Abstract
Plants sequester intermediates of metabolic pathways into different cellular compartments, but the mechanisms by which these molecules are transported remain poorly understood. Monoterpene indole alkaloids, a class of specialized metabolites that include the anti-cancer agent vincristine, anti-malarial quinine and neurotoxin strychnine, are synthesized in several different cellular locations. However, the transporters that control the movement of these biosynthetic intermediates within cellular compartments have not been discovered. Here we present the discovery of a tonoplast localized Nitrate/Peptide Family (NPF) transporter from Catharanthus roseus, CrNPF2.9, that exports strictosidine, the central intermediate of this pathway, into the cytosol from the vacuole. This discovery highlights the role that intracellular localization plays in specialized metabolism, and sets the stage for understanding and controlling the central branch point of this pharmacologically important group of compounds.
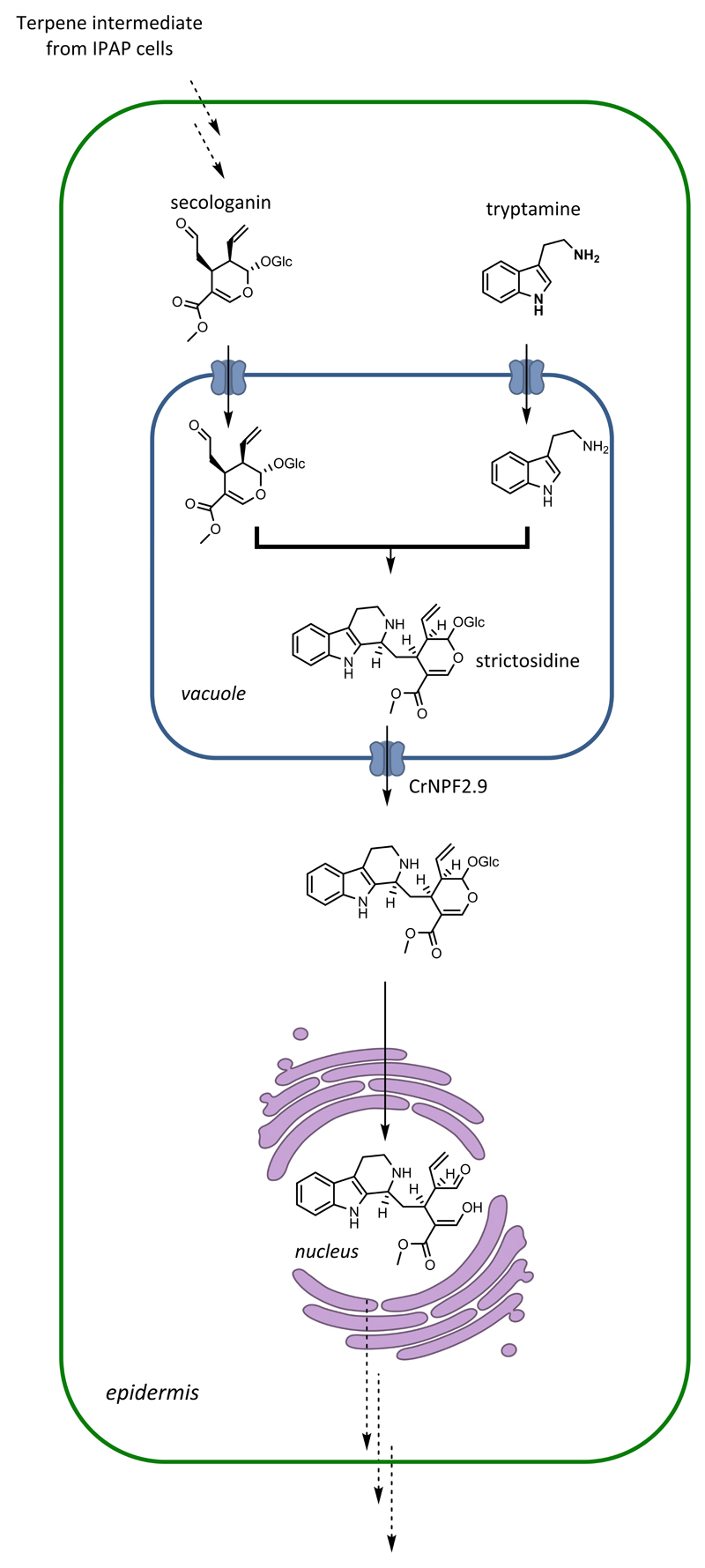
Specialized metabolic pathways of plants are often compartmentalized. The monoterpene indole alkaloids (MIAs), a structurally diverse group of ~3000 specialized metabolites that are produced across six plant families, include the anti-cancer agents vinblastine and vincristine, neurotoxin strychnine and anti-malarial quinine 1 2. While the chemical diversity of the MIAs is derived from enzymatic transformations on a central biosynthetic intermediate, strictosidine 1, the spatial distribution of these biosynthetic enzymes in planta adds an additional layer of control and complexity to this pathway (Fig. 1).
Figure 1. Localization of the MIA pathway in C. roseus.
Biosynthesis of strictosidine occurs in the vacuole of epidermal cells in C. roseus, necessitating the import of secologanin and tryptamine into the vacuole, as well as the export of strictosidine from the vacuole. CrNPF2.9, identified in this study, is annotated.
In the medicinal plant Catharanthus roseus, it has been shown that MIA synthesis takes place in at least three distinct cell types, the internal phloem associated parenchyma (IPAP), laticifers/idioblasts, and epidermal cells, with the epidermis hosting the final steps of strictosidine formation and many of the downstream reactions 3. Notably, the epidermis-located steps also display compartmentalization at the intracellular level. The direct precursors of strictosidine, tryptamine and the monoterpene secologanin, are synthesized in the cytoplasm and must be transported into the vacuole, where biosynthesis of the central intermediate strictosidine by the enzyme strictosidine synthase takes place 3 4. The next enzyme in the MIA pathway, strictosidine β-glucosidase, is sequestered in the nucleus 5. The separation of strictosidine synthase and strictosidine β-glucosidase has been hypothesized to serve as a firewall, preventing the inappropriate accumulation of strictosidine aglycone, which is a highly reactive dialdehyde that induces protein crosslinking 5. Although the reasons for specific localization to the cytoplasm, vacuole and nucleus remain unproven, this organellar separation implicates the need for transport of MIA intermediates into and out of the vacuole.
Here we identify transporter genes that are co-regulated with the early steps of the MIA pathway using a self-organizing map of C. roseus transcriptomic expression data. Three transporter candidates, two showing homology to multidrug and toxic compound extrusion (MATE) proteins (CrMATE1 and CrMATE2) and one showing homology to nitrate and peptide transport family (NPF) proteins (CrNPF2.9), were identified using this approach. Loss of function studies using virus induced gene silencing (VIGS) and in vitro characterization in Xenopus laevis oocytes strongly suggest that CrNPF2.9 is responsible for the trans-tonoplastic movement of strictosidine out of the vacuole. The transporter is localized to the tonoplast membrane, which is consistent with this function. This marks the first identification of an intracellular transporter gene in MIA biosynthesis, highlights the emerging role of NPF transporters in plant specialized metabolism and provides insight into how localization provides an additional layer of regulation in specialized metabolism.
Results
Self-organizing maps used to identify candidates
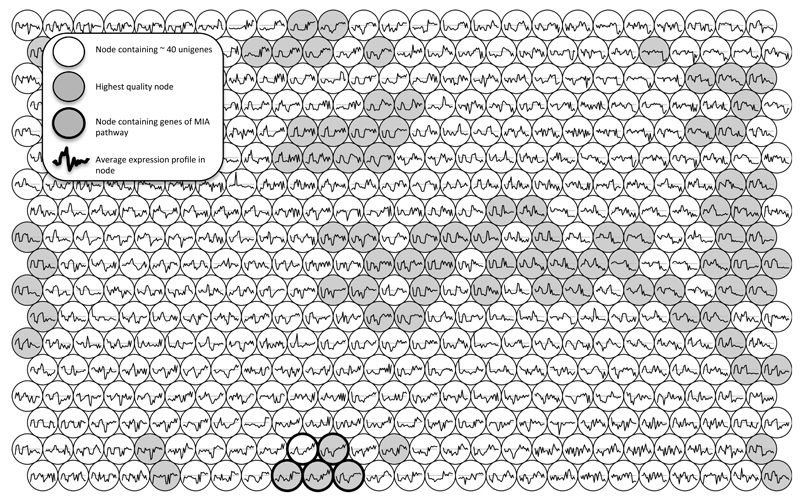
To identify candidate transport genes, we utilized a publicly available RNA-seq database 6 that has been used for identification of a number of genes in the C. roseus MIA pathway 7. The expression profiles of genes from the transcriptomic dataset were clustered using a self-organizing map. This methodology has previously been employed to cluster large datasets 8,9 as it provides an unbiased and unsupervised method for clustering. Nine contigs corresponding to the known MIA biosynthetic pathway genes (Fig. 2) were mapped to five nodes of the self-organizing map that were spatially close and of high quality (Fig. 2), indicating the co-expression of these genes. Contigs within these five nodes were considered as candidates involved in MIA metabolism. Three putative transporters, one showing homology to an NPF transporter (CrNPF2.9), and two showing homology to MATEs (CrMATE1 and CrMATE2), were observed in these nodes and were selected for further study. Given the recent evidence that NPF transporters have a much broader substrate specificity than previously recognized 10, we chose to focus on establishing whether CrNPF2.9 plays a role in MIA biosynthesis.
Figure 2. Self-organizing map of C. roseus transcriptomic data.
Each circular node represents approximately 40 unigenes with the most similar expression profile. Furthermore, neighbouring nodes are related to each other by the similarity of their expression profile. The average expression profile of genes in the node is plotted within each node. The early MIA pathway genes strictosidine synthase, secologanin synthase, tryptophan decarboxylase, strictosidine β-glucosidase, geraniol 8-hydroxylase and loganic acid methyltransferase all localize to the nodes highlighted by a bold circle. Nodes colored gray represent the highest quality nodes.
In planta function by VIGS
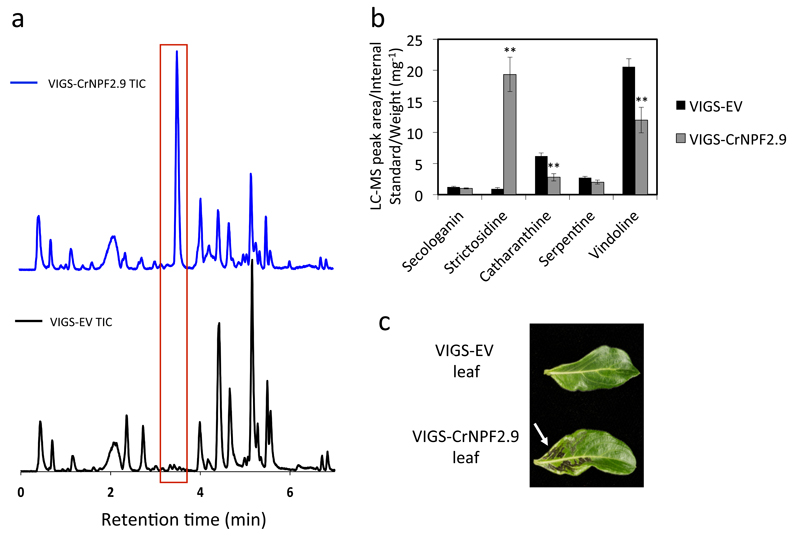
To assess the physiological relevance of this transporter in planta, we used virus induced gene silencing (VIGS) to transiently silence the expression of the CrNPF2.9 gene in C. roseus and screen for a metabolic phenotype 11 (Fig. 3, Supplementary Fig. 1). Silencing of the CrNPF2.9 transporter resulted in a dramatic increase in the level of strictosidine in C. roseus leaf tissue together with a significant decrease in vindoline and catharanthine, the end products of the MIA pathway (Fig. 3, Supplementary Fig.1). Silencing using a different fragment from a distinct region of the CrNPF2.9 gene resulted in the same phenotype (Supplementary Fig. 1). The accumulation of strictosidine is consistent with CrNPF2.9 acting as an exporter of strictosidine from the vacuole: silencing of CrNPF2.9 prevents the export of strictosidine, leading to its accumulation in the vacuole, whilst concomitantly leading to the decrease in the downstream alkaloids.
Figure 3. In planta silencing of CrNPF2.9.
A. Representative total ion chromatogram (TIC) of leaf tissue extracts from Little Bright Eyes plants transformed with empty vector (VIGS-EV, black), and a vector designed to silence CrNPF2.9 (VIGS-CrNPF2.9, blue). The red box highlights the accumulation of strictosidine. B. Alkaloid profiles for tissue transformed with the VIGS-CrNPF2.9 vector relative to the empty vector control (VIGS CrNPF2.9 (n = 12), VIGS-EV (n = 10)) ** p < 0.01. All data shown are mean ± SEM. C. Blackened leaf tissue that results upon silencing of CrNPF2.9 highlighted by a white arrow.
Additionally, VIGS of CrNPF2.9 resulted in cell death at the site of infection in the leaf tissue (Fig. 3, Supplementary Fig. 2). This was validated by confocal microscopy of the leaf tissue after staining with both DAPI and propidium iodide (Supplementary Fig. 2). This cell death was not observed in empty vector controls, and has never been reported when any other known MIA biosynthetic gene has been silenced. We hypothesize that the substantial accumulation of strictosidine is cytotoxic to the plant, leading to cell death.
To further validate the distinct role of CrNPF2.9, we performed additional VIGS experiments with the other transporters that had been identified in the co-expression analysis (Supplementary Fig. 3). Simultaneous silencing of CrNPF2.9 and CrMATE1, as well as triple silencing of CrNPF2.9, CrMATE1 and CrMATE2, also resulted in the accumulation of strictosidine (Supplementary Fig. 3). In all cases, silencing experiments were consistent with a role of CrNPF2.9 in strictosidine metabolism.
qPCR was used to confirm the silencing of the putative transporter gene. mRNA levels could not be accurately measured on CrNPF2.9 silenced plants due to the cytotoxicity that is observed upon CrNPF2.9 silencing. However, silencing of CrNPF2.9 was confirmed in the double and triple silenced plants (which displayed less necrotic tissue) (Supplementary Fig. 3). The similar metabolic profiles that result when two distinct gene fragments were tested for CrNPF2.9 (Supplementary Fig. 1) suggest that off-target silencing is not responsible for the observed metabolic phenotype.
CrNPF2.9 is located at the tonoplast membrane
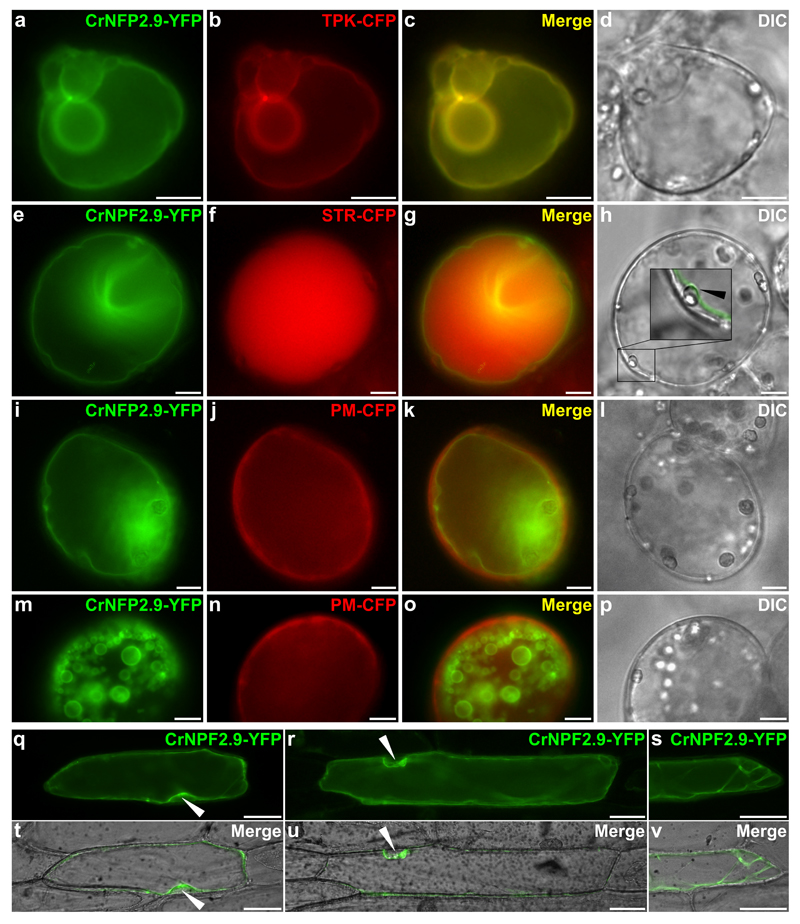
The subcellular localization of any protein directly impacts its physiological role, so a transporter involved in strictosidine biosynthesis must be localized to the tonoplast. To investigate the subcellular localization of CrNPF2.9, a C-terminal yellow fluorescent protein (YFP) fusion of CrNPF2.9 (CrNPF2.9-YFP) was co-expressed in C. roseus cell suspension cultures with a set of distinct subcellular compartment markers. These markers included C-terminal cyan fluorescent protein (CFP) fusions of three different proteins: the Arabidopsis two pore K+ channel, AtTPK1 (TPK-CFP), which is known to localize to the tonoplast 12–15, strictosidine synthase from C. roseus (STR-CFP), known to localize to the vacuole 5 and the aquaporin PIP2a (PM-CFP), which is targeted to the plasma membrane 16 (Fig. 4). In transiently transformed cells, CrNPF2.9-YFP co-localized to the same membrane as AtTPK1-CFP (Fig. 4A-D). Furthermore, CrNPF2.9-YFP was observed to surround the vacuole labelled with STR-CFP (Fig. 4E-H) and CrNPF2.9-YFP also appeared distinct from the plasma membrane labelled with PM-CFP (Fig. 4I-P). A similar pattern of localization was also observed in onion cells expressing CrNPF2.9-YFP (Fig. 4Q-V). Therefore it is highly likely that this transporter is localized to the tonoplast, a location compatible with a role in vacuolar transport.
Figure 4. Overexpression of CrNPF2.9-YFP in C. roseus and onion cells.
C. roseus cells were transiently co-transformed with the plasmid expressing CrNPF2.9-YFP and the plasmid encoding the tonoplastic AtTPK1 (TPK)-CFP marker (A-D), the plasmid encoding the vacuolar localized strictosidine synthase (STR-CFP) marker (E-H) or the plasmid containing the plasma membrane targeted aquaporin PIP2A (PM-CFP) marker (I-P). Co-localization of the fluorescence signals appears in yellow when merging the two individual (green/red) false color images (C, G, K, O). Inset in panel H highlights the YFP labelling on the inner side of plastids (white arrowhead) indicating a tonoplastic localization. Cell morphology is observed with differential interference contrast (DIC) (D, H, L, P). Bars, 10 μm Onion cells were transiently transformed with the plasmid expressing CrNPF2.9-YFP (Q-V) and fluorescent signals (Q, R, S) were merged with cell morphology observed with DIC (T, U, V). Black arrowheads highlight the YFP labelling on the inner side of nucleus indicating a tonoplastic localization. Bars, 50 µm.
A (D/E)X3-5L(L/I) motif has been implicated in the localization of plant membrane proteins to the tonoplast membrane, and mutagenesis of this motif has directly proven its role in directing targeting to the tonoplast 17 18 13 19. CrNPF2.9 contains this motif at the N-terminus (Supplementary Fig. 4). Additionally, CrNPF2.9 has a relatively acidic isoelectric point of 6.1. A lower isoelectric point has been used as a predictive indication of tonoplast localization 20,21. These data provide further support that CrNPF2.9 is likely to be tonoplastic.
CrNPF2.9 is co-expressed with MIA biosynthetic genes in leaf epidermis
Since strictosidine synthesis and deglycosylation occur in leaf epidermis, cellular localization of CrNPF2.9 was examined by analyzing its transcript distribution. A leaf epidermis-enriched fraction of mRNAs was generated and relative abundance of transporter messengers was evaluated by comparison with a whole leaf fraction. qPCR analysis showed that the epidermis fraction was strongly enriched with CrNPF2.9 transcripts. A similar enrichment was observed for strictosidine synthase and strictosidine glucosidase (Supplementary Fig. 5). The transcripts of hydroxymethylbutenyl 4-diphosphate synthase, known to accumulate in internal phloem associated parenchyma, were not enriched in the epidermis fraction. The localization of CrNPF2.9 in leaf epidermis, as observed for strictosidine synthase and strictosidine glucosidase, reinforces the proposed involvement in MIA biosynthesis.
Biochemical characterization of strictosidine export
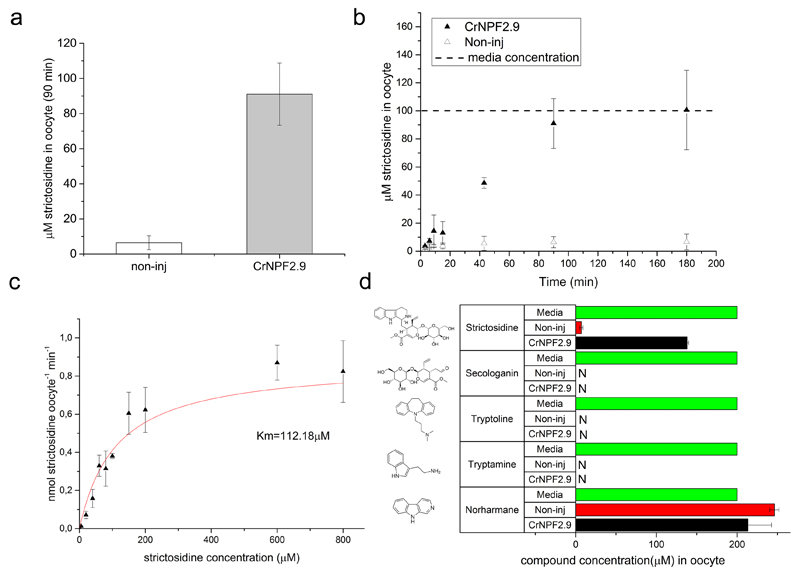
As a putative vacuolar strictosidine exporter, CrNPF2.9 would, in planta, transport strictosidine from the acidic vacuolar lumen to the neutral cytosol. To validate this hypothesis, we expressed CrNPF2.9 heterologously in Xenopus laevis oocytes and incubated the expressing oocytes in an uptake buffer containing strictosidine at pH 5. Hence, with respect to substrate and proton gradients, import of strictosidine from the acidic exterior to the neutral oocyte cytosol in principle mimics export across the plant cell tonoplast. Oocytes expressing CrNPF2.9 accumulated strictosidine to 10 fold higher levels compared to control oocytes (Fig. 5A). Incubating CrNPF2.9-expressing oocytes in 100 μM strictosidine for varying time periods showed a linearly increasing accumulation that reached a stable plateau at ~100 μM internal strictosidine concentration after approximately 90 minutes. In comparison, the concentration in control oocytes never increased above 6 μM, even after prolonged three hours of incubation (Fig. 5B). This indicates that the main rate-limiting step for strictosidine accumulation in oocytes is import across the plasma membrane. Therefore, the main contribution to strictosidine accumulation in the oocytes stems from CrNPF2.9-mediated import across the plasma membrane. As internal strictosidine concentration in CrNPF2.9-expressing oocytes did not increase above the concentration in the external assay medium (Fig. 5B), the data indicates that the transporter cannot accumulate strictosidine against a concentration gradient. Based on the time course assays, a 20-minute assay length was chosen for estimating the CrNPF2.9s affinity constant towards strictosidine. Plotting transport rates as a function of increasing strictosidine concentrations yielded a saturation curve, which was fitted to the Michaelis–Menten equation (Fig. 5C) with an apparent affinity constant towards strictosidine of ~110 μM. In addition, oocytes expressing CrNPF2.9 were incubated with the MIA intermediates secologanin and tryptamine, as well as β-carboline alkaloid norharmane and the non-natural alkaloid tryptoline. These oocytes did not import these compounds, indicating specificity of CrNPF2.9 for strictosidine (Fig. 5D). In combination, our data show that CrNPF2.9 is a high-affinity transporter with specificity for strictosidine.
Figure 5. In vitro characterization of CrNPF2.9.
A. Strictosidine uptake in non-injected oocytes and oocytes expressing CrNPF2.9. Oocytes (15) were incubated for 90 minutes in 100 μM strictosidine (pH 5) and strictosidine content in oocytes was determined in 3x5 oocytes by LC-MS (Error bars are SD, n = 3). Data is representative of five independent experiments, each using oocytes from different frogs. B. Strictosidine uptake over time in oocytes (15) injected with CrNPF2.9. Oocytes were incubated for indicated times in 100 μM strictosidine (pH 5) and content was determined in 3x5 oocytes by LC-MS (Error bars are SD, n = 3). Data is representative of two independent experiments. C. Kinetic analysis of CrNPF2.9-mediated strictosidine transport. Strictosidine transport rate (uptake measured after 20 minutes) is plotted against increasing strictosidine concentrations and yields a saturation curve, which was fitted with a Michaelis-Menten equation (solid line, error bars are SD, n = 3). Data is representative of two independent experiments. D. Substrate specificity of CrNPF2.9 determined by measuring uptake of MIA intermediates secologanin, tryptamine, as well as β-carboline alkaloids norharmane and the non-natural alkaloid tryptoline in CrNPF2.9 expressing and non-injected oocytes. Each substrate was tested individually at pH 5. Oocytes (15) were incubated in each assay in 200 μM external compound concentration (represented by the “media” bar) and uptake was determined in 3x5 oocytes by LC-MS (Error bars are SD, n = 3). N = not detected.
Discussion
There has been tremendous recent progress in the elucidation of plant biosynthetic pathways. However, the mechanisms that control the localization of many plant pathways remain less clear. For example, while many biosynthetic enzymes have been discovered and characterized for strictosidine and other MIAs 7,22–25, only two transport steps in MIA biosynthesis have been explored: ABC transporters have been shown to be involved in the export of catharanthine to the cuticle 26, and a proton-driven antiporter appears to transport vindoline, catharanthine and anyhydrovinblastine into the vacuole 27.
In strictosidine biosynthesis, tryptamine and secologanin are imported into the vacuole, whilst strictosidine, in turn, is exported for subsequent chemical steps (Fig. 1). The mechanism controlling the movement of these biosynthetic intermediates was unknown. In this work, loss-of-function studies strongly suggest a role for CrNPF2.9 in strictosidine export from the vacuole. CrNPF2.9 was clearly shown to transport strictosidine with high affinity and specificity along its concentration gradient when expressed in X. laevis oocytes. The localization of CrNPF2.9 to the tonoplast in leaf epidermis is consistent with this role.
To date the NPF transporter family has been implicated in the movement of nitrate, peptides, glucosinolates 28, auxin 29, abscisic acid 30 10 and gibberellic acid 31, and this work further expands the substrate repertoire of this family to include the alkaloid strictosidine. The characterized plant NPF transporters to date have primarily focused on substrate transport at the plasma membrane. A number of NPF transporters are present in the C. roseus transcriptome, but CrNPF2.9 appears to be the only one with a tonoplast membrane targeting sequence. Although there are Arabidopsis NPF transporters that are known to localize to the tonoplast membrane, such as AtPTR2,4 and 6 17,32, the function of these transporters has not been elucidated. The characterization of CrNPF2.9 therefore provides the first evidence of the role of NPF transporters in the intracellular movement of specialized metabolites in planta.
It has been shown that an EXXE(R/K) motif in the peptide transporter from Shewanella oneidensis, PepTSO, is essential for the coupling of solute movement to the proton gradient 33. This motif is conserved in the Arabidopsis nitrate transporter, NRT1.1, as well as in the Arabidopsis glucosinolate transporter NPF2.11, and has been shown through mutagenesis, to contribute to coupling nitrate and glucosinolate movement to the proton gradient in NRT1.1 and NPF2.11 respectively 34,35. Notably, when mutated in the EXXE(R/K) motif, AtNPF2.11 loses its ability to accumulate glucosinolates against its concentration gradient 34. The CrNPF2.9 transporter does not contain this conserved motif (Supplementary Fig. 4), which is consistent with the inability of CrNPF2.9 to accumulate strictosidine against its concentration gradient. Such a limit on transport could have been retained during evolution to avoid the formation of high amounts of the reactive strictosidine aglycone and resulting cytotoxicity 5.
The discovery and characterization of CrNPF2.9 in C. roseus provides insight into the importance that intracellular metabolite transport plays in the regulation of the MIA pathway. Notably, silencing of CrNPF2.9 and the subsequent accumulation of strictosidine causes extensive tissue death in C. roseus (Fig. 3, Supplementary Fig. 2), suggesting that the intravacuolar accumulation of strictosidine is cytotoxic, and that CrNPF2.9 plays a key role in exporting this MIA intermediate from the vacuole. Both secologanin and tryptamine must also be transported from the cytosol into the vacuole for strictosidine biosynthesis to occur, but no dedicated transporters for these steps have yet been identified.
The transport processes that control the movement of alkaloid biosynthetic intermediates are generally not well characterized. This study shows that dedicated NPF transporters play a pivotal role in shuttling alkaloid intermediates between subcellular compartments. To date, an ABC transporter has been shown to be involved in the export of the MIA catharanthine to the leaf surface in C. roseus 26, and MATE-type transporters 36–38 as well as a purine permease 39, have been implicated in nicotine transport. Thus, the discovery of CrNPF2.9 further highlights that alkaloid localization is controlled by a wide variety of transporter types. The diversity of plant transporters, and an understanding of their substrate specificities and roles in metabolism, represents a potentially untapped resource for metabolic engineering and synthetic biology. As demonstrated in Arabidopsis, genetic manipulation of transporters can be used to affect the source to sink accumulation of anti-nutritional metabolites in the edible parts of crops 28. Additionally, the heterologous expression of transport proteins may be useful for biotechnological applications for metabolic engineering. There has been a concerted effort for the engineering of complex plant metabolic pathways in yeast, as exemplified by the production of strictosidine 40 and benzylisoquinoline alkaloids in Saccharomyces cerevisiae 41. It is feasible that utilizing transport proteins for either the internal sequestration of intermediates in these metabolic pathways, or for the export of toxic intermediates to the media upon fermentation may improve titres of these important pharmaceuticals. The roles that transport proteins play in plant specialized metabolism is largely understudied. However, the decreasing costs of next generation sequencing technologies for interrogation of plant genomes and transcriptomes will allow for the mining and subsequent characterization of transport proteins with a diverse array of substrate specificities.
Methods
Generation of self-organizing maps
The filtered transcriptomic data 6 (contigs with FPKM expression values of zero for more than half of the treatments or with zero expression variance across the samples were removed) was normalized to have a mean expression level of zero and unit variance across conditions. Self-organizing maps were implemented and visualized in R (R Core Team 2013, version 3.0.2) using the kohonen package 42. The map sizes were chosen to give approximately forty contigs per node, with the map shape selected such that the ratio between the two edge lengths was the same as the ratio between the two largest eigenvalues of the data 43. A toroidal map was used so that every node had the same number of neighbours. This avoided boundary effects occurring when the neighbourhood distance metric was calculated. To assess how well the generated self-organizing map fitted the data, two quality metrics were analyzed. The first was the within-node distance, which is defined as the mean distance from the weight vector of a node to the samples mapped to it. Therefore, the smaller the within-node distance, the more accurately the node's weight vector represents the samples mapped to the node. The other quality metric used was the inter-nodal distance, defined as the sum of the distances from a node's weight vector to the weight vectors of its neighbouring nodes. The smaller the value, the more similar a node's weight vector is to its surrounding nodes' weight vectors. In order for a node to be classed as high quality in this analysis, both of the described quality metrics for that node had to be in the lowest quartile compared to all nodes.
Virus Induced Gene Silencing (VIGS)
The VIGS constructs for CrNPF2.9, as well as the double silencing vector CrNPF2.9-CrMATE1 and the triple silencing vector CrNPF2.9-CrMATE1-CrMATE2, were generated using a USER compatible virus induced gene silencing vector pTRV2u. The USER compatible virus induced gene silencing vector pTRV2u 7 was digested with AsiI and Nt.BbvCI, and an approximately 200-500 bp fragment of the desired gene, amplified from C. roseus cDNA using the primers in Supplementary Table 1, was USER cloned into the cut vector. For generation of the double (VIGS-CrNPF2.9-CrMATE1 double) and triple (VIGS-CrNPF2.9-CrMATE1-CrMATE2 triple) fusion vectors, USER fusion cloning was employed 44,45. Agrobacterium GV3101 strains containing pTRV1 (ABRC), pTRV2u-empty vector, pTRV2u-MgChl (for silencing magnesium chelatase) and pTRV2u-gene of interest, were grown overnight in 5 ml LB supplemented with rifampicin, gentamycin and kanamycin at 28 °C. These cultures were pelleted at 3,000 g, resuspended in Agrobacterium inoculation solution (10 mM MES, 10 mM MgCl2, 200 µM acetosyringone) to an OD600 of 0.7, and incubated at 28 °C for 2 h. Transformants were confirmed by PCR using the gene specific primers used to amplify the gene fragment. Strains containing pTRV2 constructs were mixed 1:1 with pTRV1 culture and this mixture was used to inoculate plants (Little Bright Eyes cultivar) by the pinch wounding method using a pair of finely bent forceps 7,46. Plants (8-12, 2 months old) were inoculated for each construct, and the plants were grown at 25 °C in a 12 h photoperiod. The pTRV2u was used as a negative (empty vector) control, whilst the pTRV2-MgChl plants were used as a visual marker of the silencing response, with bleaching of the leaves occurring 21-25 days post inoculation. Upon silencing the plant material was harvested, ground in a Retsch ball mill under liquid nitrogen and stored at -80 °C before analysis by qPCR and liquid chromatography mass spectrometry (LC-MS). All of the VIGS experiment were replicated a minimum of three times per construct. All data were analyzed by a pairwise Student’s t-test against the EV control tissue.
qPCR of VIGS tissue
Approximately 100 mg of plant tissue was used for extraction of RNA from each replicate in a VIGS experiment using a Qiagen RNeasy plant mini kit in accordance with the manufacturer’s instructions. For each set of pTRV2u plants, a minimum of 8 plants were selected, including the plants that yielded the most pronounced metabolic phenotype, for RNA extraction and qPCR. RNA quality was assessed on a 1% agarose gel and the concentration was measured on a NanoDrop® ND-1000. cDNA for qPCR from each replicate was synthesized using the Biorad iScript cDNA synthesis kit.
For each qPCR experiment, cDNA from eight replicates of pTRV2-EV and eight replicates of pTRV2-gene of interest were used, and the qPCR reaction was performed in technical duplicates. The relative quantification of gene expression was performed using the delta delta cycle method using the 40S ribosomal protein 9 (Rps9) as a reference gene for normalization 7,22,24,46. Each primer pair was used in a standard PCR reaction against C. roseus cDNA to ensure only one target was amplified. For each primer pair, a standard curve was generated to ensure amplification efficiency had a linear relationship with cDNA concentration, with only primer pairs giving a linear regression (R2) value of 0.99 used. The primer efficiency values generated in this study were between 99-101%.
Liquid Chromatography Mass Spectrometry (LC-MS) of VIGS tissue
Ground leaf tissue was weighed and collected into 200 µl methanol containing 40 µM caffeine or 10 µM esculin as an internal standard and incubated at 60 °C for 2 hours. After a 30 minute centrifugation step at 5000 g, an aliquot of the supernatant (25 µl) was mixed with an equal volume of water and analyzed on a Thermo-Finnigan instrument equipped with a Deca XP ion trap detector or a Shimadzu IT-TOF. In both systems the column used was a Phenomenex Kinetic 5 µ C18 100A (100 x 2.10 mm x 5 µm) and the binary solvent system consisted of acetonitrile (ACN) and 0.1% formic acid in water. The elution program was as follows: 1 minute isocratic at 12% ACN, 3.5 minute gradient up to 25% ACN, 2.5 minute gradient up to 50% ACN, 1 minute gradient up to 100% ACN, 6 minute isocratic at 100% ACN, 1 minute gradient down to 12% ACN, and 2.5 minute isocratic at 12% ACN. Peak areas were calculated using the ICIS algorithm in Finnigan’s Xcalibur software or LCMS solutions and normalized by leaf mass (fresh weight) and the peak area of the internal standard.
Confocal microscopy of CrNPF2.9 silenced leaves
For DAPI staining, C. roseus leaves were vacuum-infiltrated and fixed in a solution of Ethanol/Acetic acid (3:1 v/v) for 1-2 hours and then washed in phosphate buffered saline (PBS) pH 7.0. Tissue was cut into pieces (3 mm x 3 mm) that included both blackened regions and normal tissue. Material was incubated in a 1 µg/ml solution of DAPI (4’, 6-diamidino-2-phenyl-indole) for 1 hour, then washed in water and mounted in Vectashield (Vector laboratories) and imaged on a Zeiss LSM780 confocal microscope using a x40 oil immersion lens. For visualisation of DAPI staining, excitation was at 405 nm and detected at 410-585 nm. Z-stacks with 1 µm spacing were used to generate Maximum projection images using ImageJ software (http://rsb.info.nih.gov/ij/). For propidium iodide staining, fresh unfixed tissue was mounted in a 10 µg/ml solution of propidium iodide and viewed on a Zeiss LSM780 confocal microscope using a x40 oil immersion lens. Excitation was at 488 nm and the signal detected at wavelengths of 535-660 nm. Z-stacks with 1 µm spacing were used to generate Maximum projection images using ImageJ software (http://rsb.info.nih.gov/ij/).
Preparation of epidermis enriched fraction and transcript distribution analysis
Epidermis-enriched fraction was obtained from a modified version of a procedure previously described by 47. Briefly, a cotton swab was dipped into a carborundum powder (particle size <300 grit, Fischer) and used to abrade both lower and upper epidermis layers of young C. roseus leaves. Abraded leaves were dipped in 4 ml of Trizol (Life Technologies) for 5 seconds in a 15 ml centrifuge tube. A total of 3 x 10 leaves were abraded and RNAs were extracted according to the manufacturer's protocol. The RNA pellet resulting from the isopropanol precipitation was washed with 70% ethanol and re-suspended in 100 µl of RNAse-free water. Excess sugars were removed by precipitation with 10% ethanol for 5 minutes on ice and centrifugation for 5 minutes at 15,000g and 4°C. The supernatant was recovered and precipitated with 0.5 volume of sodium acetate 3M pH 5.2 and 2.5 volumes of 100% ethanol. The pellet was washed with 70% ethanol and re-suspended with 20 µl of RNase free water. Total RNAs from whole young leaves were also extracted with Trizol (Life Technologies) according to the manufacturer's protocol. RNA from both fractions was quantified using a NanoDrop® ND-1000 and 1 µg was retro-transcribed with the RevertAid First Strand cDNA Synthesis Kit according to provider’s instructions (ThermosFisher Scientific). Gene expression levels were measured by qPCR run on a CFX96 Touch Real-Time PCR System (Bio-Rad). Each reaction was performed in a total reaction volume of 25 μL containing an equal amount of cDNAs (1/3 dilution), 0.05 μM forward and reverse specific primers (Supplemental Table 4), and 1 × DyNAmo™ ColorFlash Probe qPCR Kit (Thermo Fisher Scientific). The amplification program was 95 °C for 7 min (polymerase heat activation), followed by 40 cycles containing 2 steps, 95 °C for 10 sec and 60 °C for 40 sec. Quantification of transcript copy number was performed with calibration curves and normalization with the C. roseus 40S Ribosomal protein S9 reference gene and expressed relatively to the amount of transcript measured in the whole leaf fraction. All amplifications were performed in triplicate and repeated at least on two independent biological repeats.
Production and purification of strictosidine
Strictosidine synthase was purified as in 48. Strictosidine was generated enzymatically by incubation of 4 mM tryptamine (Sigma), 2 mM purified secologanin (Sigma) and 2 µM strictosidine synthase in 50 mM phosphate buffer pH 7.4 buffer, in a total volume of 50 ml, overnight at 37 °C. This reaction was monitored for strictosidine production and consumption of secologanin by LC-MS. The solution was purified by solid phase extraction, eluted in 100% MeOH, and dried under vacuum. The dried product was resuspended in H2O, filtered with 2 µm filters and purified by preparative HPLC in 2 ml aliquots. The HPLC used was a Dionex Ultimate 3000 pump, with a variable wavelength UV detector. The column used for chromatographic separation was a Phenomenex Luna 5µ C18(2) 100 A (250 x 30 mm), with the binary solvent system consisting of 0.1% TFA and acetonitrile. The elution program was the following: 1 min isocratic at 10% ACN, 8 min gradient up to 30% ACN, 4 min gradient up to 100% ACN, 6 min isocratic at 100% ACN, 1 min gradient down to 10% ACN, and 6 min isocratic at 10% ACN. The compounds were monitored by UV 241 nm, and the fractions collected at the retention time 13.5 min. The resulting fractions were lyophilized and analyzed by NMR.
Subcellular localization
The full length CrNPF2.9 gene was subcloned into the pSCA-cassette vector as a YFP fusion, and the Arabidopsis AtTPK1 was subcloned into the into the pSCA-cassette vector as a CFP fusion. The vacuolar STR-CFP and the plasma membrane PM-CFP (CD3-1002) markers were described previously 5,16. All vectors used a 35S promoter for expression. Transient transformation of C. roseus cells by particle bombardment and fluorescence imaging were performed following the procedures previously described 5,49. C. roseus cells were bombarded with DNA-coated gold particles (1 μm) and 1,100 psi rupture disc at a stopping-screen-to-target distance of 6 cm, using the Bio-Rad PDS1000/He system and 100 ng of each plasmid per transformation. For the transient transformation of onion cells, internal epidermis of fresh onion purchased from local producer (Les Vergers de la Breteche, St-Paterne Racan), were peeled and placed on solid vitamin-free MS medium and bombarded following the same protocol. Both cell-types were cultivated for 16 h to 38 h before being harvested and observed. The subcellular localization was determined using an Olympus BX-51 epifluorescence microscope equipped with an Olympus DP-71 digital camera and a combination of YFP and CFP filters.
Expression and functional characterization of CrNPF2.9 in Xenopus laevis oocytes
The full length coding sequence of the CrNPF2.9 gene was cloned into the USER cloning compatible Xenopus laevis expression vector, pNB1u 50 and verified by sequencing. Primers (SI) were used to amplify CrNPF2.9 CDS, including the surrounding T7 promoter and Xenopus-β-tubulin gene’s 5’ and 3’ UTRs via PCR from the Xenopus pNB1u expression vector. cRNA was in vitro transcribed from the purified linear PCR fragments using the mMESSAGE mMACHINE® T7 Transcription Kit (ThermoFisher Scientific). The resulting cRNA was purified using LiCl precipitation and adjusted with RNase free water to a concentration of 0.5 µg/µl.
Defoliculated Xenopus laevis oocytes at maturation stage V-VI were purchased from Ecocyte bioscience, Germany. Oocytes were injected the day after surgery with 25 ng cRNA and were incubated for 3-4 days at 18 °C before assaying.
For transport assays, oocytes were pre-incubated for 5 minutes in Kulori Buffer (90 mM NaCl, 1 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5mM MES, pH 5) to ensure intracellular steady state pH before transferral to Kulori buffer pH 5 containing one of the tested compounds (strictosidine, secologanin, tryptamine, norharmane and tryptoline). Assays were generally stopped after 90 minutes unless otherwise stated by washing in 4 x 20 ml Kulori buffer without substrate. Each assay consisted of 15 oocytes and in pools of five oocytes were homogenized in 50 μl 50% MeOH. Following precipitation (20,000 g for 10 min), supernatants were frozen overnight and centrifuged (20,000 g for 10 min). For analysis by UHPLC/TQ-MS, supernatants were additionally filtered through 96-well filter plates (Millipore, cat. no. MAHVN 4550) and diluted 3 times in H2O before LC-MS analyses (see below). Compound concentration in oocytes was calculated by relating peak areas to standard dilution curves and assuming an oocyte volume of 1 µl.
LC-MS analyses of compounds used in Xenopus oocyte uptake experiments
In Figures 5A-C, strictosidine was measured using UHPLC/TQ-MS on an AdvanceTM-UHPLC/EVOQTMElite-TQ-MS instrument (Bruker) equipped with a C-18 reversed phase column (Kinetex 1.7 u XB-C18, 10 cm x 2.1 mm, 1.7 µm particle size, Phenomenex) by using a 0.05% formic acid in water (v/v) (solvent A)-0.05% formic acid in acetonitrile (v/v) (solvent B) gradient at a flow rate of 0.4 ml/min at 40 °C. The gradient applied was as follows: 2 % B (0.9 min), 2-100 % (0.1 min), 100 % B (0.5 min), 100-2 % B (0.1 min), and 2 % B (1.4 min). Strictosidine was ionized by ESI with a spray voltage of +3500 V and a heated probe temperature of 360 °C. The following MRM transitions were monitored: (+)531>514 [19V] (quantifier), (+)531>144 [34V], and (+)531>352 [25V]. Quantification was based on external standard curves for strictosidine measured in control oocyte extracts.
In Figure 5D, strictosidine, secologanin, tryptamine, norharmane and tryptoline were measured by LC-MS using Agilent 1100 Series LC (Agilent Technologies) coupled to a Bruker HCTUltra ion trap mass spectrometer (Bruker Daltonics). A Zorbax SB-C18 column (Agilent;1.8 μM, 2.1 × 50 mm) was used at a flow rate of 0.2 ml/min. The oven temperature was maintained at 35 °C. The mobile phases were as follows: A, water with 0.1% (v/v) HCOOH and 50 μM NaCl; and B, acetonitrile with 0,1% (v/v) HCOOH. The gradient program was following: 0 to 0.5 min, isocratic 2% B; 0.5 to 7.5 min, linear gradient 2 to 40% B; 7.5 to 8.5 min, linear gradient 40% to 90% B; 8.5 to 11.5 isocratic 90% B; 11.6 to 17 min, isocratic 2% B. The flow rate was increased to 0.3 ml/min in the interval 11.2 to 13.5 min. The MS was run in positive electrospray mode. For processing of the LC-MS data, DataAnalysis 4.1. (Bruker Daltonics) was used.
Data Availability
Sequences for the three transporters mentoned in the study have been deposited in NCBI (CrMATE1 (KX372304), CrMATE2 (KX372305) and CrNPF2.9 (KX372303)). Transcriptomic data used in this study are available at http://medicinalplantgenomics.msu.edu.
Supplementary Material
Acknowledgements
This work was supported by grants from the European Research Council (311363), BBSRC (BB/J004561/1) (S.E.O.), Danish National Research Foundation (DNRF99 for D.X., V.N., M.B., H.H.N., B.A.H. and 10-082858 for F.G.-F.), the Innovation Fund Denmark (j.nr. 76-2014-3) (H.H.N.) and from the Région Centre, France (ABISAL grant; E.F., A.O., T.D.D.B., V.C.). R.M.E.P. was supported by a BBSRC Studentship.
Footnotes
Author Contributions
R.M.E.P. made the initial discovery of CrNPF2.9 and designed and carried out all silencing experiments; D.X. and H.H.N-E designed and carried out kinetic experiments; E.F., M.I.S.T.C., T.D.B. and V.C. designed and carried out localization experiments; F.G-F., S.E.O., H.H.N-E, V.C., B.A.H. contributed to the conception and design of the experiments; R.M.E.P., D.X., E.F., M.I.S.T.C., A.O., T.D.B, V.N., M.B., C-E.O, D.M.J., E.C.T., A.P. contributed to the acquisition of data; R.M.E.P., D.X., E.F., M.I.S.T.C., F.G-F., V.C., H.H.N-E., S.E.O. contributed to data analysis and interpretation; R.M.E.P. and S.E.O. drafted the manuscript and all authors critically revised and approved the final version of the manuscript for publication.
References
- 1.O' Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Natural Product Reports. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 2.Keglevich P, Hazai L, Kalaus G, Szantay C. Modifications on the basic skeletons of vinblastine and vincristine. Molecules. 2012;17:5893–5914. doi: 10.3390/molecules17055893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courdavault V, et al. A look inside an alkaloid multisite plant: the Catharanthus logistics. Current Opinion in Plant Biology. 2014;19:43–50. doi: 10.1016/j.pbi.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 4.McKnight TD, Roessner CA, Devagupta R, Scott AI, Nessler CL. Nucleotide sequence of a cDNA encoding the vacuolar protein strictosidine synthase from Catharanthus roseus. Nucleic Acids Research. 1990;18:4939–4939. doi: 10.1093/nar/18.16.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guirimand G, et al. Strictosidine activation in Apocynaceae: towards a "nuclear time bomb"? BMC Plant Biology. 2010;10:182. doi: 10.1186/1471-2229-10-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gongora-Castillo E, et al. Development of Transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLOS One. 2012;7:e52506. doi: 10.1371/journal.pone.0052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geu-Flores F, et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- 8.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim DH, Grun D, van Oudenaarden A. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nature Genetics. 2013;45:1337–1344. doi: 10.1038/ng.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leran S, et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends in Plant Science. 2014;19:5–9. doi: 10.1016/j.tplants.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Padmanabhan M, Dinesh-Kumar SP. Virus-induced gene silencing as a tool for delivery of dsRNA into plants. Cold Spring Harbor Protocols. 2009 doi: 10.1101/pdb.prot5139. pdb.prot5139. [DOI] [PubMed] [Google Scholar]
- 12.Gomez C, et al. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiology. 2009;150:402–415. doi: 10.1104/pp.109.135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larisch N, Schulze C, Galione A, Dietrich P. An N-Terminal dileucine motif directs two-pore channels to the tonoplast of plant cells. Traffic. 2012;13:1012–1022. doi: 10.1111/j.1600-0854.2012.01366.x. [DOI] [PubMed] [Google Scholar]
- 14.Latz A, et al. TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant Journal. 2007;52:449–459. doi: 10.1111/j.1365-313X.2007.03255.x. [DOI] [PubMed] [Google Scholar]
- 15.Marinova K, et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 2007;19:2023–2038. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 17.Komarova NY, Meier S, Meier A, Grotemeyer MS, Rentsch D. Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic. 2012;13:1090–1105. doi: 10.1111/j.1600-0854.2012.01370.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamada K, et al. Functional analysis of an Arabidopsis thaliana abiotic stress-inducible facilitated diffusion transporter for monosaccharides. Journal of Biological Chemistry. 2010;285:1138–1146. doi: 10.1074/jbc.M109.054288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfenstetter S, Wirsching P, Dotzauer D, Schneider S, Sauer N. Routes to the tonoplast: The sorting of tonoplast transporters in Arabidopsis mesophyll protoplasts. Plant Cell. 2012;24:215–232. doi: 10.1105/tpc.111.090415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter C, et al. The vegetative vacuole proteorne of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell. 2004;16:3285–3303. doi: 10.1105/tpc.104.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomes D, et al. Aquaporins are multifunctional water and solute transporters highly divergent in living organisms. Biochimica et Biophysica Acta-Biomembranes. 2009;1788:1213–1228. doi: 10.1016/j.bbamem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Asada K, et al. A 7-deoxyloganetic acid glucosyltransferase contributes a key step in secologanin biosynthesis in Madagascar Periwinkle. Plant Cell. 2013;25:4123–4134. doi: 10.1105/tpc.113.115154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miettinen K, et al. The seco-iridoid pathway from Catharanthus roseus. Nature Communications. 2014;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salim V, Wiens B, Masada-Atsumi S, Yu F, De Luca V. 7-Deoxyloganetic acid synthase catalyzes a key 3 step oxidation to form 7-deoxyloganetic acid in Catharanthus roseus iridoid biosynthesis. Phytochemistry. 2014;101:23–31. doi: 10.1016/j.phytochem.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Salim V, Yu F, Altarejos J, De Luca V. Virus-induced gene silencing identifies Catharanthus roseus 7-deoxyloganic acid-7-hydroxylase, a step in iridoid and monoterpene indole alkaloid biosynthesis. Plant Journal. 2013;76:754–765. doi: 10.1111/tpj.12330. [DOI] [PubMed] [Google Scholar]
- 26.Yu F, De Luca V. ATP-binding cassette transporter controls leaf surface secretion of anticancer drug components in Catharanthus roseus. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15830–15835. doi: 10.1073/pnas.1307504110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carqueijeiro I, Noronha H, Duarte P, Geros H, Sottomayor M. Vacuolar transport of the medicinal alkaloids from Catharanthus roseus Is mediated by a proton-driven antiport. Plant Physiology. 2013;162:1486–1496. doi: 10.1104/pp.113.220558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nour-Eldin HH, et al. NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature. 2012;488:531–534. doi: 10.1038/nature11285. [DOI] [PubMed] [Google Scholar]
- 29.Krouk G, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell. 2010;18:927–937. doi: 10.1016/j.devcel.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Kanno Y, et al. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tal I, et al. The Arabidopsis NPF3 protein is a GA transporter. Nature Communications. 2016;7:11. doi: 10.1038/ncomms11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weichert A, et al. AtPTR4 and AtPTR6 are differentially expressed, tonoplast-localized members of the peptide transporter/nitrate transporter 1 (PTR/NRT1) family. Planta. 2012;235:311–323. doi: 10.1007/s00425-011-1508-7. [DOI] [PubMed] [Google Scholar]
- 33.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO Journal. 2011;30:417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen ME, et al. A functional EXXEK motif is essential for proton coupling and active glucosinolate transport by NPF2.11. Plant and Cell Physiology. 2015;56:2340–2350. doi: 10.1093/pcp/pcv145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature. 2014;507:68–72. doi: 10.1038/nature13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morita M, et al. Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2447–2452. doi: 10.1073/pnas.0812512106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shitan N, et al. Involvement of the Leaf-specific multidrug and toxic compound extrusion (MATE) Transporter Nt-JAT2 in Vacuolar sequestration of nicotine in Nicotiana tabacum. PLOS One. 2014;9 doi: 10.1371/journal.pone.0108789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoji T, et al. Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiology. 2009;149:708–718. doi: 10.1104/pp.108.132811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hildreth SB, et al. Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18179–18184. doi: 10.1073/pnas.1108620108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown S, Clastre M, Courdavault V, O' Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galanie S, Thodey K, Trenchard IJ, Interrante MF, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wehrens R, Buydens LMC. Self- and super-organizing maps in R: The kohonen package. Journal of Statistical Software. 2007;21:1–19. [Google Scholar]
- 43.Vesanto J, Alhoniemi E. Clustering of the self-organizing map. Ieee Transactions on Neural Networks. 2000;11:586–600. doi: 10.1109/72.846731. [DOI] [PubMed] [Google Scholar]
- 44.Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research. 2007;35:e55. doi: 10.1093/nar/gkm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nour-Eldin HH, Geu-Flores F, Halkier BA. USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods in Molecular Biology. 2010;643:185–200. doi: 10.1007/978-1-60761-723-5_13. [DOI] [PubMed] [Google Scholar]
- 46.Liscombe DK, O' Connor SE. A virus-induced gene silencing approach to understanding alkaloid metabolism in Catharanthus roseus. Phytochemistry. 2011;72:1969–1977. doi: 10.1016/j.phytochem.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murata J, Roepke J, Gordon H, De Luca V. The leaf epidermome of Catharanthus roseus reveals its biochemical specialization. Plant Cell. 2008;20:524–542. doi: 10.1105/tpc.107.056630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy E, Galan MC, O'Connor SE. Substrate specificity of strictosidine synthase. Bioorganic & Medicinal Chemistry Letters. 2006;16:2475–2478. doi: 10.1016/j.bmcl.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 49.Guirimand G, et al. Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Reports. 2009;28:1215–1234. doi: 10.1007/s00299-009-0722-2. [DOI] [PubMed] [Google Scholar]
- 50.Nour-Eldin HH, Hansen BG, Norholm MHH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Research. 2006;34:8. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences for the three transporters mentoned in the study have been deposited in NCBI (CrMATE1 (KX372304), CrMATE2 (KX372305) and CrNPF2.9 (KX372303)). Transcriptomic data used in this study are available at http://medicinalplantgenomics.msu.edu.