Class I dicots, Populus (421814, 1168734, 9998899, and 7239226), Nicotiana (231676 and 231675), Medicago (399168), Aralia (1168727), Zinnia (1944403), Eucalyptus (1705554, 10281656, 399165, 10719920, and 3913185); Class I monocots, Saccharum (10719916), Zea (3913182 and 7430938), Lolium (3913181), Festuca (15428276, 15428278, 15428280, and 15428282); gymnosperm CAD, Picea (584872 and 10719915), Pinus (107623, 3334135, 1168733, and 3372645); Class II dicots, Stylosanthes (3913194), Apium (12643507), Petroselinum (1168732), Lycopersicon (8099340 and 7430935), Mesembryanthemum (10720090), Fragaria (10720093, 13507210), and Populus (14279694); and Class III dicots: Stylosanthus (3913193) and Medicago (10720088). Arath, Arabidopsis.
Class II CADs (CAD3, CAD4, and CAD5) cluster with a number of alcohol dehydrogenases with diverse substrate preferences, such as the poplar (Populus tremuloides) sinapyl alcohol dehydrogenase (Li et al., 2001), the celery (Apium graveolens) mannitol dehydrogenase (Williamson et al., 1995), and the parsley ELI3/CAD proteins (Kiedrowski et al., 1992; Logemann et al., 1997). CAD4 (AtELI3-1) and CAD5 (AtELI3-2) have been identified previously as responsive to elicitor treatments and Pseudomonas syringae infection (Kiedrowski et al., 1992). Moreover, CAD5 has a substrate specificity distinct from “true” CADs, mannitol dehydrogenase, and aromatic alcohol: NADP+ oxidoreductase and was, therefore, named benzyl alcohol dehydrogenase (BAD; Somssich et al., 1996).
Class III CADs (CAD1, CAD7, and CAD8) cluster in a group with an alcohol dehydrogenase from alfalfa (Medicago sativa), which is able to catalyze the reduction of cinnamaldehyde, sinapaldehyde, and coniferaldehyde, but also several aliphatic aldehydes and various substituted benzaldehydes (Brill et al., 1999). Being very divergent from class I “true” CADs, this class also represents a group of multisubstrate alcohol dehydrogenases.
All CAD genes, except CAD2, CAD4, and CAD5, are expressed in all stages of inflorescence stem development (Fig. 3). Moreover, CAD2 and CAD6 are expressed in the inflorescence stem close to the bundle and interfascicular cambium, as revealed by promoter::GUS constructs (Sibout et al., 2003). Expression of most CAD genes is documented by ESTs, except for CAD7 and CAD8, which are nevertheless expressed, as indicated in the RT-PCRs (Fig. 3; supplemental Table Xs).
The promoter analysis revealed that CAD6 from class I and CAD5 from class II contain AC elements (supplemental Table Xs). In addition, an A box was detected in the CAD6 promoter. The fact that only one gene in the pathway contains both an AC element and an A box casts doubt on the previous assumption that an A box works in conjunction with AC elements (Logemann et al., 1995).
Based on the fact that they cluster with other well-characterized “true” CAD genes in the phylogenetic tree, CAD2 and CAD6 are the most likely candidates for the monolignol pathway in Arabidopsis. Of these two, only CAD6 has an AC element. Moreover, only the CAD6 gene mutant, but not that of CAD2, showed altered lignin content and structure (Sibout et al., 2003). The function of class II and class III CAD genes remains less clear. However, CAD3, CAD4, and CAD5 of class II are the closest homologs of the poplar SAD (Li et al., 2001). Possibly, these proteins show a preference for sinapyl alcohol or sinapaldehyde, turning them into S branch-specific enzymes.
DISCUSSION
Toward the Core Monolignol Biosynthesis Gene Set for Developmental Lignification
Lignification is a process that occurs predominantly in cells of the vascular tissue, found in almost all organs, but most abundantly in stems and roots. A strong expression of monolignol biosynthesis genes in stems and roots is documented in numerous publications (supplemental Tables Is–Xs, and refs. therein). Possibly, lignification cDNAs are relatively highly represented in root libraries because of the absence of other very abundant processes, such as photosynthesis, or, as could be concluded from AtC4H::GUS analysis (Nair et al., 2002), the phenylpropanoid pathway in roots is active in more cells than the vascular ones to generate compounds not destined for lignification. In addition, flowers, seeds, and siliques accumulate significant amounts of other phenylpropanoid-derived compounds, such as sinapate esters and flavonoids (Chapple et al., 1994; Chen and McClure, 2000).
All 34 genes, annotated from the Arabidopsis genome sequence for their potential involvement in monolignol biosynthesis, are expressed at some stage of inflorescence stem development, a tissue with a prominent portion of lignifying cells (Table I and supplemental Tables Is–Xs; Dharmawardhana et al., 1992). Of these genes, 23 are expressed throughout stem development (Table II). Furthermore, the expression of many genes increases during the later stages of inflorescence stem development, when lignification is more prominent (Fig. 3; Dharmawardhana et al., 1992).
Table II.
Summary of expression characteristics and occurrence of AC elements in monolignol biosynthesis genes
Characteristics are listed for each gene (marked with x): the corresponding mutants with lignification-related phenotypes, the clustering with certified proteins of other species in the phylogenetic analysis, a constitutive expression in the inflorescence stem (as determined by our RT-PCR analysis), ESTs of the different relevant categories in total nos., and the occurrence of AC elements. Genes marked with asterisks are predicted to be membrane associated. The position of AC elements found with stringent parameters is given in base pairs from ATG and the strand within parentheses. Underlined numbers indicate an overrepresentation of ESTs in this particular tissue or condition as compared with the presence of this gene in all ESTs. Overrepresentation was judged by comparing the relative occurrence of a gene in all ESTs with that of the same gene in a particular tissue or condition (see supplemental Tables Is–Xs for values). When fewer than three ESTs were detected in a particular tissue or condition, no overrepresentation was calculated. ESTs from inflorescence stem (presented in the supplemental Tables Is–Xs) are not presented here due to the small no. of ESTs available from this tissue (see “Materials and Methods”).
| Gene Family
|
Genes with AC Element
|
Gene family
|
Genes without AC Element
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Corresponding mutants | Phylogenetic clustering | Constitutively in stem | EST total | EST aboveground organs | EST root | EST seed | EST stress, wound, pathogen | AC element (+/-) strand (core 0.9; matrix 0.9) | Gene | Corresponding mutants | Phylogenetic clustering | Constitutively in stem | EST total | EST aboveground organs | EST root | EST seed | EST stress, wound, pathogen | ||
| PAL | PAL1 | pal1a | x | x | 41 | 1 | 13 | 2 | 21 | 483 (+) | PAL | PAL3 | x | x | 1 | |||||
| PAL2 | pal2a | x | x | 50 | 2 | 17 | 12 | 17 | 246 (+), 495 (+) | PAL4 | x | x | 28 | 1 | 5 | 16 | 4 | |||
| C4H | C4H | C4H* | ref3b | x | x | 29 | 2 | 5 | 15 | |||||||||||
| 4CL | 4CL1 | x | x | 8 | 1 | 4 | 159 (+) | 4CL | 4CL3 | 8 | 1 | 2 | 3 | |||||||
| 4CL2 | x | x | 13 | 6 | 2 | 2 | 124 (+), 233 (+) | 4CL4 | x | 2 | 2 | |||||||||
| HCT | HCT | x | x | 57 | 13 | 11 | 8 | 15 | 132 (-) | HCT | ||||||||||
| C3H | C3H1* | ref8 | x | x | 36 | 5 | 5 | 7 | 16 | 145 (+) | C3H | C3H2 | 2 | |||||||
| C3H3 | 0 | |||||||||||||||||||
| CCoAOMT | CCoAOMT1 | x | x | 45 | 19 | 9 | 10 | 174 (+), 651 (+) | CCoAOMT | CCoAOMT2 | 0 | |||||||||
| CCoAOMT3* | 6 | 3 | ||||||||||||||||||
| CCoAOMT4 | x | 2 | 1 | |||||||||||||||||
| CCoAOMT5 | x | 1 | ||||||||||||||||||
| CCoAOMT6 | 2 | |||||||||||||||||||
| CCoAOMT7 | x | 4 | 3 | 1 | ||||||||||||||||
| CCR | CCR1 | irx4 | x | x | 43 | 8 | 8 | 8 | 10 | 269 (+) | CCR | CCR2 | x | 4 | 3 | 1 | ||||
| F5H | F5H | F5H1* | fah1 | x | x | 2 | ||||||||||||||
| F5H2* | x | 0 | ||||||||||||||||||
| COMT | COMT | COMT* | comt | x | x | 99 | 16 | 28 | 22 | 20 | ||||||||||
| CAD | CAD5 | 2 | 1 | 256 (-) | CAD | CAD1 | x | 32 | 4 | 10 | 10 | |||||||||
| CAD6 | cad-D | x | x | 23 | 5 | 5 | 6 | 515 (+) | CAD2 | cad-C | x | 33 | 4 | 11 | 2 | 8 | ||||
| CAD3 | x | 1 | 1 | |||||||||||||||||
| CAD4 | 26 | 5 | 8 | |||||||||||||||||
| CAD7 | x | 0 | ||||||||||||||||||
| CAD8 | x | 0 | ||||||||||||||||||
| CAD9 | x | 9 | 5 | 3 | ||||||||||||||||
a A. Rohde and W. Boerjan, unpublished data.
b C. Chapple, personal communication.
A constitutive expression in the inflorescence stem (Fig. 3) and the phylogenetic classification in groups with functionally characterized proteins of other species were used as the first two criteria to delineate those family members that are the most likely to be involved in monolignol biosynthesis during developmental lignification (Table II). These criteria are fulfilled for 14 genes: PAL1, PAL2, PAL3, PAL4, C4H, 4CL1, 4CL2, HCT, C3H1, CCoAOMT1, CCR1, F5H1, COMT, and CAD6. Of these 14, eight genes have been already certified for their involvement in monolignol biosynthesis through the characterization of the corresponding mutants: PAL1 (pal1), PAL2 (pal2), C4H (ref3), C3H1 (ref8), CCR1 (irx4), F5H1 (fah1), COMT (comt1), and CAD6 (cad-D; Chapple et al., 1992; Jones et al., 2001; Franke et al., 2002a, 2002b; Goujon et al., 2003; Sibout et al., 2003; C. Chapple, personal communication; A. Rohde and W. Boerjan, unpublished data). Except for CAD6, these 14 genes have the highest expression level in their respective gene families, as judged from the number of ESTs (Table II). In conclusion, this set of 14 genes is through their expression and phylogeny eligible for being involved in the developmental monolignol biosynthesis in Arabidopsis.
AC Elements Signpost a Number of G-Branch Monolignol Biosynthesis Genes
AC elements, originally identified in the promoters of the parsley PAL1 gene, the bean PAL2 and PAL3 genes, and the parsley 4CL1 gene (Cramer et al., 1989; Lois et al., 1989; Hauffe et al., 1991; Leyva et al., 1992), are thought to enhance the expression of genes in xylem and at the same time to prevent their expression in the adjacent phloem and cortical cells. Because the deletion of the AC element results in derepression of phloem expression within the vascular tissue, it has been suggested that a (possibly phloem-specific) repressor is normally bound to the AC element, preventing expression in cells other than xylem cells. In contrast, in xylem cells, the repressor would be released to give rise to typically high expression levels (Hauffe et al., 1993; Hatton et al., 1995). A number of MYB and other transcription factors regulate the expression of monolignol biosynthesis genes and, moreover, bind to AC elements resulting in trans-activation of the respective promoters (e.g. Sablowski et al., 1995; Séguin et al., 1997; Jin et al., 2000; Sugimoto et al., 2000). Overexpression of specific MYB factors leads to lignin-related phenotypes (Tamagnone et al., 1998; Borevitz et al., 2000).
Given the importance of AC elements in specifying vascular expression, the presence of an AC element in the promoters of the 34 annotated genes has been examined. In the past, most AC elements were identified by consensus sequences built from both experimentally verified AC elements and AC elements detected by sequence similarity. Often on top of such a consensus, a number of mismatches were allowed. Moreover, AC elements were often subdivided into ACI and ACII boxes, despite the fact that they align perfectly and were shown to be functionally redundant with respect to vascular expression (see supplemental data; Hatton et al., 1995). In view of the limited knowledge on the binding specificity of AC elements by transcription factors in vivo, we have built one unifying matrix for element identification based on the five experimentally verified and delineated elements (see supplemental data) with very stringent parameters in the search. To illustrate the power of matrix versus consensus approach, the statistical significance of both methods was evaluated on 1,000 random intergenic regions distributed uniformly throughout the Arabidopsis genome. The consensus approach used, for example, by Wanner et al. (1995), has a probability to find an AC element by chance of once every 1,200 bp, whereas with our approach, it is once every 37,000 bp. With our matrix approach, some of the AC elements that had been identified previously based on similarity to a consensus were not detected, such as the AC element in the PAL3, C4H, and 4CL3 promoters (Wanner et al., 1995; Mizutani et al., 1997; Ehlting et al., 1999). Note that the elements in these promoters have not been verified experimentally.
By searching all 29,787 Arabidopsis genes predicted with EuGene (Schiex et al., 2001), AC elements on either DNA strand were found in 780 promoters (2.6%). In the set of 34 monolignol biosynthesis genes, 10 of 34 promoters have AC elements (29%; Table II): eight on the positive and two on the negative strand.
Seven gene families have at least one family member with an AC element in their promoter (Table II). Genes with an AC element do not simply correspond with genes that are highly expressed as estimated from the number of ESTs (Table II). Rather, AC elements coincide with those gene family members that were assigned to be involved in developmental lignification based on expression and phylogeny (see above): of these 14 genes, nine contain an AC element (Table II). Thus, within their respective gene families, the following genes are extra-qualified for playing a role in developmental lignification in vascular tissues: PAL1, PAL2, 4CL1, 4CL2, HCT, C3H1, CCoAOMT1, CCR1, and CAD6. CAD5 has an AC element but did not cluster with the true CAD clade in the phylogenetic tree (Fig. 12). In contrast, no AC elements were found in the gene families C4H, F5H, and COMT. Of these three gene families, C4H and COMT are single genes that, contrary to multigene families, may have acquired a more relaxed promoter organization compatible with expression in a broader range of cells and conditions. Maybe these genes contain more degenerated AC elements that were not picked up under the stringent search parameters used. The F5H family consists of two genes that are not functionally redundant because F5H2 fails to compensate for the loss of F5H1 in the fah1 mutant (Meyer et al., 1998). In this rationale, F5H1 also probably has to be considered as a single gene. However, this hypothesis, explaining why C4H, F5H, and COMT promoters lack an AC element, is not in agreement with HCT, which is a single gene as well, but has an AC element, albeit on the negative strand.
A tantalizing alternative hypothesis starts out from the notion that all AC element-containing monolignol biosynthesis genes code for enzymes acting in the G branch of the pathway (Fig. 1; Table II). None of the 14 other promoter elements analyzed, including stress- and elicitor-responsive elements, could be linked in a similarly meaningful way to particular groups of genes (supplemental Tables Is–Xs), underscoring how important the presence of AC elements may be for a common regulation of G-branch genes. A separate regulation of S-branch genes is a valid option to explain why the latter lack AC elements, given the spatio-temporal differences in deposition of S and G lignin (Dharmawardhana et al., 1992; Dixon et al., 2001; Donaldson, 2001; Jones et al., 2001). Young tissues accumulate preferentially G lignin, whereas the content of S lignin increases with tissue maturity (Meyer et al., 1998). At the level of individual cells, G-branch enzymes are involved in the lignin deposition during earlier stages of cell wall formation than S-branch enzymes (Terashima et al., 1986). Within the vascular tissue, xylem vessels contain G lignin, whereas fibers and parenchyma cells contain a mixture of G and S lignin, with the latter predominating in fibers (Donaldson, 2001, and refs. therein). Maybe the profound induction of G-branch enzymes suffices to achieve the required extra-production of G monolignols for secondary cell wall formation, typical of a lignifying xylem vessel cell. This suggestion is in line with the previously proposed mode of action of AC elements within the vascular tissue: AC elements drive high expression in xylem vessels, whereas in phloem (consisting primarily of fibers), they repress it (Hauffe et al., 1993; Hatton et al., 1995). The only G-branch gene family lacking a member with AC element is the single C4H. However, C4H might be regulated separately. Its transcriptional regulation was shown to be distinct from other monolignol genes in Arabidopsis and in pine (Jin et al., 2000; Anterola et al., 2002).
If this scenario were true, AC elements correlate with a strong expression of G-lignin genes. Furthermore, COMT and F5H would have been recruited specifically into the S branch during the evolution of angiosperms because no S lignin is made in gymnosperms. As a consequence, a putatively S-specific alcohol dehydrogenase, as identified in poplar (Li et al., 2001), might also exist in Arabidopsis (Fig. 12).
Putative Membrane Localization of Six Enzymes
Growing evidence suggests that cytochrome P450 enzymes provide membrane anchors in the ER for assembling multienzyme complexes involved in metabolic channeling within the phenylpropanoid pathway (Wagner and Hrazdina, 1984; Chapple, 1998; Rasmussen and Dixon, 1999; Winkel-Shirley, 1999). Metabolic channeling has been reported from Phe to p-coumarate with a possible association of PAL and C4H on microsomal membranes (Czichi and Kindl, 1977; Wagner and Hrazdina, 1984; Rasmussen and Dixon, 1999).
Among the three P450 enzyme families of the pathway (C4H, C3H, and F5H), C4H, C3H1, F5H1, and F5H2 have a well-conserved membrane-anchoring region, in agreement with their proposed localization in the ER membrane (Ro et al., 2001; Achnine et al., 2002; Franke et al., 2002b). C3H2 and C3H3 are not predicted to contain an ER-targeting peptide and do not comply to the amino acid features of the membrane anchor.
In addition, CCoAOMT3 contains also a putative ER-targeting signal. Besides membrane association, this could also imply a vacuolar or extracellular localization of this enzyme. Sinapoyl-Glc:malate sinapoyltransferase (SMT) and sinapoyl-Glc:choline sinapoyltransferase, involved in modification of sinapoyl-Glc, have been identified as proteins with an ER-targeting peptide (Lehfeldt et al., 2000; Shirley et al., 2001). These enzymes were suggested to be localized in the vacuole based on previous studies showing SMT activity in vacuoles (Strack and Sharma, 1985). Whether CCoAOMT3 shares this localization needs experimental verification.
Finally, a putative myristoylation site was detected in COMT, possibly involved in membrane anchoring. In agreement with this finding, a fraction of COMT from alfalfa stem was shown to be associated with the microsomal membranes, and channeling by COMT and F5H was suggested from coniferaldehyde to sinapaldehyde in the S branch of the monolignol pathway (Guo et al., 2002). This observation can be interpreted as coupling of COMT with the membrane-anchored F5H, although our data do not exclude that COMT itself could be anchored into the membrane by myristoylation.
Evolutionary Note
The number of candidate monolignol biosynthesis genes found in the Arabidopsis genome varies greatly among the gene family studied, ranging from single genes to large gene families. A complex history of gene duplications has caused the expansion and diversification of the respective gene families. Interestingly, the polyploidy event, estimated to have occurred 24 to 86 million years ago, that marks the evolutionary history of Arabidopsis (Simillion et al., 2002; Blanc et al., 2003; Bowers et al., 2003; Ermolaeva et al., 2003; Raes et al., 2003) did not create new classes within any of the investigated families. In all cases, this event, together with several small-scale duplications, was responsible only for a greater within-class diversity (data not shown). Classes must have originated at an earlier time in evolution, i.e. before 24 to 86 million years ago.
In conclusion, the genome-wide analysis of monolignol biosynthesis genes, as presented here, provides the foundation of the next steps in unraveling the monolignol pathway. The combination of reverse genetics with transcript and metabolite profiling analyses of the respective mutants will profoundly enlarge our understanding of this pathway and its relation with plant development.
MATERIALS AND METHODS
Annotation
For each of the 10 enzymes of the monolignol biosynthetic pathway, the corresponding genes were annotated in four steps: (a) experimentally certified family members were collected from a variety of species, and a family-specific profile was created; (b) an Arabidopsis protein database was scanned with this profile; (c) true family members were selected; and (d) prediction on the selected genes was improved with information from different sources, such as cDNA and EST sequences and within-family sequence similarity.
More specifically, from a ClustalW protein alignment of experimentally certified family members from different plant species, a hidden Markov model-based profile was created using the HMMER package (Thompson et al., 1994; Eddy, 1998). This profile was used to scan an Arabidopsis protein database that was constructed through a Genemark.hmm prediction (Lukashin and Borodovsky, 1998) on the complete Arabidopsis genome sequence (The Institute for Genomic Research, release 4, March 21, 2003, available at ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/). In a second scan, the complete genome sequence was searched with TBLASTN to detect genes that were not or wrongly annotated and would have been missed by using the protein database.
To delineate the gene family, several factors were taken into account. First, only HMMER hits with an E-value score below the default cutoff value (E = 10.0) were considered. Second, in most cases, a clear “drop” in the E-value score could be detected, indicating that sequences below this threshold did not fulfill the family model as well as did those above, thus providing a means to distinguish potential family members from false positives or—in the case of large superfamilies—genes of other subfamilies. This approach can potentially lead to wrong conclusions because of incomplete or biased sampling of the family. For this reason, a third method was applied, based on a phylogenetic analysis of the (super)family using the detected genes, close homologs, and more distantly related members of distinct, well-known (sub)families, retrieved from GenBank (Benson et al., 2003) and the literature. The resulting tree, showing the relations within the complete (super)family, was used to decide whether a protein belonged to the investigated family or not. As a rule, genes that clustered together with experimentally certified family members were considered to be part of the family. (Groups of) genes that did neither belong to the family nor cluster with other known families within the superfamily were considered as “likes” when they formed a sister group to the family investigated.
For the family members selected through these three criteria, the automatic annotation was improved by using information from different sources. First, the public databases were searched using BLASTN (Altschul et al., 1997) for ESTs and full-length cDNAs (percentage identity > 95% and further manual inspection of hits), and transcripts were aligned to the genomic region to verify intron-exon borders (Sim4; Florea et al., 1998). Second, the deduced protein sequences were aligned with the other family members to detect prediction errors (for example, missed exons). Third, predictions for candidate genes were verified with an alternative gene prediction tool called EuGene (specificity = 0.63 and sensitivity = 0.74 at the gene level; Schiex et al., 2001), available at http://www.inra.fr/bia/T/EuGene. This information was compiled with ARTEMIS (Rutherford et al., 2000) and was used to decide on a final gene structure.
For the annotation of the C4H, C3H, and F5H families, a substantial amount of information from the P450 databases (at http://www.biobase.dk/P450/p450.shtml and http://drnelson.utmem.edu/CytochromeP450.html) was used to improve the annotation. Prediction of myristoylation sites was done with the algorithm of Maurer-Stroh et al. (2002), available at http://mendel.imp.univie.ac.at/myristate/. Small Perl scripts were written to detect putative C-terminal farnesylation and geranylgeranylation sites (CaaX, CCXX, XCXC, and XXCC with a, aliphatic; C, Cys; and X, any amino acid; Nambara and McCourt, 1999; Randall and Crowell, 1999; Thompson and Okuyama, 2000). Signals for subcellular localization were predicted with the TargetP server (http://www.cbs.dtu.dk/services/TargetP/; specificity cutoff of >0.90; Emanuelsson et al., 2000). The annotation results were submitted to The Arabidopsis Information Resource and Munich Information Center for Protein Sequences databases for public access and are also accessible at http://www.psb.ugent.be/bioinformatics/lignin/.
Phylogenetic Analysis
The nonredundant protein database was scanned for homologous sequences using BLASTp (Altschul et al., 1997), and the results were inspected manually. Sequences were aligned with ClustalW version 1.84 (Thompson et al., 1994), and alignments were improved by eye. Trees were constructed on conserved positions of the alignment with the neighbor-joining algorithm, as implemented in TREECON (Van de Peer and De Wachter, 1994), and by maximum-likelihood analysis (quartet puzzling) with TREE-PUZZLE (Schmidt et al., 2002). Alignments were edited and reformatted with ForCon (Raes and Van de Peer, 1999; available at http://www.psb.ugent.be/~jerae/ForCon/) and BioEdit (Hall, 1999). Statistical significance of nodes in the neighbor-joining approach was tested by using 500 bootstrap replicates.
Promoter Analysis
Both strands of upstream regions (1,000 bp before the ATG codon or the distance between the previous gene and the ATG) and first and second introns of the genes were analyzed for regulatory elements with MatInspector (Quandt et al., 1995). To avoid false positives, we opted for a conservative approach with very strict parameters (core similarity = 0.9 and matrix similarity = 0.9). Furthermore, 1,000 random intergenic regions uniformly distributed throughout the Arabidopsis genome were searched with these parameters to have a rough estimate of the random occurrence of the motifs.
A list of potentially interesting motifs was compiled on the basis of the following three criteria: the motif had to be (a) experimentally characterized, (b) implicated in transcriptional regulation of known genes in the monolignol biosynthesis pathway, and/or (c) involved in elicitor, wound, or pathogen response. The motifs (and their respective calculated random occurrences in the Arabidopsis genome) that passed these criteria were: for Arabidopsis, GCC box (1/73,000 bp), jasmonate- and ethylene-responsive element (1/1,239,000 bp), W box (1/2,300 bp; withdrawn from results because of its high random occurrence), and S box (1/24,000 bp), all responsive to elicitation, wounding, and pathogens (Rushton et al., 2002); for parsley (Petroselinum crispum) FP56 (not detected in the random set; enhanced 4CL expression; Neustaedter et al., 1999) and E box (1/31,000 bp; elicitation; Grimmig and Matern, 1997); for pea (Pisum sativum), AT-rich sequence (1/26,000 bp; elicitation; Seki et al., 1996); for tobacco (Nicotiana tabacum), salicylic acid-responsive element (1/18,000 bp; Shah and Klessig, 1996) and hypersensitive-response element (1/92,000 bp; pathogen; Pontier et al., 2001). Furthermore, the joint presence of the Arabidopsis OBP-1-binding site (1/38,000 bp; Chen et al., 1996) with an As-1 box (not detected in the random set; salicylic acid, hydrogen peroxide; Yu et al., 1993; Krawczyk et al., 2002), or a common bean (Phaseolus vulgaris) H box (1/7,700 bp; elicitation; Lindsay et al., 2002; also considered without G box) with a G box (1/3300 bp; Loake et al., 1992; in conjunction with an H box responsible for induction by p-coumaric acid), respectively, were tested. For the AC I and AC II elements, one unifying profile was built from all experimentally confirmed AC I and AC II elements from different species to increase sensitivity (see supplemental data). The following AC elements were used: Eucalyptus gunnii AC I (Lacombe et al., 2000), common bean AC I and II (Hatton et al., 1995), parsley AC II (Hauffe et al., 1993), and an AC II element (CTCACCAACCCCCAC) from the poplar (Populus trichocarpa) gPtCCoAOMT1 promoter (Chen et al., 2000; C. Chen and W. Boerjan, unpublished data). The occurrence of an AC element at random using this matrix was once per 37,000 bp. In addition, the A box, suggested to work in conjunction with AC elements in parsley, was included, even though not experimentally verified (1/11,000 bp; Logemann et al., 1995). Motifs used were retrieved from or submitted to the PlantCARE database (Lescot et al., 2002; http://intra.psb.ugent.be:8080/PlantCARE/).
Experimental Verification of Annotation and Expression Study
Expression analysis was carried out in Arabidopsis ecotype Columbia plants. Seeds were surface sterilized and placed on Murashige and Skoog medium supplemented with 10 g L–1 Suc. After the seeds had undergone a cold treatment for homogenous germination (overnight at 4°C), they were exposed to 20°C, 50 μmol m–2 s–1 light intensity, and 70% relative humidity, under a 16-h-light/8-h-dark cycle. Fourteen days after germination, plants were transferred to soil and cultivated in a greenhouse. Conditions were as follows: 23°C, 50 μmol m–2 s–1 light intensity at plant level (MBFR/U 400 W incandescent lamps; Philips, Eindhoven, The Netherlands), 40% relative humidity, and a 16-h-light/8-h-dark cycle, without shielding from incident day light. Material was harvested from a number of plants (within brackets) and pooled: seedling leaves and roots of 14-d-old in vitro plants (n = 100); rosette leaves, flowers, and green siliques of 7-week-old plants (n = 50); and inflorescence stems at 1-, 3-, 5-, 10-, 15-, and 20 cm length (n = 20 for 1, 3, and 5 cm; n = 10 for all later stages). At 20 cm, the stems were fully grown.
RNA Extraction, Primer Design, and RT-PCR
Total RNA was extracted with a LiCl method according to Goormachtig et al. (1995) and digested with DNase I to eliminate residual genomic contamination. Subsequently, 5 μg of total RNA was reverse-transcribed into double-stranded cDNA (cDNA Synthesis System Plus, Amersham Biosciences, Little Chalfont, UK). Primers were designed either with the SPADS program that selects specific primers for a particular gene from the Arabidopsis genome (21 genes; available at http://intra.psb.ugent.be:8080/SPADS/) or manually (PAL1, PAL2, PAL3, PAL4, C4H, 4CL2, 4CL3, HCT, CCoAOMT1, COMT, F5H1, CAD7, and CAD8). Primers were designed to span at least one intron for reliable distinction of amplification from cDNA (except for C3H2 and C3H3, which are single exon genes). In RT-PCR experiments, 25 μL of reaction buffer supplied with the Taq polymerase and 50 ng of each primer contained a modified nucleotide mix: 200 pmol of dCTP, dTTP, and dGTP, whereas dATP was reduced to 20 pmol. To each reaction, 0.1 μL of 33P-labeled dATP (10 mCi mL–1 or 2,500 Ci mmol–1) was added, resulting in a hot:cold dATP ratio of 1:2,500. Products were separated on 3% or 4.5% (w/v) polyacrylamide gels and visualized on dried gels through autoradiography. To increase the reliability of the assays, the PCR reaction was run with at least two template concentrations (1 μL of 1:10 diluted cDNA and 1 μL of undiluted cDNA). These expression categories for a particular gene apply only for comparison of different tissues but not between genes because of the different PCR dynamics of shorter or longer amplification products.
EST Analysis
Data on size and nature of EST libraries were obtained from http://www.ncbi.nlm.nih.gov/UniLib/, http://www.ncbi.nlm.nih.gov/Entrez/, and additionally for the RIKEN Arabidopsis full-length cDNA clones from Seki et al. (2002). A total of 160,776 Arabidopsis ESTs were grouped into 11 categories: whole plant (35,544; 22.1%), aboveground organs (17,934; 11.2%), seedlings (3,207; 2.0%), roots (20,332; 12.6%), flowers (6,814; 4.2%), inflorescence stems (1,384; 0.9%), siliques and seeds (25,043; 15.6%), pathogen infection (2,366; 1.5%), wounded leaves (707; 0.4%), various stresses (44,007; 27.4%), and yet unclassified ESTs (542; 0.3%). Stress ESTs are from subtracted, normalized, and nonsubtracted, nonnormalized libraries. The whole-plant category includes whole plants, whole rosettes, and cell suspensions as starting material. Aboveground organs include, next to libraries that are described as such, libraries from mixed aboveground sources, such as whole inflorescences. Each EST was assigned to one class only. Although inevitably arbitrary and subjective, this classification was done to create clarity and to allow an easier interpretation of the results. Full details on classes and a complete list of ESTs found for each gene is available as supplemental data and at http://www.psb.ugent.be/bioinformatics/lignin/.
Supplementary Material
Acknowledgments
The authors would like to thank Vincent Thareau for the gene-specific primer design; Pierre Rouzé for helpful discussions; Gunnar von Heijne for help in the interpretation of the TargetP results; Clint Chapple, Lise Jouanin,Michael Walter, and Carine Serizet for sharing unpublished data; Vanessa Hostyn for excellent technical assistance; Stephane Rombauts for help with promoter analysis; and Martine de Cock for help in preparing the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026484.
Note Added in Proof
During the review process, another study on lignification genes in Arabidopsis was published by Goujon et al. (Goujon T, Sibout R, Eudes A, MacKay J, Jouanin L (2003) Genes involved in the biosynthesis of lignin precursors in Arabidopsis thaliana. Plant Physiol Biochem 41: 677–687).
This work was supported in part by the European Commission programs EDEN (grant no. QLK5–CT–2001–00443) and by the Fund for Scientific Research–Flanders (postdoctoral fellowship to A.R.).
The online version of this article contains Web-only data.
References
- Achnine L, Rasmussen S, Blancaflor E, Dixon RA (2002) Metabolic channeling at the entry point into the phenylpropanoid pathway: physical association between l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase. In I El Hadrami, ed, Proceedings of the XXI International Conference on Polyphenols, Imprimerie El-Watania, Marrakech, Morocco, pp 7–8
- AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola AM, Jeon J-H, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J Biol Chem 277: 18272–18280 [DOI] [PubMed] [Google Scholar]
- Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 [DOI] [PubMed] [Google Scholar]
- Bate NJ, Orr J, Ni W, Meromi A, Nadler-Hassar T, Doerner PW, Dixon RA, Lamb CJ, Elkind Y (1994) Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc Natl Acad Sci USA 91: 7608–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C (1997) Cinnamate-4-hydroxylase expression in Arabidopsis. Plant Physiol 113: 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2003) GenBank. Nucleic Acids Res 31: 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, McCollum TG, Mayer RT (2001) Differential expression of two cinnamate 4-hydroxylase genes in “Valencia” orange (Citrus sinensis Osbeck). Plant Mol Biol 46: 741–748 [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Brill EM, Abrahams S, Hayes CM, Jenkins CLD, Watson JM (1999) Molecular characterisation and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne (Medicago sativa L.). Plant Mol Biol 41: 279–291 [DOI] [PubMed] [Google Scholar]
- Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311–343 [DOI] [PubMed] [Google Scholar]
- Chapple CCS, Shirley BW, Zook M, Hammerschmidt R, Somerville SC (1994) Secondary metabolism in Arabidopsis. In EM Meyerowitz, CR Somerville, eds, Arabidopsis, Cold Spring Harbor Monograph Series, Vol 27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 989–1030 [Google Scholar]
- Chapple CCS, Vogt T, Ellis BE, Somerville CR (1992) An Arabidopsis mutant defective in the general phenylpropanoid pathway. Plant Cell 4: 1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Meyermans H, Burggraeve B, De Rycke RM, Inoue K, De Vleesschauwer V, Steenackers M, Van Montagu MC, Engler GJ, Boerjan WA (2000) Cell-specific and conditional expression of caffeoyl-CoA O-methyltransferase in poplar. Plant Physiol 123: 853–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Kota P, Blount JW, Dixon RA (2001) Chemical syntheses of caffeoyl and 5-OH coniferyl aldehydes and alcohols and determination of lignin O-methyltransferase activities in dicot and monocot species. Phytochemistry 58: 1035–1042 [DOI] [PubMed] [Google Scholar]
- Chen M, McClure JW (2000) Altered lignin composition in phenylalanine ammonia-lyase-inhibited radish seedlings: implications for seed-derived sinapoyl esters as lignin precursors. Phytochemistry 53: 365–370 [DOI] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh KB (1996) The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J 10: 955–966 [DOI] [PubMed] [Google Scholar]
- Cramer CL, Edwards K, Dron M, Liang X, Dildine SL, Bolwell GP, Dixon RA, Lamb CJ, Schuch W (1989) Phenylalanine ammonia-lyase gene organization and structure. Plant Mol Biol 12: 367–383 [DOI] [PubMed] [Google Scholar]
- Cukovic D, Ehlting J, VanZiffle JA, Douglas CJ (2001) Structure and evolution of 4-coumarate:coenzyme A ligase (4CL) gene families. Biol Chem 382: 645–654 [DOI] [PubMed] [Google Scholar]
- Czichi U, Kindl H (1977) Phenylalanine ammonia-lyase and cinnamic acid hydroxylase as assembled consecutive enzymes on microsomal membranes of cucumber cotyledons: co-operation and subcellular distribution. Planta 134: 133–143 [DOI] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE (1992) Characterization of vascular lignification in Arabidopsis thaliana. Can J Bot 70: 2238–2244 [Google Scholar]
- Dixon RA, Chen F, Guo D, Parvathi K (2001) The biosynthesis of monolignols: a “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57: 1069–1084 [DOI] [PubMed] [Google Scholar]
- Donaldson LA (2001) Lignification and lignin topochemistry: an ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Büttner D, Wang Q, Douglas CJ, Somssich IE, Kombrink E (1999) Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Ermolaeva MD, Wu M, Eisen JA, Salzberg SL (2003) The age of the Arabidopsis thaliana genome duplication. Plant Mol Biol 51: 859–866 [DOI] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8: 967–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Hemm MR, Denault JW, Ruegger MO, Humphreys JM, Chapple C (2002a) Changes in secondary metabolism and deposition of an unusual lignin in the ref8 mutant of Arabidopsis. Plant J 30: 47–59 [DOI] [PubMed] [Google Scholar]
- Franke R, Humphreys JM, Hemm MR, Denault JW, Ruegger MO, Cusumano JC, Chapple C (2002b) The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Valerio-Lepiniec M, Szczyglowski K, Van Montagu M, Holsters M, de Bruijn FJ (1995) Use of differential display to identify novel Sesbania rostrata genes enhanced by Azorhizobium caulinodans infection. Mol Plant-Microbe Interact 8: 816–824 [DOI] [PubMed] [Google Scholar]
- Goujon T, Sibout R, Pollet B, Maba B, Nussaume L, Bechtold N, Lu F, Ralph J, Mila I, Barrière Y et al. (2003) A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase: I. Impact on lignins and on sinapoyl esters. Plant Mol Biol 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Grimmig B, Matern U (1997) Structure of the parsley caffeoyl-CoA O-methyltransferase gene, harbouring a novel elicitor responsive cis-acting element. Plant Mol Biol 33: 323–341 [DOI] [PubMed] [Google Scholar]
- Guo D, Chen F, Dixon RA (2002) Monolignol biosynthesis in microsomal preparations from lignifying stems of alfalfa (Medicago sativa L.). Phytochemistry 61: 657–667 [DOI] [PubMed] [Google Scholar]
- Guo D, Chen F, Wheeler J, Winder J, Selman S, Peterson M, Dixon RA (2001) Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res 10: 457–464 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98 [Google Scholar]
- Harding SA, Leshkevich J, Chiang VL, Tsai C-J (2002) Differential substrate inhibition couples kinetically distinct 4-coumarate:coenzyme A ligases with spatially distinct metabolic roles in quaking aspen. Plant Physiol 128: 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton D, Sablowski R, Yung M-H, Smith C, Schuch W, Bevan M (1995) Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J 7: 859–876 [DOI] [PubMed] [Google Scholar]
- Hauffe KD, Lee SP, Subramaniam R, Douglas CJ (1993) Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL1 expression in transgenic tobacco. Plant J 4: 235–253 [DOI] [PubMed] [Google Scholar]
- Hauffe KD, Paszkowski U, Schulze-Lefert P, Hahlbrock K, Dangl JL, Douglas CJ (1991) A parsley 4CL1 promoter fragment specifies complex expression patterns in transgenic tobacco. Plant Cell 3: 435–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Maury S, Martz F, Geoffroy P, Legrand M (2003) Purification, cloning and properties of an acyltransferase controlling shikimate and quinate ester intermediates in phenylpropanoid metabolism. J Biol Chem 278: 95–103 [DOI] [PubMed] [Google Scholar]
- Hu W-J, Kawaoka A, Tsai C-J, Lung J, Osakabe K, Ebinuma H, Chiang VL (1998) Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc Natl Acad Sci USA 95: 5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys JM, Chapple C (2002) Rewriting the lignin roadmap. Curr Opin Plant Biol 5: 224–229 [DOI] [PubMed] [Google Scholar]
- Humphreys JM, Hemm MR, Chapple C (1999) New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 96: 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ennos AR, Turner SR (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J 11: 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk S, Thurow C, Niggeweg R, Gatz C (2002) Analysis of the spacing between the two palindromes of activation sequence-1 with respect to binding to different TGA factors and transcriptional activation potential. Nucleic Acids Res 30: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnl T, Koch U, Heller W, Wellmann E (1987) Chlorogenic acid biosynthesis: characterization of a light-induced microsomal 5-O-(4-coumaroyl)-d-quinate/shikimate 3′-hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch Biochem Biophys 258: 226–232 [DOI] [PubMed] [Google Scholar]
- Lacombe E, Van Doorsselaere J, Boerjan W, Boudet AM, Grima-Pettenati J (2000) Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein-DNA complex formation. Plant J 23: 663–676 [DOI] [PubMed] [Google Scholar]
- Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J (2001) Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Lee D, Ellard M, Wanner LA, Davis KR, Douglas CJ (1995) The Arabidopsis thaliana 4-coumarate:CoA ligase (4CL) gene: stress and developmentally regulated expression and nucleotide sequence of its cDNA. Plant Mol Biol 28: 871–884 [DOI] [PubMed] [Google Scholar]
- Lee D, Meyer K, Chapple C, Douglas CJ (1997) Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehfeldt C, Shirley AM, Meyer K, Ruegger MO, Cusumano JC, Viitanen PV, Strack D, Chapple C (2000) Cloning of the SNG1 gene of Arabidopsis reveals a role for a serine carboxypeptidase-like protein as an acyltransferase in secondary metabolism. Plant Cell 12: 1295–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NG, Yamamoto E (1990) Lignin: occurrence, biogenesis, and biodegradation. Annu Rev Plant Physiol Plant Mol Biol 41: 455–496 [DOI] [PubMed] [Google Scholar]
- Leyva A, Jarillo JA, Salinas J, Martinez-Zapater JM (1995) Low temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol 108: 39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A, Liang X, Pintor-Toro JA, Dixon RA, Lamb CJ (1992) cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell 4: 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL (2001) The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. Plant Cell 13: 1567–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Osakabe Y, Joshi CP, Chiang VL (1999) Secondary xylem-specific expression of caffeoyl-coenzyme A 3-O-methyltransferase plays an important role in the methylation pathway associated with lignin biosynthesis in loblolly pine. Plant Mol Biol 40: 555–565 [DOI] [PubMed] [Google Scholar]
- Li L, Popko JL, Umezawa T, Chiang VL (2000) 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J Biol Chem 275: 6537–6545 [DOI] [PubMed] [Google Scholar]
- Li L, Popko JL, Zhang X-H, Osakabe K, Tsai C-J, Joshi CP, Chiang VL (1997) A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc Natl Acad Sci USA 94: 5461–5466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Fliegmann J, Ebel J (2003) Deletion of a single amino acid residue from different 4-coumarate-CoA ligases from soybean results in the generation of new substrate specificities. J Biol Chem 278: 2781–2786 [DOI] [PubMed] [Google Scholar]
- Lindsay WP, McAlister FM, Zhu Q, He X-Z, Dröge-Laser W, Hedrick S, Doerner P, Lamb C, Dixon RA (2002) KAP-2, a protein that binds to the H-box in a bean chalcone synthase promoter, is a novel plant transcription factor with sequence identity to the large subunit of human Ku autoantigen. Plant Mol Biol 49: 503–514 [DOI] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA (1992) Combination of H-box [CCTACC(N)7CT] and G-box [CACGTG] cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc Natl Acad Sci USA 89: 9230–9234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Parniske M, Hahlbrock K (1995) Modes of expression and common structural features of the complete phenylalanine ammonialyase gene family in parsley. Proc Natl Acad Sci USA 92: 5905–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Reinold S, Somssich IE, Hahlbrock K (1997) A novel type of pathogen defense-related cinnamyl alcohol dehydrogenase. Biol Chem 378: 909–913 [DOI] [PubMed] [Google Scholar]
- Lois R, Dietrich A, Hahlbrock K, Schulz W (1989) A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J 8: 1641–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM (2002) An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128: 1008–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, Eisenhaber F (2002) N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol 317: 541–557 [DOI] [PubMed] [Google Scholar]
- Maury S, Geoffroy P, Legrand M (1999) Tobacco O-methyltransferases involved in phenylpropanoid metabolism: the different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol 121: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CA, Harrison MJ, Dixon RA (1993) Molecular characterization and expression of alfalfa isoliquiritigenin 2′-O-methyltransferase, an enzyme specifically involved in the biosynthesis of an inducer of Rhizobium meliloti nodulation genes. Plant J 4: 971–981 [DOI] [PubMed] [Google Scholar]
- Meyer K, Cusumano JC, Somerville C, Chapple CCS (1996) Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc Natl Acad Sci USA 93: 6869–6874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Shirley AM, Cusumano JC, Bell-Lelong DA, Chapple C (1998) Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc Natl Acad Sci USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyermans H, Morreel K, Lapierre C, Pollet B, De Bruyn A, Busson R, Herdewijn P, Devreese B, Van Beeumen J, Marita JM et al. (2000) Modification in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J Biol Chem 275: 36899–36909 [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta S, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 113: 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RB, Joy RW IV, Kurylo E, Shi X, Schnaider J, Datla RSS, Keller WA, Selvaraj G (2000) Identification of a CYP84 family of cytochrome P450-dependent mono-oxygenase genes in Brassica napus and perturbation of their expression for engineering sinapine reduction in the seeds. Plant Physiol 123: 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RB, Xia Q, Kartha CJ, Kurylo E, Hirji RN, Datla R, Selvaraj G (2002) Arabidopsis CYP98A3 mediating aromatic 3-hydroxylation: developmental regulation of the gene, and expression in yeast. Plant Physiol 130: 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, McCourt P (1999) Protein farnesylation: a greasy tale. Curr Opin Plant Biol 2: 388–392 [DOI] [PubMed] [Google Scholar]
- Nedelkina S, Jupe SC, Blee KA, Schalk M, Werck-Reichert D, Bolwell GP (1999) Novel characteristics and regulation of a divergent cinnamate 4-hydroxylase (CYP73A15) from French bean: engineering expression in yeast. Plant Mol Biol 39: 1079–1090 [DOI] [PubMed] [Google Scholar]
- Neustaedter D, Lee SP, Douglas CJ (1999) A novel parsley 4CL cis-element is required for developmentally regulated expression and protein-DNA complex formation. Plant J 18: 77–88 [DOI] [PubMed] [Google Scholar]
- Ohl S, Hedrick SA, Chory J, Lamb CJ (1990) Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell 2: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Tsao CC, Li L, Popko JL, Umezawa T, Carraway DT, Smeltzer RH, Joshi CP, Chiang VL (1999) Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc Natl Acad Sci USA 96: 8955–8960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakusch A-E, Matern U, Schiltz E (1991) Elicitor-inducible caffeoyl-coenzyme A 3-O-methyltransferase from Petroselinum crispum cell suspensions: purification, partial sequence, and antigenicity. Plant Physiol 95: 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathi K, Chen F, Guo D, Blount DW, Dixon RA (2001) Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J 25: 193–202 [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Geoffroy P, Fritig B, Legrand M (1993) Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotiana tabacum L.) leaves by infection or elicitor treatment. Plant Physiol 103: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinçon G, Maury S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2001) Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57: 1167–1176 [DOI] [PubMed] [Google Scholar]
- Pontier D, Balagué C, Bezombes-Marion I, Tronchet M, Deslandes L, Roby D (2001) Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J 26: 495–507 [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MtaInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes J, Van de Peer Y (1999) ForCon: a software tool for the conversion of sequence alignments. EMBnet.news 6; http://www.ebi.ac.uk/embnet.news/vol6_1
- Raes J, Vandepoele K, Saeys Y, Simillion C, Van de Peer Y (2003) Investigating ancient duplication events in the Arabidopsis genome. J Struct Funct Genom 3: 117–129 [PubMed] [Google Scholar]
- Ralph J, Lapierre C, Marita JM, Kim H, Lu F, Hatfield RD, Ralph S, Chapple C, Franke R, Hemm MR et al. (2001) Elucidation of new structures in lignins of CAD- and COMT-deficient plants by NMR. Phytochemistry 57: 993–1003 [DOI] [PubMed] [Google Scholar]
- Randall SK, Crowell DN (1999) Protein isoprenylation in plants. Crit Rev Biochem Mol Biol 34: 325–338 [DOI] [PubMed] [Google Scholar]
- Rasmussen S, Dixon RA (1999) Transgene-mediated and elicitor-induced perturbation of metabolic channeling at the entry point into the phenylpropanoid pathway. Plant Cell 11: 1537–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Mah N, Ellis BE, Douglas CJ (2001) Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol 126: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Meyer K, Cusumano JC, Chapple C (1999) Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol 119: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945 [DOI] [PubMed] [Google Scholar]
- Sablowski RWM, Baulcombe DC, Bevan M (1995) Expression of a flower-specific Myb protein in leaf cells using a viral vector causes ectopic activation of a target promoter. Proc Natl Acad Sci USA 92: 6901–6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiex T, Moisan A, Rouzé P (2001) EuGÈne: an eukaryotic gene finder that combines several sources of evidence. Lect Notes Comput Sci 2066: 111–125 [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504 [DOI] [PubMed] [Google Scholar]
- Schneider K, Hovel K, Witzel K, Hamberger B, Schomburg D, Kombrink E, Stuible HP (2003) The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc Natl Acad Sci USA 100: 8601–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch G, Goepfert S, Morant M, Hehn A, Meyer D, Ullmann P, Werck-Reichhart D (2001) CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J Biol Chem 276: 36566–36574 [DOI] [PubMed] [Google Scholar]
- Séguin A, Laible G, Leyva A, Dixon RA, Lamb CJ (1997) Characterization of a gene encoding a DNA-binding protein that interacts in vitro with vascular specific cis elements of the phenylalanine ammonia-lyase promoter. Plant Mol Biol 35: 281–291 [DOI] [PubMed] [Google Scholar]
- Seki H, Ichinose Y, Kato H, Shiraishi T, Yamada T (1996) Analysis of cis-regulatory elements involved in the activation of a member of chalcone synthase gene family (PsChs1) in pea. Plant Mol Biol 31: 479–491 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sewalt VJH, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb C, Dixon RA (1997) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of l-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol 115: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Klessig DF (1996) Identification of a salicylic acid-responsive element in the promoter of the tobacco pathogenesis-related β-1,3-glucanase gene, PR-2d Plant J 10: 1089–1101 [DOI] [PubMed] [Google Scholar]
- Shirley AM, McMichael CM, Chapple C (2001) The sng2 mutant of Arabidopsis is defective in the gene encoding the serine carboxypeptidase-like protein sinapoylglucose:choline sinapoyltransferase. Plant J 28: 83–94 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Seguin A, Lapierre C, Jouanin L (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis: isolation and characterization of the corresponding mutants. Plant Physiol 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu M, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE, Wernert P, Kiedrowski S, Hahlbrock K (1996) Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol:NADP+ oxidoreductase. Proc Natl Acad Sci USA 93: 14199–14203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack D, Sharma V (1985) Vacuolar localization of the enzymatic synthesis of hydroxycinnamic acid esters of malic acid in protoplasts from Raphanus sativus leaves. Physiol Plant 65: 45–50 [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H (2000) MYB-related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 12: 2511–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita N, Fujiwara H, Mimura H, Fitchen JH, Yamada Y, Sato F (1995) Molecular cloning and characterization of S-adenosyl-l-methionine: scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol 36: 29–36 [PubMed] [Google Scholar]
- Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K, Martin C (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R, Aubourg S, Lecharny A, Kreis M (2000) Organization and structural evolution of four multigene families in Arabidopsis thaliana: AtLCAD, AtLGT, AtMYST and AtHD-GL2. Plant Mol Biol 42: 703–717 [DOI] [PubMed] [Google Scholar]
- Terashima N, Fukushima K, Tsuchiya S (1986) Heterogeneity in formation of lignin: VII. An autoradiographic study on the formation of guaiacyl and syringyl lignin in poplar. J Wood Chem Technol 6: 495–504 [Google Scholar]
- Thompson GA Jr, Okuyama H (2000) Lipid-linked proteins in plants. Prog Lipid Res 39: 19–39 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272: 19176–19186 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10: 569–570 [DOI] [PubMed] [Google Scholar]
- Vernon DM, Bohnert HJ (1992) A novel methyl transferase induced by osmotic stress in the facultative halophyte Mesembryanthemum crystallinum. EMBO J 11: 2077–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GJ, Hrazdina G (1984) Endoplasmic reticulum as a site of phenylpropanoid and flavonoid metabolism in Hippeastrum. Plant Physiol 74: 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner LA, Mittal S, Davis KR (1993) Recognition of the avirulence gene avrB from Pseudomonas syringae pv. glycinea by Arabidopsis thaliana. Mol Plant-Microbe Interact 6: 582–591 [DOI] [PubMed] [Google Scholar]
- Williamson JD, Stoop JMH, Massel MO, Conkling MA, Pharr DM (1995) Sequence analysis of a mannitol dehydrogenase cDNA from plants reveals a function for the pathogenesis-related protein ELI3. Proc Natl Acad Sci USA 92: 7148–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107: 142–149 [Google Scholar]
- Ye Z-H, Kneusel RE, Matern U, Varner JE (1994) An alternative methylation pathway in lignin biosynthesis in Zinnia. Plant Cell 6: 1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LM, Lamb CJ, Dixon RA (1993) Purification and biochemical characterization of proteins which bind to the H-box cis-element implicated in transcriptional activation of plant defense genes. Plant J 3: 805–816 [DOI] [PubMed] [Google Scholar]
- Zhang X-H, Chinnappa CC (1997) Molecular characterization of a cDNA encoding caffeoyl-coenzyme A 3-O-methyltransferase of Stellaria longipes. J Biosci 22: 161–175 [Google Scholar]
- Zhong R, Morrison WH III, Negrel J, Ye Z-H (1998) Dual methylation pathways in lignin biosynthesis. Plant Cell 10: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.