Abstract
Generalist and specialist species differ in the breadth of their ecological niche. Little is known about the niche width of obligate human pathogens. Here we analyzed a global collection of Mycobacterium tuberculosis Lineage 4 clinical isolates, the most geographically widespread cause of human tuberculosis. We show that Lineage 4 comprises globally distributed and geographically restricted sublineages, suggesting a distinction between generalists and specialists. Population genomic analyses showed that while the majority of human T cell epitopes were conserved in all sublineages, the proportion of variable epitopes was higher in generalists. Our data further support a European origin for the most common generalist sublineage. Hence, the global success of Lineage 4 reflects distinct strategies adopted by different sublineages and the influence of human migration.
Introduction
Ecologists distinguish between generalists and specialists depending on the width of an organism’s ecological niche1. In infectious diseases, the niche of a given pathogen is determined by host range and the agent’s capacity to survive in the environment2. Some microbes are obligate pathogens restricted to one or several host species3,4, others are mainly free-living and only occasionally pathogenic5. Little is known on the niche width of obligate human pathogens3. The causative agent of tuberculosis, known as the Mycobacterium tuberculosis complex (MTBC), is an obligate pathogen that comprises seven phylogenetic lineages adapted to humans and two lineages adapted to various wild and domestic animal species6. Some human-adapted MTBC lineages have received particular attention. For example, Lineage 2, which includes the Beijing family of strains, has repeatedly been associated with drug resistance7. Lineage 2 likely originated in East Asia8,9, and has recently been expanding in some parts of the world10. By contrast, Lineages 5 and 6 (also known as Mycobacterium africanum West Africa I and II), and Lineage 7 are largely restricted to West- and East Africa, respectively11,12. The observation that the human-adapted MTBC population is phylogeographically structured has led to the hypothesis that the different lineages might be adapted to particular human populations13. Support for this notion comes from the observation that sympatric host-pathogen associations in human tuberculosis remain stable over time, even in metropolitan settings where host and pathogen populations intermix14–17. Moreover, sympatric host-pathogen associations are perturbed in HIV coinfected patients14, indicating that in the context of reduced host immune-competence, the different lineages can successfully infect and cause disease irrespective of the host genetic background.
Contrary to the other main human-adapted MTBC lineages, Lineage 4 occurs at significant frequencies on all inhabited continents18. It is hence geographically the most widespread cause of human tuberculosis19. Yet, the reasons for this global success are unknown. Lineage 4 has been shown to exhibit enhanced virulence in macrophage and animal models of infection, albeit with much variation between different Lineage 4 strains19,20. Moreover, molecular epidemiological studies have reported considerable variation in the transmission success of different Lineage 4 strains in clinical settings19. These observations suggest that Lineage 4 is genetically and phenotypically diverse, and this diversity might determine the epidemiology of different Lineage 4 subtypes in different parts of the world. The purpose of this study is to get a better understanding of the global population structure of Lineage 4 and the evolutionary forces that have contributed to the success of Lineage 4 across the world. For this we combined large-scale single nucleotide polymorphism (SNP)-typing with targeted whole-genome sequencing of a global collection of Lineage 4 clinical isolates.
Results
MTBC Lineage 4 comprises 10 separate sublineages
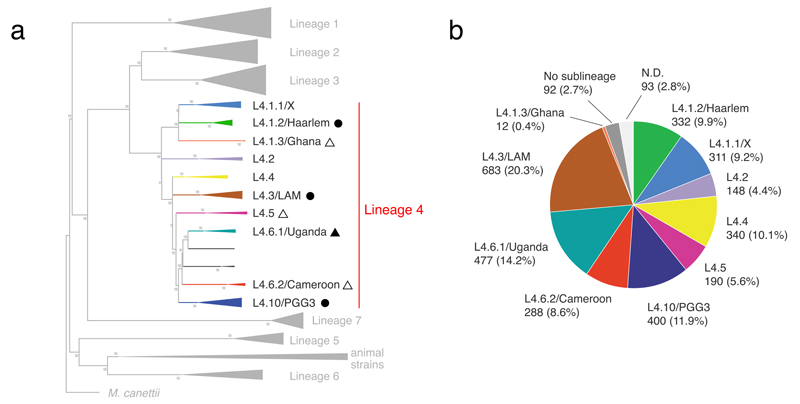
We first analyzed 72 published genome sequences of Lineage 4 clinical strains from global sources21,22. These strains harbored 9,455 variable single nucleotide positions which divided Lineage 4 into 10 sublineages (L4.1.1 to L4.10 in Fig. 1a and Supplementary Fig. 1). We used four complementary approaches to validate these sublineages. First, we performed a principal component analysis, which showed a clear separation of seven sublineages (L4.1.1, L4.1.3, L4.1.2, L4.2, L4.3, L4.4, L4.10; Supplementary Fig. 2). Sublineages L4.5, L4.6.1/Uganda and L4.6.2/Cameroon were less clearly separated. Second, we found that the mean pairwise genetic distance between pairs within the sublineages was significantly lower than between sublineages (276 SNPs versus 602 SNPs, Wilcoxon rank sum test, p < 0.0001, Supplementary Fig. 3). Overall, the mean pairwise SNP distance between any two strain pairs was 565 SNPs. Third, we calculated pairwise fixation indexes (FST) to evaluate the degree of population differentiation. All FST values between the sublineages were larger than 0.33 (Supplementary Table 1), indicating that these populations are separated. Fourth, we mapped previously reported phylogenetic markers onto our genome-based phylogenetic tree15,23–28. Most of these markers were congruent with our sublineage definition (Supplementary Fig. 1).
Figure 1. Definition and global frequency of Lineage 4 sublineages.
(a) We defined 10 sublineages based on the analysis of 72 MTBC Lineage 4 genome sequences published previously21,22. Sublineages were labeled according to Coll et al.27 (whenever possible) and previous designations based on spoligotyping (see Supplementary Fig. 1). Black triangles indicate sublineages identified as specialists, black circles indicate generalists. Filled shapes indicate sublineages, for which we performed deep genomic analyses. (b) Global proportion of each sublineage. A total of 3,366 MTBC Lineage 4 isolates were screened for sublineage-specific SNPs. L4.3/LAM was the most frequent sublineage globally.
Sublineages differ in their phylogeographic distribution
Because the MTBC exhibits limited sequence variation and no signficant ongoing horizontal gene exchange, SNP homoplasies are extremely rare, making SNPs ideal phylogenetic markers29. We further scrutinized the 9,455 variable positions among the 72 MTBC Lineage 4 genomes, and found 51 to 277 specific for one of each of the 10 sublineages. All of these variable positions were mutually exclusive, i.e. they showed no homoplasy. We selected a subset of these sublineage-specific SNPs and used these to screen a global collection of 3,366 Lineage 4 clinical isolates from 100 countries using various genotyping platforms30–35. First, we developed a novel sublineage-specific multiplexed SNP-typing assay using the Luminex platform as previously reported36, and used that method to screen 2,001 isolates (Supplementary Table 2). In addition, we screened 741 isolates using the Sequenom MassARRAY platform (Supplementary Table 3)37, and 624 isolates by PCR and Sanger sequencing (Supplementary Table 4). Overall, 3,181/3,366 (94.5%) Lineage 4 isolates were successfully assigned to a sublineage (Supplementary Table 5). An additional 92/3,366 (2.7%) isolates harbored the reference allele for all sublineages, indicating they belonged to one or several additional and unknown sublineages. For the remaining 93/3,366 (2.8%) isolates, no classification could be obtained for various technical reasons. Among the 3,181 Lineage 4 isolates assigned to one of the 10 sublineages, L4.3/LAM was the most frequent, accounting for 20.3%, followed by L4.6.1/Uganda (14.2%), L4.10/PGG3 (11.9%), L4.4 (10.1%), and L4.1.2/Haarlem (9.9%) (Fig. 1b).
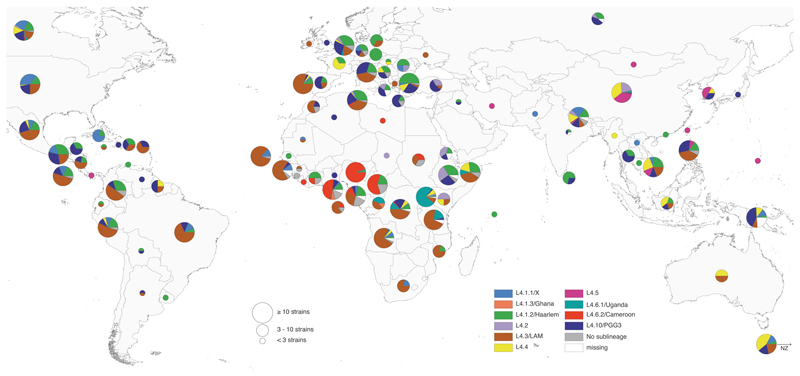
Mapping the proportion of each sublineage by country showed that the sublineages differed in their geographical distribution (Fig. 2). Specifically, L4.1.2/Haarlem, L4.3/LAM and L4.10/PGG3 occurred globally (Fig. 3a, Supplementary Fig. 4). By contrast, L4.1.3/Ghana, L4.5, L4.6.1/Uganda and L4.6.2/Cameroon occurred at high frequencies in specific regions of Africa or Asia, and were almost completely absent from Europe and the Americas (Fig. 3b). The geographical spread of the three remaining sublineages was intermediate (Fig. 2, Supplementary Figs. 4 and 5). L4.1.1/X mainly occurred in the Americas and in lower proportions in few countries of Southern Africa, Asia and Europe. L4.2 and L4.4 occurred in high proportions among isolates from particular countries in Asia and Africa, but were largely absent from the Americas (Fig. 2, Supplementary Figs. 4 and 5). A similar pattern of sublineage distribution was observed when normalizing by TB prevalence38 and country surface area (Supplementary Fig. 6).
Figure 2. Global distribution of Lineage 4 sublineages.
Pie charts showing proportions of the 10 Lineage 4 sublineages among all MTBC Lineage 4 isolates in each country. Circle sizes correspond to the number of isolates analyzed per country. A total of 3,366 MTBC Lineage 4 isolates were included. Color codes are as in Fig. 1.
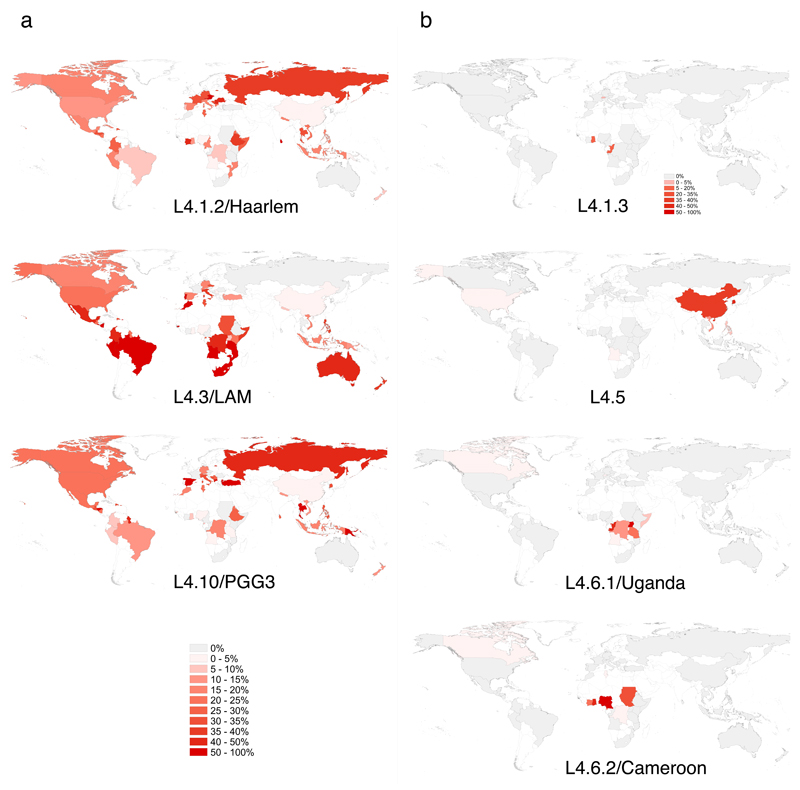
Figure 3. Country-specific proportions of sublineages reveal generalists and specialists.
(a) The generalist sublineages L4.1.2/Haarlem, L4.3/LAM and L4.10/PGG3 were found globally at high proportions. (b) The locally restricted specialist sublineages L4.1.3/Ghana, L4.5, L4.6.1/Uganda and L4.6.2/Cameroon occurred at high frequencies in only a few countries and were restricted to certain geographical regions. Intensity of red indicates proportion of the sublineage among all Lineage 4 isolates in each country. Countries with fewer than three isolates in total are shown as “no data” and are filled white. A total of 3,366 Lineage 4 isolates were included in this analysis. The color scale for all sublineages is as indicated in Panel a, except for sublineage L4.1.3/Ghana (separate scale shown).
Populations that occupy a broader variety of environments may exhibit a wider geographic distribution. Humans differ in their susceptibility to TB39, and human genetic diversity may thus determine the width of the ecological niche accessible to different MTBC genotypes40,41. The geographical restriction of particular MTBC genotypes might reflect local adaption of these pathogen variants to the corresponding human host populations13,15. Such a sympatric host-pathogen association in human TB is compatible with the “local” sublineages observed here, and supports the notion that these sublineages represent ecological specialists. By contrast, the three “global” sublineages could represent generalists capable of infecting and causing disease in many different human populations. This notion was supported by the fact that the three generalist sublineages L4.1.2/Haarlem, L4.3/LAM and L4.10/PGG3 were observed in 49, 47 and 47 countries, respectively, whereas the specialist sublineages L4.1.3/Ghana, L4.5, L4.6.1/Uganda and L4.6.2/Cameroon were only found in few countries each (3, 7, 9 and 10 countries, respectively). The country frequencies of the remaining three sublineages L4.1/X, L4.2/Ural and L4.4 were intermediate (27, 14 and 26 countries, respectively) (Supplementary Fig. 4).
The different geographical distribution of generalist and specialist sublineages could be due to intrinsic biological factors, extrinsic factors such as human migration, or both. Hence we next performed various population genomic analyses to explore the genomic characteristics of these Lineage 4 generalists and specialists, as well as the role of human migration in the global spread of the most successful generalist sublineage.
Genomic features of generalist and specialist sublineages
The geographic and niche distribution of populations can be correlated with their genetic variability or with that of their ancestors. One possible reason for the restricted host range of the specialist sublineages might be historical, i.e. the ancestor populations of the extant specialist populations may have harbored more deleterious mutations, restricting their host range. To assess this possibility, we characterized the mutations which contributed to the divergence of the different sublineages; these mutations are variants that have become fixed during the evolution of these sublineages. We focused on the substitutions that occurred in all isolates of any of the generalist sublineages (L4.1.2/Haarlem, L4.3/LAM and L4.10/PGG3) and compared them to the substitutions that occurred in all isolates of any of the three specialist sublineages (L4.6.1/Uganda, L4.5 and L4.6.2/Cameroon). We identified nonsynonymous SNPs predicted to have a functional effect using SIFT42. We found that overall, the specialist and generalist sublineages showed a similar proportion of fixed substitutions (among all substitutions) predicted to impact gene function (23.0% versus 20.6%, χ2 test p=0.62; Supplementary Table 6), suggesting that the mutational load of the ancestor populations did not differ significantly between generalists and specialists.
Small populations with restricted geographic ranges are expected to have reduced levels of genetic diversity43. Thus, one possible restriction to niche expansion by specialist sublineages could be that these sublineages have low genetic diversity precluding adaptation to new hosts. We characterized the genetic diversity associated with the process of diversification in Lineage 4 generalists and specialists, focusing on L4.3/LAM and L4.6.1/Uganda, globally the most frequent generalist and specialist sublineages of Lineage 4 in our dataset, respectively (Fig. 1b). We analyzed the whole-genome sequences of 293 L4.3/LAM clinical strains representing the global diversity of this sublineage. These were selected from a global collection of 2,132 L4.3/LAM isolates based on standard genotyping data (Supplementary Table 7, Supplementary Figs. 7-9). For L4.6.1/Uganda, we analyzed whole-genome sequences of 203 clinical strains from Uganda and several neighboring countries (Supplementary Table 7, Supplementary Figs. 10 and 11). This sample included 28 L4.6.1/Uganda strains identified through screening of 13,067 publically available MTBC whole genome sequences (see Online Methods)44–56. Comparing the genetic diversity between these two bacterial populations showed that L4.3/LAM was significantly more diverse than L4.6.1/Uganda (mean number of 395 SNPs between pairs compared to 215 SNPs, respectively; Wilcoxon rank sum test p<0.0001), consistent with the expected difference between generalists and specialists43.
Antigenic diversity in Lineage 4 sublineages
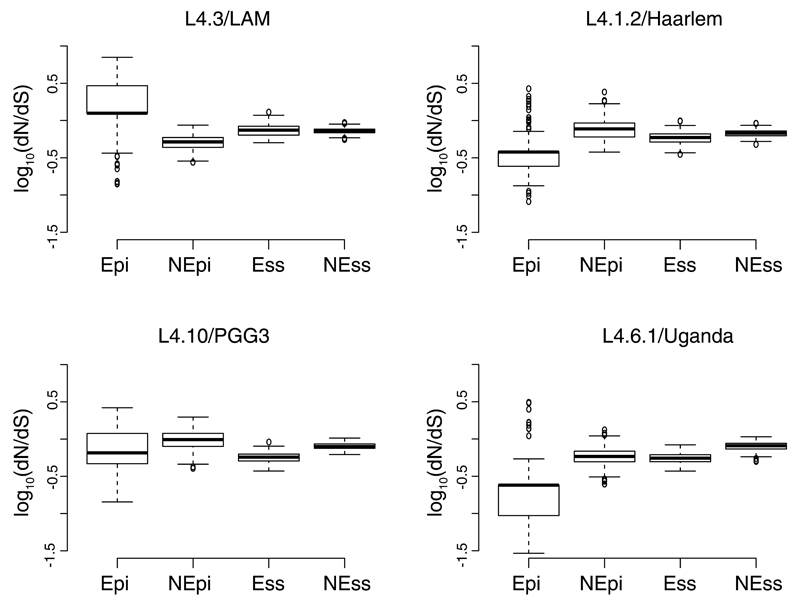
We previously reported that in the human-adapted MTBC, experimentally confirmed human T cell epitopes were conserved57,58. This is unlike many other pathogens where genomic regions encoding antigens tend to be diverse as a result of antigenic variation linked to immune escape59. When we assessed the evolutionary conservation of 1,226 experimentally confirmed human T cell epitopes60 in L4.6.1/Uganda by calculating their dN/dS, we found that these epitopes were significantly more conserved than the non-epitope regions of the corresponding T cell antigens (Wilcoxon rank sum test, p<0.0001, Fig. 4). This result was consistent with our previous findings for the MTBC overall57,58. However for L4.3/LAM, we saw the opposite, i.e. the T cell epitopes showed a significantly higher dN/dS than the non-epitope regions (Wilcoxon rank sum test, p<0.0001, Fig. 4). To test whether a high dN/dS in T cell epitopes is characteristic of the generalist sublineages, we analyzed the genomes of 228 L4.2/Haarlem strains and 301 L4.10/PGG3 strains identified by screening of 13,067 publically available genomes (Supplementary Table 7, Supplementary Figs. 12 and 13). We found that in contrast to L4.3/LAM, the epitope regions in these generalist sublineages were more conserved than the corresponding non-epitope regions, i.e. similar to L4.6.1/Uganda and the MTBC overall57,58 (Fig. 4). Consistent with previous reports57,58,61, essential genes62 were significantly more conserved than nonessential genes in all sublineages, except L4.3/LAM in which the dN/dS of essential and nonessential genes were not significantly different (Fig. 4)
Figure 4. Pair-wise ratios of rates of nonsynonymous to synonymous substitutions (dN/dS) in generalist and specialist sublineages for different gene categories.
Abbreviations: Epi – experimentally confirmed human T cell epitopes; nEpi – non-epitope regions of T-cell antigens, both obtained from the Immune Epitope Database60; Ess – essential genes62; nEss – non-essential genes62. Wilcoxon rank sum tests: L4.6.1/Uganda (N=203) Epi vs nEpi, W=4952, p<0.001; L4.6.1/Uganda (N=203) Ess vs nEss, W=1415, p<0.001; L4.3/LAM (N=293) Epi vs nEpi, W=74540, p<0.001, L4.3/LAM (n=293) Ess vs nEss W=45067, p-value=0.29; L4.1.2/Haarlem (N=228) Epi vs nEpi, W=6561, p<0.001, L4.1.2/Haarlem (N=228) Ess vs nEss W=13369, p<0.001; L4.10/PGG3 (N=301) Epi vs nEpi, W= 27335, p<0.001, L4.10/PGG3 (N=301) Ess vs nEss W= 3103, p<0.001.
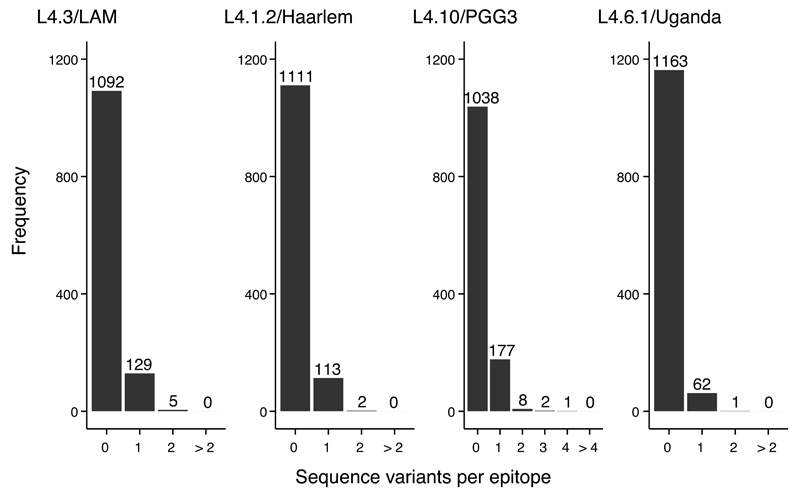
One of the limitations of our dN/dS analyses was that despite a large number of genomes analyzed, within individual sublineages, the mean number of pair-wise differences in regions encoding T cell epitopes was very small (Supplementary Table 8), limiting the accuracy of dN/dS inferences for epitopes. Hence, we assessed T cell epitope diversity by comparing the number of epitopes affected by nonsynonymous variants in the different sublineages (Fig. 5). We found that in all four sublineages, the majority of epitopes were completely conserved, consistent with our previous findings for the MTBC overall57,58. However, each of the three generalist sublineages showed significantly more epitopes harboring at least one amino acid change when compared to the specialist sublineages L4.6.1/Uganda (Fig. 5, χ2 tests p<0.0001 for all comparisons). It is possible that this comparably higher epitope diversity in generalists might reflect interactions with broader host populations.
Figure 5. Frequency distribution of the number of epitopes with nonsynonymous variants in generalist and specialist sublineages.
A total of 1,226 T cell epitopes were included in the analysis. The number above each bar corresponds to epitope counts. Generalist sublineages L4.3/LAM, L4.1.2/Haarlem and (L4.10/PGG3. Specialist sublineage L4.6.1/Uganda. Tests: L4.6.1/Uganda vs L4.3/LAM Χ2= 27.04, p<0.001; L4.6.1/Uganda vs L4.1.2/Haarlem Χ2=15.75, p<0.001; L4.6.1/Uganda vs L4.1.2/PGG3 Χ2= 68.24, p<0.001.
The epitopes interrogated in our analysis were encoded by a total of 304 antigens. The number of antigens containing nonsynonymous variation in epitopes was 60, 26, 46 and 48 antigens in L4.3/LAM, L4.6.1/Uganda, L4.2/Haarlem and L4.10/PGG3, respectively. When excluding nonsynonymous mutations present only in one strain in each sublineage (which likely represent transient mutations), the number of antigens dropped to 20, 11, 12, 24 in L4.3/LAM, L4.6.1/Uganda, L4.2/Haarlem and L4.10/PGG3, respectively (Supplementary Table 9). Interestingly, 10 of those antigens exhibited independent parallel, nonsynonymous variation in epitope regions in the different sublineages (Supplementary Table 9). Of those antigens, Pst1, an adhesin promoting phagocytosis63, and FbpB, the precursor of the secreted antigen 85-B64, had already been pointed out as encoding diverse epitopes by a previous study, in which several MTBC lineages were compared57. (Supplementary Table 9). Other antigens exhibiting parallel nonsynonymous changes by different sublineages include known immunodominant, secreted antigens such as Mpb6464, MPT3265 and MPT7066 and three latency-associated antigens (Rv1733c67, Rv3034c, Rv262868, Supplementary Table 9).
Origin and global spread of the L4.3/LAM sublineage
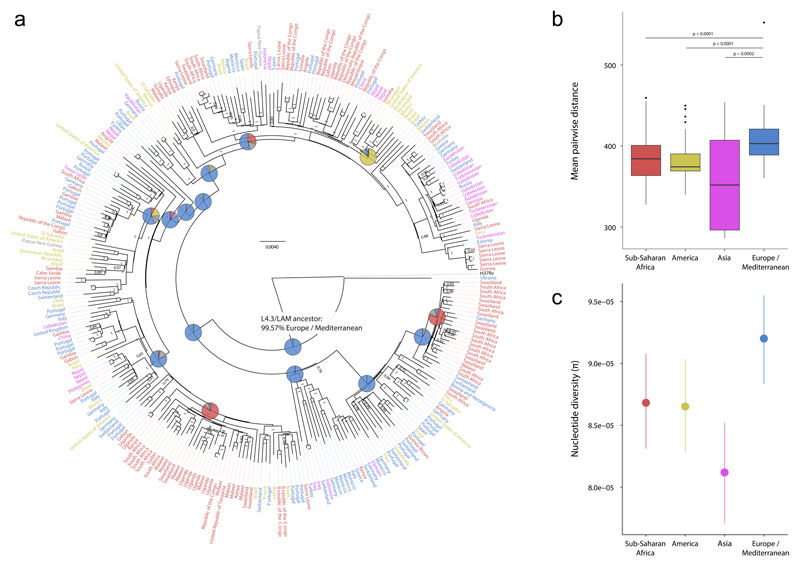
Irrespective of the putative biological differences between the Lineage 4 sublineages, human migration could also have led to variation in the global distribution of MTBC lineages. Because the most successful sublineage of Lineage 4 was also frequently found in Europe, we hypothesized that the global success of L4.3/LAM was driven by European migration and colonization. To test this hypothesis, we first determined the most likely geographical origin of the most recent common ancestor of L4.3/LAM using two methods for reconstruction of ancestral states69. By both methods, Europe was predicted as the most likely place of origin of L4.3/LAM (100% and 99.6%, respectively) (Fig. 6a, Supplementary Fig. 14). Moreover, the ancestral geographical regions reconstructed for subsequent nodes in the phylogeny were consistent with the spread of L4.3/LAM from Europe to other parts of the world (Fig. 6a). Finally, we found that L4.3/LAM strains from Europe were genetically more diverse than L4.3/LAM strains from other continents, which further supports a European origin for this sublineage (Fig. 6b, Kruskall-Wallis test p<0.0001; Fig. 6c).
Figure 6. Genome-based phylogeny and diversity by continent of 293 strains of the L4.3/LAM sublineage.
(a) Bayesian phylogeny with label colors indicating continent of strain origin: blue, Europe/Mediterranean; red, Sub-Saharan Africa; yellow, America; pink, Asia. Numbers on nodes indicate posterior probabilities. Pie charts indicate reconstructed ancestral geographical regions of the internal nodes. The hypothetical L4.3/LAM-ancestor is labeled and a European origin for this ancestor was supported using a Bayesian Method (shown) and a Maximum Parsimony method (Supplementary Fig. 14). The pie colors correspond to the colors of the taxa labels. (b) Boxplot of pairwise genetic distances (number of polymorphisms) of L4.3/LAM strains by continent (p-values from Wilcoxon rank sum test). (c) Nucleotide diversity per site (π), measured by continent. Error bars indicate 95% confidence intervals. MTBC isolates from countries of the continent group “Oceania“ (UN category; including Australia and New Zealand, Melanesia, Micronesia and Polynesia) were excluded for the genetic diversity analysis in panels B and C due the low number of samples.
Discussion
Our findings show that the global success of Lineage 4 is a consequence of both biological and social phenomena. Specifically, we found that Lineage 4 is genetically diverse, and that this diversity is phylogeographically structured. The phylogeography of Lineage 4 supports an ecological distinction between globally represented generalists and geographically restricted specialists. Our in-depth population genomic analyses of one specialist and three generalist sublineages showed that even though the majority of human T cell epitopes were completely conserved in all four sublinages, the proportion of epitopes with amino acid substitutions was significantly higher in generalists. Finally, we demonstrate a likely European origin for L4.1/LAM, the most frequent and globally widespread generalist sublineage of Lineage 4.
Our observation that Lineage 4 is phylogenetically diverse is in line with previous findings27,70, and highlights the importance of large and globally representative samples when studying the population structure of human pathogens. We found that Lineage 4 comprises at least 10 sublineages, which differ in their geographical distribution. The phylogeography of these sublineages is consistent with an ecological separation into specialists and generalists, with some sublineages showing an intermediate geographical distribution. Our phylogenetic analyses also showed that the three generalist sublineages identified within Lineage 4 were not monophyletic (Fig. 1a), indicating that generalism was acquired multiple times independently during the evolution of Lineage 4. Specialist sublineages also emerged multiple times, which is consistent with local adaptation to separate human populations13.
One could argue that the reason for specialist sublineages being geographically restricted is they diverged later than the generalist sublineages during the evolution of Lineage 4, and thus had insufficient time to spread globally. However, based on recent findings by Comas et al.71, the African specialist sublineages already existed at least several centuries ago, perhaps even several millennia ago, depending on the age of the most recent common ancestor of the MTBC that has been estimated between 70’000 years9,21 and 6’000 years72,73. Thus, this timespan should have offered ample opportunity for the specialist sublineages to become more geographically widespread.
The genetic diversity of the specialist sublineage L4.6.1/Uganda was significantly lower than that of the generalist L4.3/LAM, as expected from populations with restricted geographical ranges43. Concomitantly, the diversity of T cell epitopes in the specialist sublineage L4.6.1/Uganda was also significantly lower than in any other of the three generalist sublineages analyzed. Whether the low genetic diversity of the specialist sublineage has hindered the adaptation of these strains to other human populations or reflects a restricted niche due to the lack of opportunity for spreading will need to be explored in future studies.
In all sublineages analyzed, the large majority of T cell epitopes were completely conserved, which is in agreement with previous reports for the MTBC overall57,58. This suggests that both these generalists and specialists do not use antigenic variation as a main mechanism of immune evasion. Despite this general trend, we found that some antigens have acquired nonsynonymous mutations in parallel in the different sublineages, suggesting that variation in these particular antigens might be beneficial. For example, acquiring nonsynonymous variation may allow particular antigens to be recognized by T cell receptors of different human populations, which might be beneficial in the presence of different human HLA alleles58. This could also provide an explanation for the differences in the degree of variation in T cell epitopes of the generalist and specialist sublineages, as generalist sublineages are expected to interact with a broader range of HLA alleles. Alternatively, some nonsynonymous mutations in epitopes might reflect escape from human T cell recognition58. More work is needed to determine if and how the limited diversity in T cell epitopes in the MTBC is linked to adaption to different host populations and/or immune escape.
Two independent phylogeographic analyses predicted Europe as the most likely geographical origin for the most recent common ancestor of L4.3/LAM. A European origin for L4.3/LAM was further supported by our finding that strains belonging to this sublineage were more genetically diverse in Europe compared to Africa, Asia and America. Taken together, these results suggest a role for Europeans for the spread of L4.3/LAM across the globe. Given the high frequency of L4.3/LAM in Europe (Fig. 2, Fig. 3a), particularly in TB patients from the Iberian Peninsula and in Latin America74,75, Portuguese and Spanish exploration, trade and conquest over the last centuries may have contributed to the global dissemination of this sublineage76.
Of note, the Americas lack specialist sublineages, including the three African specialist sublineages, despite centuries of slave trade. Importantly, this also applies to MTBC Lineage 5 and 6 (i.e. M. africanum) which today are largely limited to parts of West Africa11, the source of most of African slaves shipped to the Americas. Even if these lineages did reach the Americas at the time, they later might have been replaced by generalist sublineages from Europe including L4.3/LAM, following the massive influx of Europeans to the Americas during the 19th and early 20th centuries77, a time when the European TB epidemic was at its peak73,78. Importantly, these human migrations can be viewed as natural experiments, in which diverse human populations came into contact with different MTBC genotypes. As mentioned above, there is evidence that the African specialist sublineages of Lineage 4 already existed in sub-Saharan Africa centuries ago71. Following European contact, generalist sublineages were introduced to Africa and today, a significant proportion of human tuberculosis in Africa is caused by L4.1/LAM and other generalists (Figs. 2 and 3a). By contrast, no significant spill-over of African specialist sublineages has occurred into Europe or American populations of European ancestry.
Three of the 10 sublineages showed an intermediate pattern of geographical distribution. Independent of the open question as to whether they might represent generalists or specialists, it is interesting to note that none of these three sublineages were found at significant frequency and proportion in Europe. They might therefore represent generalist of a non-European origin. Deeper analyses are needed to shed more light on these sublineages.
Our study is limited in that many of the MTBC isolates analyzed come from convenience samples and might therefore not be representative of a particular country. However, for the analysis of sublinage distributions by SNP-genotyping, we included more than 3,000 clinical isolates from 100 countries, which should reduce any potential selection bias. For the deep genomic analyses, we selected strains basesd on a large and diverse collection of classical genotyping patterns, and in addition, screened <13,000 MTBC whole genome sequences available in public repositories. As a further limitation, some isolates in our collection were obtained from patients who recently emigrated from a high tuberculosis incidence region into a low-incidence country. However, we excluded cases from ongoing transmission and focused on immigrants with reactivation disease, i.e. they were most likely infected in their country of origin before moving abroad. Moreover, we used country of birth for all analyses as opposed to country of tuberculosis diagnosis.
In conclusion, our findings indicate that the global success of Lineage 4 partly results from the different evolutionary strategies adopted by different sublineages. These strategies reflect an ecological distinction between specialists and generalists. The specialist sublineages are adapted to their sympatric host populations and geographically restricted. The generalist sublineages exhibit a broader ecological niche and are geographically widespread. Moreover, Europeans contributed to the global spread of the most successful generalist sublineage of Lineage 4. Our results highlight the ecological and epidemiological relevance of the deep phylogenetic diversity within the MTBC79. More generally, exploring potential differences between specialists and generalists in other pathogens will improve our understanding of the biology and epidemiology of infectious diseases.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Sequencing reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) under the study accession number PRJEB11460.
Online Methods
Mycobacterial isolates
For the definition of Lineage 4 sublineages, we used 72 whole genome sequences of MTBC Lineage 4 and reference sequences of the other MTBC lineages published previously21,22 (Supplementary Table 7). These represented the largest collection of Lineage 4 whole-genome sequences available at that time. For the SNP-screening of clinical isolates for sublineage-classification, we used a retrospective global collection of 3,366 MTBC Lineage 4 isolates from 100 countries (Supplementary Table 5)15,30–35. All isolates had previously been identified as MTBC Lineage 4 by SNP-typing, genomic deletion analysis or spoligotyping. Approximately one third of these isolates were from patients who migrated to another country (1,106; 32.9%), and we used country of birth of the patient as a proxy for the origin of the MTBC strains. Two thirds of the isolates (2,260; 67.1%) were from countries where both country of isolation and country of birth were identical. Isolates of L4.6.1/Uganda from Uganda were genotyped in our previous work34. For the in-depth population genomic analysis of L4.3/LAM, we included previously published genomes21,27,44, and generated whole genome sequences of additional strains selected from a large collection of 2,132 MIRU-VNTR-genotyped isolates representing the global diversity of L4.3/LAM (Supplementary Fig. 7). Starting from 500 whole genome sequences, we excluded sequences with bad quality (sequencing coverage < 15x, proportion of homozygous variant calls <85%), isolates in transmission clusters (defined as isolate pairs differing by ≤12 SNPs) and strains with unknown country of origin, resulting in whole genome sequencing (WGS) data for 293 L4.3/LAM strains, which were included in the final analysis (Supplementary Table 7). For the in-depth population genomic analyses of L4.6.1/Uganda, we generated WGS data from 175 isolates of the L4.6.1/Uganda genotype, selected for maximal geographic diversity and from previous studies34. Moreover, to further increase geographic coverage and genetic diversity among L4.6.1/Uganda strains, we analyzed all available WGS data from several published studies8,45–56,75 and other whole genome data available in the public domain. We used KvarQ80 to screen for the L4.6.1/Uganda-specific SNPs described below. Starting from 13,067 genome sequences and excluding all clustered isolates except for one representative of each cluster, we identified 28 additional L4.6.1/Uganda genome sequences which we included in our analysis of a total of 203 genomes (Supplementary Table 7). For genomic analysis of L4.1.2/Haarlem and L4.10/PGG3 strains, we screened the same 13,067 isolates (plus our own collection) for clade-specific SNPs of these two sublineages. We identified 505 genome sequences of strains of L4.1.2/Haarlem and 748 sequences of L4.10/PGG3. After excluding problematic sequences and strains in transmission clusters (criteria see above), we used 228 strains of L4.1.2/Haarlem and 301 strains of L4.10/PGG3. H37Rv was used as outgroup for all sublineage phylogenies except L4.10/PGG3, for which an isolate of L4.1.2/Haarlem was used (H37Rv belong to L4.10/PGG3).
Whole genome sequencing, variant calling and filtering
WGS of new MTBC isolates was performed using Illumina chemistry (MiSeq, HiSeq2000/2500, NextSeq; paired end or single end). Illumina MiSeq-generated sequencing reads were clipped for adapters with Trimmomatic81 before mapping. We used a previously described pipeline for the mapping of short sequencing reads to the reference genome (a reconstructed hypothetical MTBC ancestor) with BWA 0.6.221. SNPs were called with SAMtools 0.1.19, and excluded if the coverage was less than 10% or more than 200% of the average coverage of the genome, if not supported by at least two reads on each strand, or if the quality was less than 30. All SNPs were then annotated using H37Rv reference annotation (AL123456.2) with Annovar82 and customized scripts. SNPs in regions annotated as “PE/PPE/PGRS”, “maturase”, “phage”, “insertion sequence” were excluded. Additionally, we excluded SNPs in genes with previously identified repetitive regions58. Small insertions and deletions called by BWA/SAMtools as “INDEL” were not considered for the analyses. The presence of large genomic deletions reported previously15,28,74 was assessed by manually inspecting BAM alignment files from BWA mappings in Artemis for the presence of reads at the genomic regions with described deletions. Alternatively, we used a new testsuite in KvarQ80 to check for reads aligning to 25 bp query sequences of the corresponding deletion.
Phylogenetic and population genetic analyses for the definition of sublineages
A phylogenetic tree was generated with all Lineage 4 genomes, plus several reference genomes from all other MTBC lineages. Pairwise SNP distances were calculated using MEGA583 and the ape-package in R84. Fixation indices (FST; estimation of population separation) were calculated using Arlequin 3.5.85. Statistical significance was obtained by permutating haplotypes between sublineages. Principal Component Analysis (PCA) was done with Jalview86. Naming of Lineage 4 sublineages was adapted to Coll et al.27 whenever possible. However, in that publication, no criteria for the definition of sublineages were given, and not all sublineages were identified as such. We therefore added continuous numbers for the clades which were not defined by Coll et al. The new sublineages defined in this study are L4.1.3/Ghana and L4.10/PGG3 (the latter including L4.5, L4.8 and L4.9 according to nomenclature by Coll et al.). The full phylogenetic tree, including previous markers and spoligotyping family names is shown in Supplementary Fig. 1.
Identification of sublineage-specific SNPs
The alignment of all SNPs from the initial 72 MTBC Lineage 4 strains was imported into Mesquite87, in parallel with the phylogenetic tree generated from the same data in MEGA5. We used the “Trace Character History” module of Mesquite to map polymorphisms to clades. The full dataset of reconstructed positions was exported, and sublineage-specific SNPs were extracted as nucleotide differences between internal nodes of the phylogeny.
SNP-typing to screen for MTBC Lineage 4 sublineages
We developed a new SNP-typing assay to screen clinical isolates for the defined Lineage 4 sublineages. For this, we selected one “diagnostic” SNP per sublineage using previously defined methods and criteria36. Oligonucleotides were designed for a 10-plex MOL-PCR assay based on the Luminex xTag platform (Luminex, Austin, USA) (Supplementary Table 2)36. DNA extracts from clinical MTBC isolates were then screened with either i) the new MOL-PCR assay, ii) standard PCR amplification and subsequent Sanger sequencing of the region up- and downstream of the sublineage-specific SNP (see Supplementary Table 4 for PCR and sequencing primers), iii) a real-time PCR melting curve assay using the same SNPs (Supplementary Table 2), or iv) the MassARRAY platform (Sequenom, San Diego CA, USA) using phylogenetically redundant SNPs (Supplementary Table 3). The set of SNPs used in the MassARRAY typing scheme covered only six of 10 sublineages. Hence, all Lineage 4 isolates without any of the six SNPs with the mutant allele defined by MassARRAY typing (n=49) were subjected to the Luminex-assay described above. For all isolates, patient place of birth was used as country information. We obtained sublineage-classification data for 3,273 (97.2%) of a total of 3,366 isolates (Supplementary Table 5).
Spatial analysis and data presentation
For each country with Lineage 4 sublineage data available, sublineage proportions (compared to all Lineage 4 isolates from the same country) were calculated and mapped to a world map with ArcGIS ArcMap 10.0 (Esri, Redland, USA). A shapefile with country boundaries was used from DIVA-GIS, which is freely available. Categories for number of countries were defined as 0, 1-3, 4-10 and >10 countries. For individual sublineage „heat maps“, countries with less than 3 isolates were not included. For the additional maps shown in Supplementary Fig. 6, sublineage proportions were normalized by the TB prevalence in the country as estimated by WHO38, and the area of the country. Other figures were generated with the ggplot2 library in R and GraphPad Prism 6.02 (GraphPad Software, San Diego, USA). Statistical analyses were performed with R or GraphPad Prism.
SIFT-analyses of functional effects of fixed sublineage SNPs
Analysis of SNPs fixed in each of the 10 sublineages were assessed for predicted functional consequence with the „Sorting Intolerant From Tolerant“ (SIFT) in the software SIFT4G (v2.1)42 and the pre-compiled Mycobacterium tuberculosis database „GCA_000195955.2.22“. Conservation levels of SNPs in the pre-compiled database had been obtained by comparing Mycobacterium tuberculosis H37Rv proteins to all proteins in the UniRef90 database. We pooled SNPs fixed in the generalist sublineages and the specialist sublineages, respectively, and excluded the L4.1.2/Ghana sublineage, as whole genome sequences of only two, very closely related isolates were available. Gene categories were analyzed based on the classification by Tuberculist88.
Phylogenetic reconstruction and population genetic analyses
The final alignment of polymorphic positions in all strains was used to estimate phylogenies with Bayesian methods using MrBayes 3.2.589 for L4.3/LAM and L4.6.1/Uganda sublineages (Fig. 6, Supplementary Fig. 10). For the Bayesian analysis we used a gamma rate distribution estimated from our dataset and a burn-in equal to 1/10 the number of generations; after the burn-in phase every 100th tree was saved. Two parallel Markov chains were run in each of two runs. Tree length, log-likelihood score and alpha value of the gamma distribution were inspected for stationarity before termination of MrBayes. Trees were generated with standard parameters. A consensus tree was used for further analyses. Additionally, we used MEGA583 to generate Maximum Likelihood phylogenetic trees (Supplementary Figs. 9, 11, 12 and 13). We used the general time reversible (GTR) model of evolution, and 500 pseudoreplicates for bootstrapping confidence levels. Positions with gaps in more than 50% of taxa were ignored. Tree figures were generated using FigTree version 1.4.2. Pairwise SNP distances were calculated with the ape-package and the dna.dist function in R version 3.2.2, using raw counts of mutations and pairwise deletions for sites with gaps. For the comparison of pairwise number of SNP distributions overall (L4.3/LAM and L4.6.1/Uganda) and between continents for L4.3/LAM, a mean SNP distance to all isolates of the same population was calculated for each isolate, and a distribution of the mean pairwise distance plotted. Wilcoxon rank sum and Kruskall-Wallis tests were used to test for differences between continents as data were assumed to not be normally distributed. Average pairwise nucleotide diversities per site (π) were calculated as the average number of pairwise mismatches among a set of sequences divided by the total length of the interrogated sequences in base pairs (equation 4.21 in Ref.90). Confidence intervals for π were obtained by bootstrapping (1000 replicates) by re-sampling with replacement the nucleotide sites of the original alignments of polymorphic positions using the function sample in R. Lower and upper levels of confidence were obtained by calculating the 2.5th and the 97.5th quantiles of the π distribution obtained by bootstrapping. Code details are available upon request.
Antigenic diversity in human T cell epitopes
Experimentally confirmed human MTBC T cell epitope sequences were retrieved from the Immune Epitope Database on the 24th of April 2015. Only linear epitopes from the MTBC (ID: 77643) tested in human T cell assays, with no MHC restrictions were selected (1,730 epitopes). The sequence of each epitope was blasted using blastP91 against the reference strain (H37Rv) to obtain genomic coordinates. Epitopes with no coordinates in H37Rv or for which no accurate coordinates could be determined (due to multiple hits) and epitopes in repetitive regions such as PE/PPE genes, phages-related genes and transposases were excluded, rendering a final set of 1,226 epitopes. Those epitopes are distributed across 304 antigens and have some overlapping sequences. In order to proceed with the sequence analysis, alignments were obtained by concatenating all epitope sequences after excluding sequence redundancy. Alignments of non-epitope containing antigens were obtained by excluding the regions described as epitopes from each respective antigen. To assess how other regions of the genome are evolving, alignments for essential and non-essential genes were also obtained62.
Alignments of epitopes and non-epitope containing antigens, essential and nonessential genes, were used to calculate pairwise dN/dS ratios for L4.3/LAM, L4.6.1/Uganda, L4.10/PGG3 and L4.1.2/Haarlem sublineages. The dN/dS measures were calculated using all polymorphic sites within each sublineage and reflect therefore both within-sublineage substitutions and transient polymorphisms. Pairwise dN and dS values within each sublineage were calculated using the R package seqinr using the kaks function. To avoid having undetermined pairwise dN/dS values due to dN or dS being zero, a mean dN/dS was then calculated per sequenced isolate by dividing its mean pairwise dN by its mean pairwise dS with respect to all other sequenced isolates within each sublineage. The statistical differences between epitopes and non-epitope regions of antigens within each sublineage were accessed by using Wilcoxon rank sum tests with continuity correction implemented in R version 3.2.2.
Reconstruction of geographical origin of L4.3/LAM
The software RASP69 was used to reconstruct the hypothetical geographic origin of the MTBC L4.3/LAM ancestor genotype. The Bayesian phylogeny of 294 isolates (including H37Rv as outgroup) and the corresponding continent of birth of the patient were loaded as distribution. We used the S-DIVA (a parsimony based method) as well as the Bayesian Binary Method (BBM) implementation in RASP. A set of trees from MrBayes89 was used to correct for phylogenetic uncertainty in the S-DIVA analysis. Populations were defined according to country of birth of the patients and according to the United Nations definition. The isolates from Turkey, Libya, Algeria and Morocco were in the category “Europe and Mediterranean”. RASP reconstruction was done without the outgroup (H37Rv). As we observed a single strain (from Ukraine) with a distinct, basal position in the phylogeny, we also performed a sensitivity analysis by excluding that isolate for the RASP analysis. With both methods, BBM as well as S-DIVA, the changes in proportions of continents were minor. With BBM, the proportion of “Europe/Mediterranean” for the “L4.3/LAM ancestor” decreased to 98.8%, and with S-DIVA, the proportion of “Europe/Mediterranean” decreased to 99.0% when excluding this basal isolate.
Supplementary Material
Acknowledgements
We thank Sophie Lecher, Sarah Li, and Julia Zallet for technical support. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing core facility at the University of Basel. This work was supported by the Swiss National Science Foundation (grants 310030_166687 (S.G.) and 320030_153442 (M.E.), Swiss HIV Cohort Study grant 740 (L.F.)), the European Research Council (309540-EVODRTB, S.G.), TB-PAN-NET (FP7-223681, S.N.), PathoNgenTrace (FP7- 278864-2, S.N.) projects, SystemsX.ch (S.G.), the German Center for Infection Research (DZIF, S.N.), the Novartis Foundation (S.G.), and the Natural Science Foundation of China (91631301, Q.G.).
Footnotes
Accession Codes
Sequencing reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) under the study accession number PRJEB11460. Sample and run accession numbers for individual strains can be found in Supplementary Table 7.
Author Contributions
D.S., D.B., L.J., S.N. and S.G. planned the experiments. D.S., L.J., D.B., A.T., L.F., L.R., S.B., M.Ba., Q.L., T.L., Q.G., M.K., M.Bo., M.E., R.M., H.M., M.M., G.T., J.F., M.G., J.T., F.J., J.G., A.A., D.Y., E.W., W.S., M.J., W.H.B., I.B., J.B., M.S., S.V., P.Suf., A.K., R.W., L.G., B.M., S.L., H.B., B.C.J., K.T., E.S., M.B., A.G., M.F., V.N.P., K.E., I.A., P.W.N., G.R., F.G., S.Ak., F.N., L.S., N.N., A.R., M.H., D.C., G.S., S.V., D.B., P.St., R.D., V.C., O.M., S.Al., L.O., F.B., J.C., L.D., P.Sup. and I.C. contributed reagents and performed the experiments. D.S., L.J., D.B., M.C., S.N. and S.G. analysed the data. D.S., L.J., D.B., S.N. and S.G. wrote the manuscript. All authors critically reviewed the manuscript.
Competing Financial Interests Statement
The authors have no competing interests as defined by Springer Nature, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
References
- 1.Futuyma DJ, Moreno G. The Evolution of Ecological Specialization. Annual Review of Ecology and Systematics. 1988;19:207–233. [Google Scholar]
- 2.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–77. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 3.Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–12. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 4.Kirzinger MW, Stavrinides J. Host specificity determinants as a genetic continuum. Trends Microbiol. 2012;20:88–93. doi: 10.1016/j.tim.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Vouga M, Greub G. Emerging bacterial pathogens: past and beyond. Clin Microbiol Infect. 2016;22:12–21. doi: 10.1016/j.cmi.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brites D, Gagneux S. Co-evolution of Mycobacterium tuberculosis and Homo sapiens. Immunol Rev. 2015;264:6–24. doi: 10.1111/imr.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13:1456–1466. [PubMed] [Google Scholar]
- 8.Merker M, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47:242–9. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo T, et al. Southern East Asian origin and coexpansion of Mycobacterium tuberculosis Beijing family with Han Chinese. Proc Natl Acad Sci U S A. 2015;112:8136–41. doi: 10.1073/pnas.1424063112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowley D, et al. Recent and rapid emergence of W-Beijing strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis. 2008;47:1252–9. doi: 10.1086/592575. [DOI] [PubMed] [Google Scholar]
- 11.de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum--review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis. 2010;4:e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firdessa R, et al. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis, Ethiopia. Emerg Infect Dis. 2013;19:460–463. doi: 10.3201/eid1903.120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux S. Host-pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol Sci. 2012;367:850–9. doi: 10.1098/rstb.2011.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenner L, et al. HIV infection disrupts the sympatric host-pathogen relationship in human tuberculosis. PLoS Genet. 2013;9:e1003318. doi: 10.1371/journal.pgen.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–2873. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker L, Brown T, Maiden MC, Drobniewski F. Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis. 2004;10:1568–77. doi: 10.3201/eid1009.040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed MB, et al. Major Mycobacterium tuberculosis lineages associate with patient country of origin. J Clin Microbiol. 2009;47:1119–28. doi: 10.1128/JCM.02142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demay C, et al. SITVITWEB - A publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–66. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Coscolla M, Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol. 2014;26:431–44. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coscolla M, Gagneux S. Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech. 2010;7:e43–e59. doi: 10.1016/j.ddmec.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–82. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coscolla M, et al. Novel Mycobacterium tuberculosis complex isolate from a wild chimpanzee. Emerg Infect Dis. 2013;19:969–76. doi: 10.3201/eid1906.121012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abadia E, et al. Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs-based method. Infect Genet Evol. 2010;10:1066–74. doi: 10.1016/j.meegid.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Filliol I, et al. Global Phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol. 2006;188:759–72. doi: 10.1128/JB.188.2.759-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sreevatsan S, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–74. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homolka S, et al. High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS One. 2012;7:e39855. doi: 10.1371/journal.pone.0039855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coll F, et al. A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat Commun. 2014;5:4812. doi: 10.1038/ncomms5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci U S A. 2004;101:4865–70. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE. 2009;4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeboah-Manu D, et al. Genotypic diversity and drug susceptibility patterns among M. tuberculosis complex isolates from South-Western Ghana. PLoS One. 2011;6:e21906. doi: 10.1371/journal.pone.0021906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malla B, et al. First insights into the phylogenetic diversity of Mycobacterium tuberculosis in Nepal. PLoS One. 2012;7:e52297. doi: 10.1371/journal.pone.0052297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenner L, et al. Mycobacterium tuberculosis transmission in a country with low tuberculosis incidence: role of immigration and HIV infection. J Clin Microbiol. 2012;50:388–95. doi: 10.1128/JCM.05392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballif M, et al. Genetic diversity of Mycobacterium tuberculosis in Madang, Papua New Guinea. Int J Tuberc Lung Dis. 2012;16:1100–7. doi: 10.5588/ijtld.11.0779. [DOI] [PubMed] [Google Scholar]
- 34.Wampande E, et al. Long-term dominance of Mycobacterium tuberculosis Uganda family in peri-urban Kampala-Uganda is not associated with cavitary disease. BMC Infect Dis. 2013;13:484. doi: 10.1186/1471-2334-13-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley SD, et al. Diversity of Mycobacterium tuberculosis and drug resistance in different provinces of Papua New Guinea. BMC Microbiol. 2014;14:307. doi: 10.1186/s12866-014-0307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stucki D, et al. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS ONE. 2012;7:e41253. doi: 10.1371/journal.pone.0041253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouakaze C, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based single nucleotide polymorphism genotyping assay using iPLEX gold technology for identification of Mycobacterium tuberculosis complex species and lineages. J Clin Microbiol. 2011;49:3292–9. doi: 10.1128/JCM.00744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. Global tuberculosis control - surveillance, planning, financing. WHO; Geneva, Switzerland: 2015. [Google Scholar]
- 39.Moller M, de Wit E, Hoal EG. Past, present and future directions in human genetic susceptibility to tuberculosis. FEMS Immunol Med Microbiol. 2010;58:3–26. doi: 10.1111/j.1574-695X.2009.00600.x. [DOI] [PubMed] [Google Scholar]
- 40.Caws M, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asante-Poku A, et al. Mycobacterium africanum is associated with patient ethnicity in Ghana. PLoS Negl Trop Dis. 2015;9:e3370. doi: 10.1371/journal.pntd.0003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- 44.Lanzas F, Karakousis PC, Sacchettini JC, Ioerger TR. Multidrug-resistant tuberculosis in panama is driven by clonal expansion of a multidrug-resistant Mycobacterium tuberculosis strain related to the KZN extensively drug-resistant M. tuberculosis strain from South Africa. J Clin Microbiol. 2013;51:3277–85. doi: 10.1128/JCM.01122-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bryant JM, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013;13:110. doi: 10.1186/1471-2334-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryant JM, et al. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med. 2013;1:786–92. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardy JL, et al. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–9. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 48.Casali N, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86. doi: 10.1038/ng.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker TM, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2012 doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roetzer A, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013;10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–9. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark TG, et al. Elucidating emergence and transmission of multidrug-resistant tuberculosis in treatment experienced patients by whole genome sequencing. PLoS One. 2013;8:e83012. doi: 10.1371/journal.pone.0083012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jamieson FB, et al. Whole-genome sequencing of the Mycobacterium tuberculosis Manila sublineage results in less clustering and better resolution than mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) typing and spoligotyping. J Clin Microbiol. 2014;52:3795–8. doi: 10.1128/JCM.01726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Lago L, et al. Whole genome sequencing analysis of intrapatient microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J Infect Dis. 2014;209:98–108. doi: 10.1093/infdis/jit439. [DOI] [PubMed] [Google Scholar]
- 55.Guerra-Assuncao JA, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis. 2015;211:1154–63. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerra-Assuncao JA, et al. Large-scale whole genome sequencing of M. tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015;4 doi: 10.7554/eLife.05166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coscolla M, et al. M. tuberculosis T cell epitope analysis reveals paucity of antigenic variation and identifies rare variable TB antigens. Cell Host Microbe. 2015;18:538–48. doi: 10.1016/j.chom.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ernst JD, et al. Meeting report: NIH Workshop on the Tuberculosis Immune Epitope Database. Tuberculosis (Edinb) 2008;88:366–70. doi: 10.1016/j.tube.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pepperell CS, et al. The role of selection in shaping diversity of natural M. tuberculosis populations. PLoS Pathog. 2013;9:e1003543. doi: 10.1371/journal.ppat.1003543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang YJ, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esparza M, et al. PstS-1, the 38-kDa Mycobacterium tuberculosis glycoprotein, is an adhesin, which binds the macrophage mannose receptor and promotes phagocytosis. Scand J Immunol. 2015;81:46–55. doi: 10.1111/sji.12249. [DOI] [PubMed] [Google Scholar]
- 64.Bekmurzayeva A, Sypabekova M, Kanayeva D. Tuberculosis diagnosis using immunodominant, secreted antigens of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2013;93:381–8. doi: 10.1016/j.tube.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Nagai S, Wiker HG, Harboe M, Kinomoto M. Isolation and partial characterization of major protein antigens in the culture fluid of Mycobacterium tuberculosis. Infect Immun. 1991;59:372–82. doi: 10.1128/iai.59.1.372-382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juarez MD, Torres A, Espitia C. Characterization of the Mycobacterium tuberculosis region containing the mpt83 and mpt70 genes. FEMS Microbiol Lett. 2001;203:95–102. doi: 10.1111/j.1574-6968.2001.tb10826.x. [DOI] [PubMed] [Google Scholar]
- 67.Coppola M, et al. Synthetic long peptide derived from Mycobacterium tuberculosis latency antigen Rv1733c protects against tuberculosis. Clin Vaccine Immunol. 2015;22:1060–9. doi: 10.1128/CVI.00271-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Araujo LS, et al. Profile of interferon-gamma response to latency-associated and novel in vivo expressed antigens in a cohort of subjects recently exposed to Mycobacterium tuberculosis. Tuberculosis (Edinb) 2015;95:751–7. doi: 10.1016/j.tube.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 69.Yu Y, Harris AJ, Blair C, He X. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol Phylogenet Evol. 2015;87:46–9. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 70.Brudey K, et al. Mycobacterium tuberculosis complex genetic diversity : mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Comas I, et al. Population Genomics of Mycobacterium tuberculosis in Ethiopia contradicts the Virgin Soil hypothesis for human tuberculosis in Sub-Saharan Africa. Curr Biol. 2015;25:3260–6. doi: 10.1016/j.cub.2015.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bos KI, et al. Pre-Columbian mycobacterial genomes reveal seals as a source of New World human tuberculosis. Nature. 2014;514:494–7. doi: 10.1038/nature13591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kay GL, et al. Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat Commun. 2015;6:6717. doi: 10.1038/ncomms7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lazzarini LC, et al. Discovery of a novel Mycobacterium tuberculosis lineage that is a major cause of tuberculosis in Rio de Janeiro, Brazil. J Clin Microbiol. 2007;45:3891–902. doi: 10.1128/JCM.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perdigao J, et al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics. 2014;15:991. doi: 10.1186/1471-2164-15-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mokrousov I, et al. Latin-American-Mediterranean lineage of Mycobacterium tuberculosis: Human traces across pathogen's phylogeography. Mol Phylogenet Evol. 2016;99:133–43. doi: 10.1016/j.ympev.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 77. http://en.wikipedia.org/wiki/European_diaspora.
- 78.Bates JH, Stead WW. The history of tuberculosis as a global epidemic. Med Clin North Am. 1993;77:1205–17. doi: 10.1016/s0025-7125(16)30188-2. [DOI] [PubMed] [Google Scholar]
- 79.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- 80.Steiner A, Stucki D, Coscolla M, Borrell S, Gagneux S. KvarQ: targeted and direct variant calling from fastq reads of bacterial genomes. BMC Genomics. 2014;15:881. doi: 10.1186/1471-2164-15-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 85.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 86.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maddison W, Maddison D. Mesquite: a modular system for evolutionary analysis. 2001 [Google Scholar]
- 88.Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList--10 years after. Tuberculosis (Edinb) 2011;91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 89.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 90.Hartl D, Clarck AG. Principles of population genetics. Sinauer; 2007. [Google Scholar]
- 91.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Sequencing reads have been submitted to the EMBL-EBI European Nucleotide Archive (ENA) Sequence Read Archive (SRA) under the study accession number PRJEB11460.