Abstract
Three-dimensional (3D) tumor models are gaining traction in the research community given their capacity to mimic aspects of the tumor microenvironment absent in monolayer systems. In particular, the ability to spatiotemporally control cell placement within ex vivo 3D systems has enabled the study of tumor-stroma interactions. Furthermore, by regulating biomechanical stimuli, one can reveal how biophysical cues affect stromal cell phenotype and how their phenotype impacts tumor drug sensitivity. Both tumor architecture and shear force have profound effects on Ewing sarcoma (ES) cell behavior and are known to elicit ligand-mediated activation of the insulin-like growth factor-1 receptor (IGF-1R), thereby mediating resistance of ES cells to IGF-1R inhibitors. Here, we demonstrate that these same biophysical cues—modeled by coculturing ES cells and mesenchymal stem cells (MSCs) in 3D scaffolds within a flow perfusion bioreactor—activate interleukin-6 and transcription factor Stat3. Critically, an active Stat3 pathway drastically alters the equilibrium of IGF-1R-targeted ligands (IGF-1) and antagonists (IGFBP-3) secreted by MSCs. To elucidate how this might promote ES tumor growth under physiological shear-stress conditions, ES cells and MSCs were co-cultured by using a flow perfusion bioreactor at varying ratios that simulate a wide range of native MSC abundance. Our results indicate that ES cells and MSCs stimulate each other's growth. Co-targeting IGF-1R and Stat3 enhanced antineoplastic activity over monotherapy treatment. Although this discovery requires prospective clinical validation in patients, it reveals the power of employing a more physiological tissue-engineered 3D tumor model to elucidate how tumor cells co-opt stromal cells to acquire drug resistance.
Keywords: : tissue engineering, tumor model, Ewing sarcoma, tumor stroma, flow perfusion bioreactor, drug resistance
Introduction
An efficient drug discovery pipeline requires a robust preclinical screening program that accurately predicts each drug's antineoplastic clinical activity. The most commonly used method to screen drug candidates in vitro relies on two-dimensional (2D) culture systems (e.g., petri dishes or tissue culture flasks) that are quick and cost-effective but unable to recapitulate the complexity of the tumor microenvironment.1 As a result, the drug discovery process is stymied by preclinical screens that, all too often, fail to predict clinical activity.1,2
To address this issue, three-dimensional (3D) tumor models have been developed to emulate specific aspects of the tumor microenvironment known to contribute to cancer progression, such as stromal cells, extracellular matrix, and biophysical stimuli.3–6 Compared with 2D culture systems, 3D tumor models can help retain a tumor-like phenotype and preserve native gene expression,7 tumor growth,8 and drug resistance.8 In particular, the collection of non-cancerous cells present in the native tumor, commonly referred to as tumor stroma, is considered a hallmark of cancer biology due to its fundamental role in cancer progression.9
For instance, endothelial cells are recruited to the tumor site on release of proangiogenic signals, whereas cancer-associated fibroblasts may develop from the local parenchyma or from tumor-induced differentiation of circulating mesenchymal cell precursors.3,10,11 These observations, and many others, demonstrate that non-malignant elements of the tumor microenvironment play a central role in disease progression, and, therefore, may be considered additional targets for therapeutic interventions.3,5
Along this line of research, our laboratory seeks to better understand how biomechanical stimuli and cell–cell interactions contribute to the phenotype and drug sensitivity of bone tumors—more specifically, Ewing sarcoma (ES), an often-fatal bone tumor with a predilection for adolescents and young adults.12,13 We previously reported that culturing ES cells on 3D electrospun poly(ɛ-caprolactone) (PCL) scaffolds resulted in a more in vivo-like cell phenotype compared with 2D cultures with respect to the insulin-like growth factor-1 receptor (IGF-1R) pathway, a key mediator of tumor progression and drug resistance.8
Subsequently, we demonstrated that the implementation of flow perfusion bioreactors in this 3D model provided cells with mechanical stimuli usually experienced within the bone microenvironment, resulting in enhanced autocrine IGF-1 secretion and altered sensitivity to therapeutic antibodies against IGF-1R.6
Though integration of shear forces and tissue-engineered scaffolds clearly improves the fidelity of our model with respect to IGF-1R signaling, this model does not include other elements within the ES niche that can affect the IGF-1 signaling pathway, such as cancer-associated fibroblasts, vasculature, and connective tissue.3,11,14 In particular, cells of mesodermal origin—such as mesenchymal stem cells (MSCs)—are known to upregulate IGF-1 expression when mechanically stimulated.15,16 MSCs also secrete high levels of interleukin-6 (IL-6), a cytokine commonly found in the ES microenvironment that activates the signal transducer and activator of transcription-3 (Stat3).17–19
IL-6/Stat3 signaling pathway has been shown to protect ES cells from apoptosis and to promote cell dissemination via activation of the Stat3 signaling pathway.17,18 This interplay between IGF-1/IGF-1R and IL-6/Stat3 pathways has been further emphasized by other in vivo investigations, where enhanced Stat3 signaling was observed in ES tumors exhibiting resistance to the IGF-1R blockade.20,21 For these reasons, culturing ES cells and MSCs under flow perfusion might improve our understanding of the ES-MSC crosstalk occurring under mechanical stimulation, with emphasis on the role of IGF-1/IGF-1R and IL-6/Stat3 pathways on ES malignancy and stroma-induced drug resistance.
The aim of the current work was to investigate the effects of MSC presence and flow perfusion on 3D cultures of ES. We hypothesized that MSCs would bolster cell proliferation and affect the phenotype and drug sensitivity of ES cells, via the IGF-1/IGF-1R pathway, but only when physiological levels of shear force were applied in a flow perfusion bioreactor. Further, we postulated that drug response would depend on the ES:MSC ratio selected, which was used as a surrogate to describe the wide range of tumor/stroma ratios occurring among different patients as well as within a single patient depending on ES stage and progression.
Finally, we hypothesized that MSC-driven IL-6 secretion would influence drug response against the IGF-1R blockade in ES cells, thus providing an explanation for the role of mesodermal stroma in ES-acquired drug resistance. Evaluation of stroma-mediated activation of IGF-1R and Stat3 pathway has broad implications not only for ES but also for all those malignancies, such as lung cancer, glioma, and osteosarcoma, in which high levels of IL-6 affect tumor progression.22–24
Materials and Methods
Experimental design
At the beginning of the experiment, ES cells and MSCs were lifted from culture flasks, mixed according to five different ES:MSC ratios (1:0, 9:1, 1:1, 1:9, and 0:1, respectively), and seeded onto electrospun PCL scaffolds. The total number of cells/scaffold was kept constant among groups (250,000 cells/scaffold) and was based on previous studies.8 Cell-seeded scaffolds were cultured in static conditions (S) or in a flow perfusion bioreactor (B) and exposed to either complete alpha minimal essential medium (αMEM) or complete αMEM medium containing a monoclonal antibody against IGF-1R at a concentration of 100 μg/mL (IGR-1Ri: IGF-1R inhibition; MK-0646; Merck & Co, North Wales, PA). Samples were collected after 10 days, analyzed for cell proliferation, protein expression, cytokine production, and drug response, and imaged via immunofluorescence microscopy.
In an additional study, scaffolds were cultured under flow perfusion conditions and evaluated for their sensitivity against a Stat3 inhibitor (Stat3i). Cells were exposed to complete medium as a control, a single agent IGF-1Ri, a single agent Stat3i, and a combination of both the IGF-1Ri and Stat3i. For the Stat3i, we used small-molecule WP1722 at a concentration of 1.1 μM.21 Samples were collected after 10 days and analyzed for cell proliferation and drug response. In all the drug-treated groups, cells were first cultured for 3 days in complete αMEM medium followed by 7 days of exposure to medium containing IGF-1Ri and/or Stat3i, similar to previous studies.6,8
Scaffold preparation and characterization
Nonwoven poly(ɛ-caprolactone) (PCL; Sigma-Aldrich, St. Louis, MO) mats with an average fiber diameter of 10 μm (10.6 ± 1.5 μm, n = 90) were fabricated as in previous studies.6,8 In brief, PCL was dissolved in a 5:1 chloroform:methanol mixture at a 16% w/w concentration. The resulting solution was pumped at 21 mL/h through a blunt 18G needle in a horizontal electrospinning setup.6,8 The gauge was exposed to 25 kV, and a grounded collecting plate was positioned normally to the gauge at a distance of 40 cm. Mats were electrospun to a thickness of ∼1.00 ± 0.2 mm and imaged by scanning electron microscopy (FEI Quanta 400 Environmental, FEI, Hillsboro, OR) to measure fiber diameter. Scaffolds of 8 mm diameter were die-punched by using dermal biopsy punches and, subsequently, press-fitted into custom-made scaffold holders, the design of which is shown in Supplementary Figure S1 (Supplementary Data are available online at www.liebertpub.com/tea). Ethylene oxide was then used to sterilize both the scaffolds and the scaffold holders (Anderson Sterilizers, Haw River, NC), followed by soaking in a progressive ethanol series (100% to 25% v/v), rinsing three times in phosphate-buffered saline (Gibco, Carlsbad, CA), and, lastly, incubation overnight in complete αMEM medium (Corning Cellgro, Manassas, VA) supplemented with 20% fetal bovine serum (FBS) and antibiotics (100 μg/mL streptomycin and 100 IU/mL penicillin; Gibco, Carlsbad, CA).
Cell culture and bioreactor setup
The human ES cell line TC71 stably expressing green fluorescent protein reporter was available from the repository of sarcoma cell lines at our institution (MDACC). Cell identity was confirmed via short-tandem repeats fingerprinting according to the manufacturer's instructions (AmpFISTR Identifiler kit; Applied Biosystems, Carlsbad, CA) and was compared with the Integrated Molecular Authentication database (CLIMA) version 0.1.200808 (http://bioinformatics.istge.it/clima/) and with available ATCC fingerprints (ATCC.org).25,26 ES cells were cultured in complete RPMI 1640 medium (Corning Cellgro, Manassas, VA) supplemented with 10% FBS (Gemini Bio-Products, West Sacramento, CA) and antibiotics. Primary bone marrow-derived human MSCs from healthy donors were provided by Dr. Ian McNiece (Stem Cell Department; The University of Texas MD Anderson Cancer Center) by following an approved protocol at The University of Texas MD Anderson Cancer Center, genetic testing, and culture in αMEM medium (Corning Cellgro, Manassas, VA) supplemented with 20% FBS and antibiotics (100 μg/mL streptomycin and 100 IU/mL penicillin; Gibco, Carlsbad, CA). At the beginning of the experiment, both cell types were lifted with 0.05% trypsin-EDTA (Gibco, Carlsbad, CA), counted by using a hemocytometer, and mixed according to five different ES:MSC ratios (1:0, 9:1, 1:1, 1:9, and 0:1, respectively). Each scaffold was seeded with 250,000 cells, according to the experimental design previously shown. Scaffolds were incubated overnight to facilitate cell adhesion, on which three scaffolds (n = 3) were harvested to evaluate cell seeding efficiency (Supplementary Fig. S2). Scaffolds under static conditions (S) were then moved to 24-well ultra-low attachment plates with 2.5 mL of complete αMEM medium. Scaffolds cultured under flow perfusion condition were left in the scaffold holder (Supplementary Fig. S1) and transferred into a flow perfusion bioreactor (B), which was previously described.27 Each bioreactor unit contained six scaffolds sustained with 50 mL of complete αMEM medium. In all experimental groups, samples were maintained in a heat-jacketed incubator at 37°C and 5% CO2 (HeraCell 150i; ThermoScientific, Waltham, MA) for the duration of the study, and half of the medium was replaced daily.
DNA quantification and drug testing
Samples for DNA quantification (n = 5) were immersed in distilled water, subjected to three cycles of freeze/thawing (10 min in liquid N2/10 min in 37°C water bath), and finally sonicated for 10 min to guarantee complete extraction of the DNA from the scaffold. The concentration of double-stranded DNA was quantified with the Quant-iT PicoGreen dsDNA assay kit (Invitrogen, Eugene, OR). Following the manufacturer's instructions, the cell lysate, the dye solution, and the buffer were mixed in a flat-bottom 96-well plate in triplicate, and the resulting fluorescence was measured (FLx800 Fluorescence Microplate Reader; BioTek Instruments, Winooski, VT). DNA concentration was determined by using a λ-DNA standard curve. The same protocol was also used to quantify cell response to IGF-1Ri and/or Stat3i, where the DNA amount indicated variations in the total number of cells present in the scaffold.
Western blotting
After 10 days of culture, samples were collected (n = 3), and proteins were extracted via incubation on ice by using a lysis buffer (1% v/v Triton X-100, 50 mM Hepes, pH 7.4, 150 mM NaCl, 100 mM NaF, 1.5 mM MgCl2, 1 mM EGTA, 1 mM Na3VO4, 10 mM Na4P2O7, 10% v/v glycerol) containing a fresh mixture of protease and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN). The protein concentration was measured by using a Micro BCA protein assay kit (Thermo Fisher, Waltham, MA), and protein lysates were stored at −80°C until they were analyzed. Proteins were resolved by SDS-polyacrylamide gel electrophoresis and transferred to PVDF membranes. The membranes were blocked by using 5% w/v milk and hybridized with different primary antibodies against markers of the IGF-1R/mammalian target of rapamycin (mTOR) and of the Stat3 signaling pathways: total insulin-like growth factor-1 receptor β (IGF-1Rβ), phosphorylated IGF-1R (pIGF-1R), mTOR, Signal transducer and activator of transcription-3 (Stat3), and phosphorylated Stat3 (pStat3).17,18,21 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as the loading control. Signals were captured by using horseradish peroxidase-conjugated secondary anti-rabbit (or anti-mouse) IgG antibodies and visualized by using SuperSignal West Dura chemiluminescent substrate (Thermo Fisher, Waltham, MA). All primary and secondary antibodies were purchased from Cell Signaling Technology (Danvers, MA) except for pIGF-1R, which was provided by Abcam (Cambridge, MA). The levels of immunoreactive proteins were determined by using chemiluminescent Hyperfilm ECL and quantified by using an ImageQuant TL computing densitometer (GE Healthcare, Piscataway, NJ).
Immunofluorescence microscopy
Constructs at day 10 were fixed in 10% neutral buffered formalin (Fisher Scientific, Pittsburgh, PA) overnight at room temperature, followed by dehydration in an ethanol gradient series (70% to 100%). Samples were then embedded in Histoprep freezing medium (Fisher Scientific, Pittsburgh, PA) and sectioned with a cryostat (Leica CM1850UV; Leica Biosystems Nussloch GmbH, Germany). Cells were stained for nuclei (DAPI) and mouse antibodies against pStat3 (Cell Signaling Technology, Danvers, MA) and IGF-1Rα (Santa Cruz Biotechnology, Santa Cruz, CA). Alexa Fluor 680 antimouse (Thermo Fisher, Waltham, MA) was used as secondary antibody. Samples were then imaged with a Nikon A1-RSI confocal microscope, and images were analyzed by using NIS Elements C imaging software (Nikon Instruments, Melville, NY).
Enzyme-linked immunosorbent assay
Conditioned medium from each group of cells was collected after 10 days to quantify the concentration of IGF-1, IL-6, insulin-like growth factor binding protein-3 (IGFBP-3), stromal cell-derived factor-1 (SDF-1), and tumor necrosis factor-α (TNF-α) by using an ELISA kit (DuoSet; R&D Systems, Minneapolis, MN). Due to different volumes of medium and the different number of scaffolds present in static and flow perfusion bioreactor conditions, comparisons between groups were made considering the amount of soluble ligand/scaffold as in previous studies.6,21 For each ligand, the optical density was measured in triplicate with a microplate reader (DTX880; Beckman Coulter), and compared with a standard curve provided in the ELISA kit, according to the vendor's protocol.
Statistical analysis
Where applicable, data are expressed as mean ± standard deviation. Statistical analysis was performed by using a two-factor analysis of variance (ANOVA) test followed by Tukey's Honestly Significant Difference post hoc test. For all analyses, differences were considered significant where p < 0.05.
Results
MSCs in co-culture with ES cells and under flow perfusion altered cellular growth and abrogated IGF-1Ri efficacy
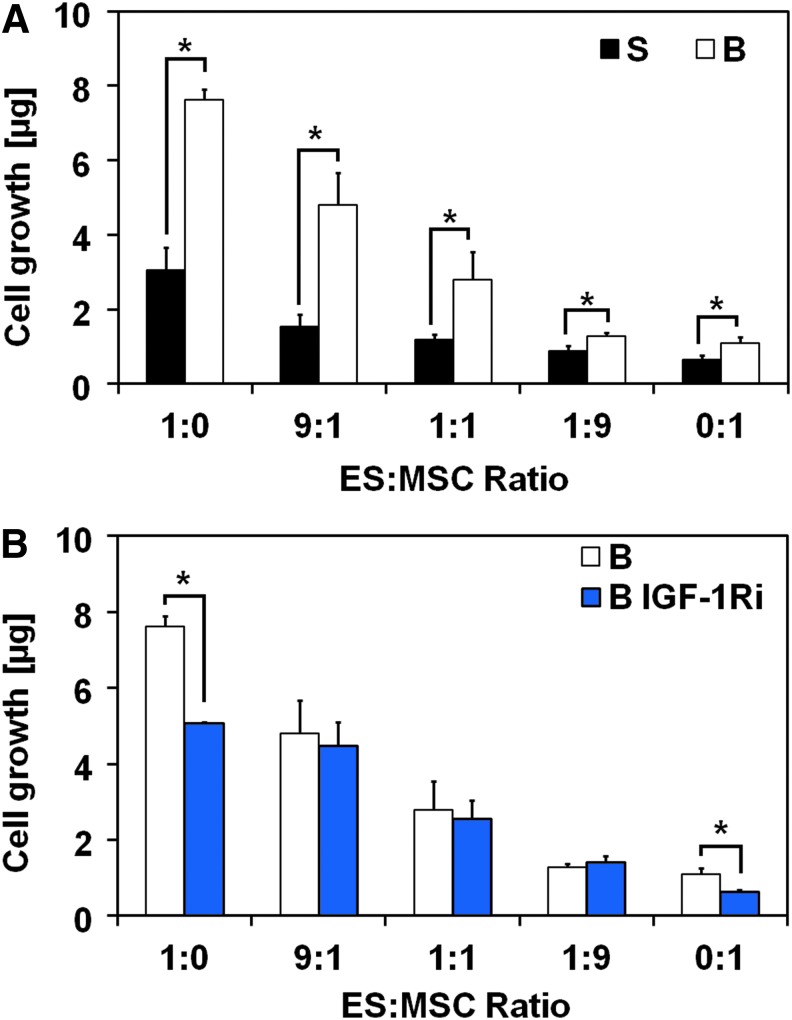
We initially determined the effect of physiological levels of shear force, regulated by controlling the flow perfusion rate, on cell proliferation by using DNA quantification. Flow perfusion conditions led to significantly faster proliferation rates for all co-culture ratios investigated when compared with static conditions (Fig. 1A). The transition from static to flow perfusion conditions led to a threefold increase in cell content in homotypic ES cultures compared with a smaller, but still statistically significant, 1.5-fold increase in homotypic MSC cultures (ES:MSC ratios of 1:0 and 0:1, respectively). This reflects the slower growth kinetics of MSCs compared with ES. Cell content within the ES:MSC co-culture groups decreased with the ES:MSC ratio in both static and flow perfusion conditions. Scanning electron microscopy images show that cells formed aggregates on the scaffold, suggesting that ES cells and MSCs were not segregated in the co-culture groups (Supplementary Fig. S3).
FIG. 1.
MSCs in coculture with ES cells and under flow perfusion altered cellular growth and abrogated IGF-1Ri efficacy. (A) DNA content after 10 days of culture for the five different ES:MSC ratios examined (1:0, 9:1, 1:1, 1:9, and 0:1) cultured in static or flow perfusion conditions (S or B, respectively). (B) Drug response to the IGF-1Ri for the five different ES:MSC ratios (1:0, 9:1, 1:1, 1:9, and 0:1) cultured under flow perfusion conditions and untreated (B) or treated with the IGF-1Ri (B IGF-1Ri). In both panels, error bars represent the standard deviation for n = 5 samples and *p < 0.05. ES, Ewing sarcoma; IGF-1R, insulin-like growth factor-1 receptor; IGF-1Ri, IGF-1R inhibitor; MSCs, mesenchymal stem cells.
Cells exposed to an IGF-1R inhibitor (IGF-1Ri) were cultured for 3 days in complete medium, followed by 7 days of IGF-1Ri-containing medium. In isolation, homotypic flow perfusion culture of ES cells or MSCs (ES:MSC ratios of 1:0 and 0:1, respectively) responded to the IGF-1R blockade with a significant decrease in cell content in IGF-1Ri-treated groups compared with control groups (Fig. 1B). Under the same flow perfusion conditions, this effect dissipated with ES:MSC co-culture, and cells acquired resistance to the IGF-1Ri regardless of the ES:MSC ratio selected. In agreement with our previous investigations,6 cells cultured under static conditions were insensitive to the IGF-1Ri action (Supplementary Fig. S4). In addition, neither flow perfusion nor drug testing appeared to have a detrimental effect on cell morphology (Supplementary Fig. S3).
ES:MSC co-cultures present different regulation of IGF-1R signaling pathway
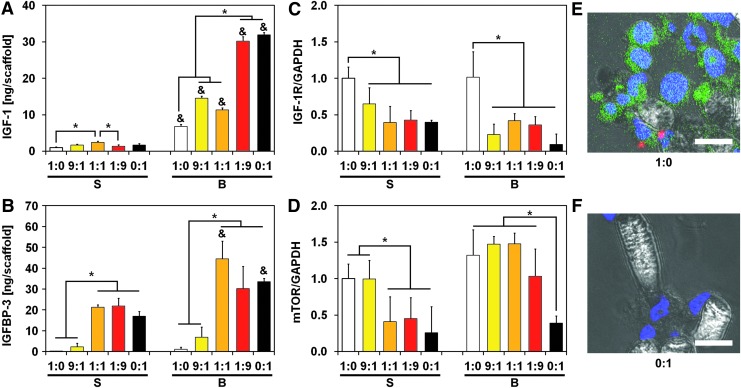
To determine how MSCs abrogated IGF-1Ri action under flow perfusion conditions, we profiled cells for biomarkers and cytokines involved in the IGF-1/IGF-1R signaling cascade. Flow-derived shear stress induced IGF-1 secretion in all experimental groups compared with static conditions (Fig. 2A). Interestingly, flow-mediated IGF-1 upregulation was more robust in MSCs than in ES cells, with a 20-fold increase in IGF-1 levels compared with a smaller 6-fold increase, respectively (Fig. 2A). Consistent with these observations, IGF-1 levels in co-culture groups amplified with increasing MSC fraction. A similar trend was observed for IGFBP-3, a soluble factor that can complex with IGF-1 and prevents its binding with IGF-1R. IGFBP-3 was primarily expressed by MSCs and stimulated under flow perfusion conditions to levels similar to those of IGF-1 (Fig. 2B). Conversely, IGFBP-3 expression levels in homotypic ES cultures were an order of magnitude lower than respective IGF-1 levels and were minimally affected by flow perfusion. Similar to IGF-1, IGFBP-3 levels amplified with a decreasing ES:MSC ratio for all experimental conditions. To account for any adsorption of IGF-1 on the constructs, ELISA analysis of cell-seeded scaffolds showed negligible amounts of IGF-1 within the constructs compared with soluble IGF-1 (Supplementary Fig. S5A).
FIG. 2.
ES:MSC cocultures present different regulation of IGF-1R signaling pathway. Protein expression within the IGF-1/IGF-1R pathway for the five different ES:MSC ratios examined (1:0, 9:1, 1:1, 1:9, and 0:1) cultured in static or flow perfusion conditions (S or B, respectively). Quantification of IGF-1 (A) and IGFBP-3 (B) ligands was evaluated via ELISA, whereas biomarkers IGF-1R (C) and mTOR (D) were analyzed via western blotting. In (A, B), ligand amount is normalized per scaffold, whereas in (C, D), signal density is normalized to GAPDH content. The error bars represent the standard deviation for n = 3 samples, *statistical difference among ES:MSC ratios and &statistical difference between static and flow perfusion conditions within each ES:MSC ratio (&*p < 0.05). Representative staining for ES:MSC coculture groups 1:0 (E) and 0:1 (F) under flow perfusion conditions. Cells were stained for nuclei (blue) and for IGF-1R expression (red). ES cells were also transfected with the GFP reporter (green), and scale bar represents 15 μm in all images. GFP, green fluorescent protein; IGFBP-3, insulin-like growth factor binding protein-3; mTOR, mammalian target of rapamycin.
To elucidate how these ligands influenced cell phenotype and drug response under flow perfusion conditions, we assessed IGF-1R-related oncoproteins of proven importance in ES survival by western blotting (see Supplementary Fig. S6 for further details). ES cells express higher levels of IGF-1R and mTOR than MSCs, and these patterns were unchanged by the use of flow perfusion conditions (Fig. 2C, D). Although IGF-1R expression increased in co-culture groups that had higher ES:MSC ratios, the levels of phosphorylated/activated IGF-1R (pIGF-1R) did not depend on the ES:MSC ratio investigated (Supplementary Fig. S7A). These findings were confirmed by immunofluorescent microscopy, where positive staining for IGF-1R qualitatively correlated with a higher ES:MSC ratio (Fig. 2E, F). Expression of mTOR, a downstream molecule of IGF-1R, mirrors the trends observed for IGF-1R (Fig. 2D). Specifically, mTOR expression levels closely matched ES:MSC ratios, whereas homotypic MSC cultures displayed the lowest levels for this protein (Fig. 2D).
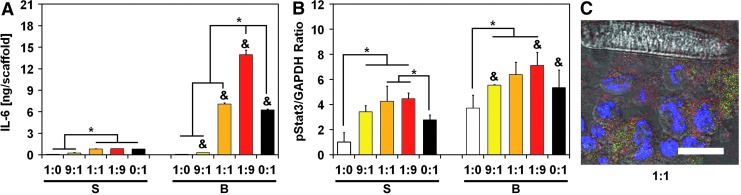
IL-6 secretion is promoted in ES:MSC co-cultures and triggers Stat3 pathway activation
Studies have shown that IL-6 is significantly expressed in the ES microenvironment, and it is primarily secreted by local stroma.17,18 Considering that IL-6/Stat3 signaling is a ubiquitous mechanism that cancer cells use to circumvent drug targeting against specific kinases, such as IGF-1R, we also profiled cells for biomarkers and cytokines along the IL-6/Stat3 pathway (see Supplementary Figs. S7 and S8 for further details). An ELISA assay indicated that flow perfusion promoted IL-6 secretion for all ES:MSC ratios except for homotypic ES cultures, which displayed the lowest levels for this ligand (Fig. 3A). IL-6 secretion was maximized in ES:MSC co-culture groups compared with either ES or MSC homotypic cultures. Strikingly, as few as 10% ES cells in the co-culture (ES:MSC ratio of 1:9) induced a 2.2-fold increase in IL-6 level compared with MSC homotypic cultures (Fig. 3A). Similar to IGF-1, a negligible amount of IL-6 was still present within the scaffold (Supplementary Fig. S5B).
FIG. 3.
IL-6 secretion is promoted in ES:MSC cocultures and triggers Stat3 pathway activation. Protein expression within the IL-6/Stat3 pathway for the five different ES:MSC ratios examined (1:0, 9:1, 1:1, 1:9, and 0:1) cultured in static or flow perfusion conditions (S or B, respectively). (A) Quantification of IL-6 ligand was evaluated via ELISA, whereas (B) pStat3 was analyzed via western blotting. IL-6 amount is normalized per scaffold, whereas pStat3 signal density is normalized to GAPDH content. The error bars represent the standard deviation for n = 3 samples, *statistical difference among ES:MSC ratios and &statistical difference between static and flow perfusion conditions within each ES:MSC ratio (&*p < 0.05). (C) Representative staining for ES:MSC coculture group 1:1 under flow perfusion conditions. Cells were stained for nuclei (blue) and for pStat3 expression (red). ES cells were also transfected with the GFP reporter (green), and scale bar represents 15 μm in all images. IL-6, interleukin-6.
Stat3 is a key target of IL-6, and we performed western blotting to analyze this transcription factor. Although total Stat3 expression remained constant among all the experimental groups according to western blotting analyses (Supplementary Figs. S7 and S8), the active form of Stat3 (pStat3) exhibits a trend that mirrors that of IL-6 (Fig. 3B). This phenomenon occurred in both static and flow perfusion conditions, where a bell-shaped curve is observed with a maximum expression of pStat3 for the 1:1 and the 1:9 ES:MSC co-culture groups (Fig. 3B). Immunofluorescent staining for pStat3 supports these data (Fig. 3C and Supplementary Fig. S9).
Although bone marrow-derived MSCs predominantly secrete IL-6, we assessed conditioned media for other cytokines that might have affected cell phenotype and drug response. Among them, interleukin-4 (IL-4), interleukin-10 (IL-10), and transforming growth factor-β were undetectable (data not shown). TNF-α and stromal cell-derived factor-1 (SDF-1) were present in small amounts (<0.5 ng/scaffold) equivalent to basal levels present in cell-free complete medium (Supplementary Fig. S5).
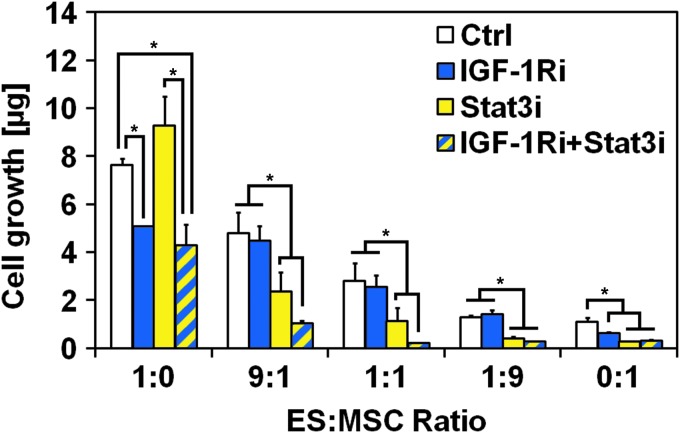
Resistance to IGF-1R blockade in ES:MSC co-cultures under flow perfusion is overcome by dual IGF-1R/Stat3 inhibition
To determine whether IL-6/Stat3 activation contributed to IGF1-Ri resistance, we assessed whether WP1722 (a Stat3 inhibitor) synergized with IGF-1Ri in ES:MSC co-cultures maintained under flow perfusion. Cells were cultured in complete medium for 3 days and then exposed for 7 days to single agent IGF-1Ri, single agent Stat3i, or an IGF-1Ri/Stat3i combination. Homotypic ES cultures were insensitive to Stat3i action, as evidenced by the similar cell content between control group and single agent Stat3i and between groups IGF-1Ri and IGF-1Ri+Stat3i (Fig. 4). Conversely, MSC cultures responded to Stat3i, which showed a greater inhibitory effect than IGF-1Ri. The combination of the two drugs had no additive effect on MSC growth inhibition, as witnessed by the comparable cell content in homotypic MSC cultures treated with either Stat3i or IGF-1Ri+Stat3i groups.
FIG. 4.
Resistance to IGF-1R blockade in ES:MSC cocultures under flow perfusion is overcome by dual IGF-1R/Stat3 inhibition. DNA content after 10 days of culture for the five different ES:MSC ratios examined (1:0, 9:1, 1:1, 1:9, and 0:1) cultured under flow perfusion conditions in complete medium (Ctrl), or treated with IGF-1R inhibitor (IGF-1Ri), Stat3 inhibitor (Stat3i), and a combination of both IGF-1R and Stat3 inhibitor (IGF-1Ri+Stat3i). Error bars represent the standard deviation for n = 5 samples and *p < 0.05.
Despite the close homology between ES and MSCs, which share a mesenchymal lineage, our data provide compelling evidence of their unique sensitivity to IGF-1R- and Stat3-targeted therapies. These cell type-specific differences in drug sensitivity help to explain why the overall effects of drug treatment observed in ES:MSC co-cultures can be diametrically opposed to the effects observed in homotypic ES cultures (Fig. 4). Cell growth was further inhibited by a combination of IGF-1Ri and Stat3i (group IGF-1Ri+Stat3i), thus indicating a concerted effect of these two drugs in inhibiting cell proliferation in ES:MSC co-cultures (Fig. 4).
Discussion
The challenges of accurately assessing tumor biology and evaluating drug-targeted therapies for cancer treatment rely on accurate modeling of tumor physiology. However, it is recognized that this requirement is poorly addressed and validated by conventional screening systems such as 2D monolayer cultures, which fail to recapitulate the basic 3D tumor architecture, or by xenograft animal models that, by definition, lack human proteins.1,3
The dearth of accurate preclinical models bolstered the development of 3D tissue-engineered models, which provide greater experimental control over culture conditions and microenvironmental cues.4–6 Toward that end, our lab has previously presented a tissue-engineered ES tumor model that makes use of 3D electrospun PCL scaffolds in concert with a flow perfusion bioreactor.6,8 Although the scaffold mimics important microarchitectural cues, the use of flow perfusion provides essential biomechanical stimuli that exist in the native bone microenvironment. ES cells responded to flow-derived shear stress by increasing IGF-1 secretion and exhibited a shear stress-dependent drug response against IGF-1R, a central player in ES progression and the object of numerous preclinical trials.6
In addition to shear stress-mediated effects, the IGF-1R cascade and ES phenotype can be impacted by local stromal cells.3,17,18 In the bone tumor microenvironment, stroma is primarily composed of cells embedded with the bone marrow (e.g., MSCs), except for later stages in which vasculature is essential for tumor extravasation and metastasis.10 Because stromal composition evolves spatially and temporally as ES tumors progress, we investigated five different ES:MSC ratios (Fig. 1) that model a wide range of tumor presentations, from a stroma-rich tumor in its early development (low ES:MSC ratio) to more advanced-stage ES nodules enriched in homogenous malignant cells (i.e., high ES:MSC ratio). Our tissue-engineered model lends itself to this endeavor and highlights likely MSC-dependent responses that cannot reliably be assessed in situ in conventional animal models. The use of flow perfusion promoted cell proliferation to a greater extent in groups with higher ES:MSC ratios due to the faster growth kinetics of ES cells compared with MSCs. At the same time, cell content decreased in an ES:MSC ratio-dependent fashion in both static and flow perfusion conditions, demonstrating that the proliferation of ES cells was unaffected by the presence of MSCs, and vice versa.
The abundance of stromal cells originating from the mesoderm (e.g., MSCs, fibroblasts, and osteoblasts) in a mechanically stimulated environment such as the ES tumor niche is particularly relevant, as mesoderm-derived cells upregulate IGF-1 secretion when exposed to biomechanical forces.15,16 This mechanism is mirrored in our model, where MSCs displayed a remarkable upregulation of IGF-1 ligand when cultured under flow perfusion (Fig. 2). Enhanced IGF-1 secretion was followed by an upregulation of IGFBP-3 secretion, a compensatory measure commonly observed in physiological conditions.28 Yet, ES cells showed an IGF-1/IGFBP-3 ratio drastically skewed toward IGF-1, highlighting how this balance is deregulated in ES cells as part of their malignant transformation.28
As IGF-1R activation is ligand dependent, we expected that the high IGF-1 state induced by flow perfusion in all culture groups would sensitize ES cells to IGF-1R blockade (IGF-1Ri). Strikingly, the introduction of as few as 10% MSCs in the system (i.e., ES:MSC group 9:1) was sufficient to induce resistance to IGF-1Ri, despite retained sensitivity by ES cells and MSCs in homotypic culture (Fig. 1). The distinctive response to IGF-1Ri among the experimental groups cannot be ascribed, however, to the different regulation of IGF-1R signaling in co-culture groups compared with homotypic cultures, as, for example, homotypic MSC cultures and the ES:MSC co-culture group 1:9 respond differently to IGF-1Ri despite the analogous regulation of IGF-1R signaling (Fig. 2). Accordingly, resistance to the IGF-1R blockade emerges through mechanisms collateral to the IGF-1/IGF-1R pathways that may be considered in the design of new therapeutic regimens for ES patients.
Among the possible mechanisms of resistance that stem from heterotypic ES:MSC interactions, we explored Stat3-mediated signaling, which is critically important in ES cell survival and whose activation is mediated primarily by the high levels of IL-6 present in the native ES microenvironment.17,18 Since MSCs retained a high degree of IL-6 production in our 3D model (Fig. 3), they appear to adequately mimic the IL-6 mediated ES-stroma crosstalk present in the native bone tumor niche. Interestingly, IL-6 secretion was further promoted under flow perfusion conditions, which supports the notion that integrin-mediated mechanotransduction can promote IL-6 secretion via kappa beta kinase/nuclear factor-kB (IKK/NF-kB) activation.29,30 Even more interesting was the super-additive release of IL-6 observed in ES:MSC co-culture groups (Fig. 3). Tumor-driven exploitation of the surrounding microenvironment has been observed already in previous work, where glioma cells coerced MSCs into secreting significantly higher amounts of IL-6.24 Our tissue-engineered model captured this aspect and, therefore, can be leveraged to assess in vitro how tumor exploitation of surrounding stroma occurs in vivo.
In our 3D tissue-engineered model, enhanced activation of Stat3 was a direct consequence of the higher levels of circulating IL-6 measured in ES:MSC co-culture groups, and this mechanism emerged not coincidentally in the same groups that showed resistance to IGF-1R blockade (Figs. 1 and 3). These results mirror our previous findings in a xenograft model, where acquired drug resistance to IGF-1Ri in ES tumors correlated with enhanced Stat3 signaling.21,31 Collectively, these data provide a compelling argument for dual targeting of IGF-1R/Stat3, a strategy already established in colorectal cancer.32 Accordingly, resistance to IGF-1Ri was overcome in ES:MSC co-cultures in the presence of Stat3i, which, on the contrary, had no efficacy in homotypic ES cultures (Figs. 4 and 5).
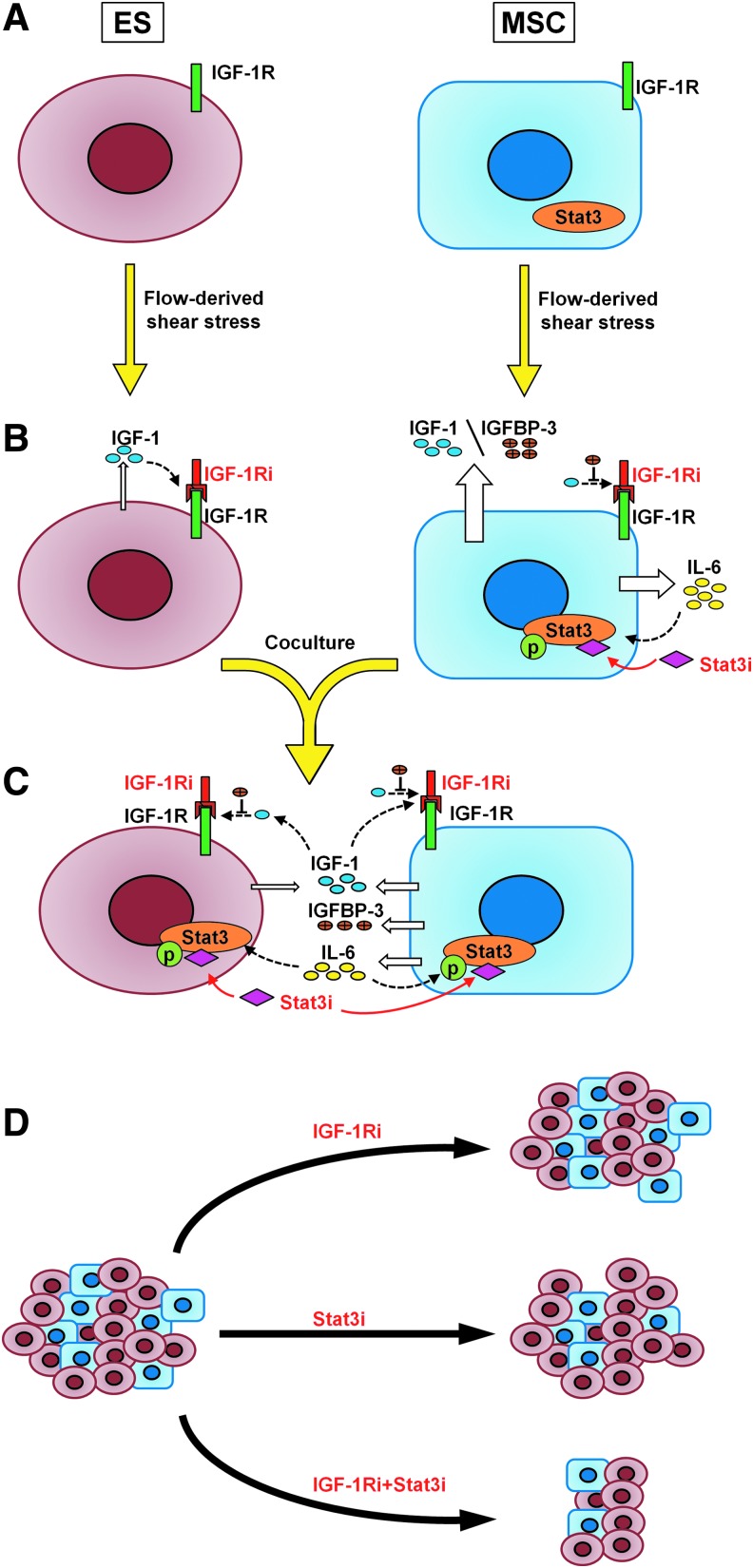
FIG. 5.
Summary mechanism. Both cells become sensitive to the IGF-1Ri in the transition from static (A) to flow perfusion conditions (B) due to the shear stress-mediated upregulation of IGF-1. Mechanosensitive promotion of IL-6 secretion also results in acquired sensitivity to Stat3i in MSCs. (C) On co-culturing, sensitivity to IGF-1Ri is lost, because tumor cells coerce MSCs into secreting higher levels of IL-6, and ES proliferation relies on IL-6/Stat3 signaling rather than IGF-1/IGF-1R signaling cascade. ES-acquired drug resistance to IGF-1R blockade is overcome only in the presence of Stat3i. (D) On a macroscopic level, IGF-1Ri will have minimal impact on solid ES tumors, Stat3i will primarily target local stroma, and significant shrinkage of tumor growth will likely occur only on dual targeting of IGF-1Ri+Stat3i.
In evaluating the importance of the ES stroma in drug sensitivity, this study reiterated the necessity of a highly controlled 3D model to perform a physiologically relevant investigation on ES-stroma interactions. Studying different ES:MSC ratios in our flow perfusion-based tumor model allowed us to understand how cell signaling evolves during ES progression, depending on the extent of the connective stroma present in the bone tumor niche. Previous in vivo studies already underlined the possible role of IL-6/Stat3 in ES tumorigenesis,17,18 but only within a tissue-engineered model could we clearly understand how ES (1) co-opts local stroma into producing IL-6 and (2) switches to Stat3 signaling on blocking the canonical IGF-1R pathway (Fig. 5).
This aspect is particularly relevant, as the interplay between IGF-1R and Stat3 pathway emerges in many other malignancies, such as ovarian cancer, lung cancer, and colorectal cancer.23,32,33 Though our research detailed the intricate interrelationship between ES and MSCs, we strongly suspect that other stromal and/or tumor-associated immune cells may also play a role in IL-6/Stat3 signaling. As just one example, several recent immunotherapy trials have shown that chimeric antigen receptor T-cells can elicit a dramatic cytokine storm mediated by release of IL-6 and other cytokines.34 Our data would suggest that this profound release of IL-6, and perhaps other cytokines, could alter tumor phenotype. Accordingly, the proposed tumor model could have far-reaching applicability and could improve our understanding of Stat3-mediated drug resistance in both non-sarcomatous malignancies and different stromal cells that are unique to those tumor types. We envision that future preclinical studies evaluating combination therapies that target the archetypal IGF-1R signaling pathway, as well as the compensatory Stat3 signaling cascade, will increase our chances of treating cancer patients who harbor tumors that are resistant to the IGF-1R blockade.
While developing our current model, a key finding is that flow perfusion conditions are crucial not only in establishing a more in vivo-like ES cell phenotype6 but also in providing MSCs with the appropriate biomechanical cues that are characteristic of the bone tumor niche (Fig. 5). Traditional 2D cell culture systems underestimate how biophysical cues, such as microarchitecture and biomechanical stimuli, affect the stromal cell phenotype, which, in turn, is indispensable for an accurate in vitro description of tumor-stroma interactions.7 We contend, therefore, that for ES as well as for other malignancies, 3D modeling should prioritize the recapitulation of biophysical cues (e.g., microarchitectural, mechanical) as a necessary prerequisite before one can adequately study how non-malignant cells contribute to tumor viability.
Conclusions
In this research, we investigated the simultaneous presence of flow perfusion and MSCs on ES phenotype and drug sensitivity. Our study focused on targeted therapies against the IGF-1/IGF-1R pathway, a central player in ES tumorigenesis, and the IL-6/Stat3 pathway, a signaling pathway heavily involved in acquired drug resistance not only in ES but also in several other malignancies.
The proposed tissue-engineered tumor model mirrored distinctive mechanisms observed in vivo, such as ES exploitation of the tumor microenvironment and stroma-mediated acquired drug resistance to biologically targeted therapeutics. Although these aspects are usually convoluted during in vivo testing, the use of a 3D tumor model enabled the detection of tumor- and/or stroma-specific mechanisms within a controlled environment. Furthermore, the culture conditions and the tumor/stroma ratios can be adjusted to determine tumor response to biologically targeted therapeutics at different stages of disease progression, in contrast to time-consuming animal experiments, where tumorigenesis occurs over a much longer timescale.
Although we primarily focused on the combined role of mechanical stimulation and stroma on ES phenotype and sensitivity, this research highlights how cells extracted from their niche, devoid of biomechanical cues, are unable to reliably model true cell–cell interaction as it exists in the tumor niche. An accurate assessment of tumor-stroma interactions can occur only if both cell types exhibit a physiologically relevant phenotype that, in turn, is dependent on the accurate recapitulation of microenvironmental cues such as microarchitecture and biomechanical stimuli. Consequently, we believe that the introduction of non-tumor cells is secondary to the description of biophysical cues when developing a tumor model.
The present study has improved our understanding of ES-stroma cross-talk in a mechanically stimulated system, but further investigations are required to elucidate the exact interplay between ES and stromal cells and the role of stroma- and/or tumor-derived extracellular matrix within this context. Ultimately, the investigation of heterotypic tumor-stroma cell signaling within our 3D tumor model is expected to: (1) advance our understanding of the interplay among different cellular and environmental cues, (2) assess the relevance of each of these aspects on tumor progression and drug resistance, and (3) develop therapeutic interventions that target both tumor cells and the local microenvironment.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (R01 CA180279) and the University of Texas MD Anderson Cancer Center's Support Grant CA016672. The authors thank Dr. Ian McNiece for providing the MSCs and Esther J. Lee and Dr. Jordan E. Trachtenberg for assistance with article preparation. This research was funded, in part, through a generous gift in honor of Faris D. Virani.
Disclosure Statement
No competing financial interests exist.
References
- 1.Breslin S., and O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 18, 240, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Kola I., and Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov 3, 711, 2004 [DOI] [PubMed] [Google Scholar]
- 3.McMillin D.W., Negri J.M., and Mitsiades C.S. The role of tumour-stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov 12, 217, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Beck J.N., Singh A., Rothenberg A.R., Elisseeff J.H., and Ewald A.J. The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination. Biomaterials 34, 9486, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhowmick N.A., Neilson E.G., and Moses H.L. Stromal fibroblasts in cancer initiation and progression. Nature 432, 332, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santoro M., Lamhamedi-Cherradi S.E., Menegaz B.A., Ludwig J.A., and Mikos A.G. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc Natl Acad Sci U S A 112, 10304, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villasante A., Marturano-Kruik A., and Vunjak-Novakovic G. Bioengineered human tumor within a bone niche. Biomaterials 35, 5785, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong E.L.S., Lamhamedi-Cherradi S.E., Burdett E., Ramamoorthy V., Lazar A.J., Kasper F.K., Farach-Carson M.C., Vishwamitra D., Demicco E.G., Menegaz B.A., Amin H.M., Mikos A.G., and Ludwig J.A. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc Natl Acad Sci U S A 110, 6500, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D., and Weinberg R.A. Hallmarks of cancer: the next generation. Cell 144, 646, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Folkman J., Bach M., Rowe J.W., Davidoff F., Lambert P., Hirsch C., Goldberg A., Hiatt H.H., Glass J., and Henshaw E. Tumor angiogenesis—therapeutic implications. New Engl J Med 285, 1182, 1971 [DOI] [PubMed] [Google Scholar]
- 11.McAllister S.S., and Weinberg R.A. Tumor-host interactions: a far-reaching relationship. J Clin Oncol 28, 4022, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Ludwig J.A. Ewing sarcoma: historical perspectives, current state-of-the-art, and opportunities for targeted therapy in the future. Curr Opin Oncol 20, 412, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Maheshwari A.V., and Cheng E.Y. Ewing sarcoma family of tumors. J Am Acad Orthop Surg 18, 94, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Stewart K.S., and Kleinerman E.S. Tumor vessel development and expansion in Ewing's sarcoma: a review of the vasculogenesis process and clinical trials with vascular-targeting agents. Sarcoma 2011, 165837, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahimic C.G., Wang Y., and Bikle D.D. Anabolic effects of IGF-1 signaling on the skeleton. Front Endocrinol (Lausanne) 4, 6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papachroni K.K., Karatzas D.N., Papavassiliou K.A., Basdra E.K., and Papavassiliou A.G. Mechanotransduction in osteoblast regulation and bone disease. Trends Mol Med 15, 208, 2009 [DOI] [PubMed] [Google Scholar]
- 17.Lissat A., Joerschke M., Shinde D.A., Braunschweig T., Meier A., Makowska A., Bortnick R., Henneke P., Herget G., Gorr T.A., and Kontny U. IL6 secreted by Ewing sarcoma tumor microenvironment confers anti-apoptotic and cell-disseminating paracrine responses in Ewing sarcoma cells. BMC Cancer 15, 552, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson J.L., Titz B., Akiyama R., Komisopoulou E., Park A., Tap W.D., Graeber T.G., and Denny C.T. Phosphoproteomic profiling reveals IL6-mediated paracrine signaling within the Ewing sarcoma family of tumors. Mol Cancer Res 12, 1740, 2014 [DOI] [PubMed] [Google Scholar]
- 19.Kuhn N.Z., and Tuan R.S. Regulation of stemness and stem cell niche of mesenchymal stem cells: implications in tumorigenesis and metastasis. J Cell Physiol 222, 268, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Lee J.S., Kang J.H., Boo H.J., Hwang S.J., Hong S., Lee S.C., Park Y.J., Chung T.M., Youn H., Lee S.M., Kim B.J., Chung J.K., Chung Y., William W.N., Jr., Shin Y.K., Lee H.J., Oh S.H., and Lee H.Y. STAT3-mediated IGF-2 secretion in the tumour microenvironment elicits innate resistance to anti-IGF-1R antibody. Nat Commun 6, 8499, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamhamedi-Cherradi S.E., Menegaz B.A., Ramamoorthy V., Vishwamitra D., Wang Y., Maywald R.L., Buford A.S., Fokt I., Skora S., Wang J., Naing A., Lazar A.J., Rohren E.M., Daw N.C., Subbiah V., Benjamin R.S., Ratan R., Priebe W., Mikos A.G., Amin H.M., and Ludwig J.A. IGF-1R and mTOR blockade: novel resistance mechanisms and synergistic drug combinations for Ewing sarcoma. J Natl Cancer Inst 108, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu B., Du L., Fan Q.M., Tang Z., and Tang T.T. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett 325, 80, 2012 [DOI] [PubMed] [Google Scholar]
- 23.Hsu H.S., Lin J.H., Hsu T.W., Su K., Wang C.W., Yang K.Y., Chiou S.H., and Hung S.C. Mesenchymal stem cells enhance lung cancer initiation through activation of IL-6/JAK2/STAT3 pathway. Lung Cancer 75, 167, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Cui X., Liu J., Bai L., Tian J., and Zhu J. Interleukin-6 induces malignant transformation of rat mesenchymal stem cells in association with enhanced signaling of signal transducer and activator of transcription 3. Cancer Sci 105, 64, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotlandi K., Manara M.C., Strammiello R., Landuzzi L., Benini S., Perdichizzi S., Serra M., Astolfi A., Nicoletti G., Lollini P.L., Bertoni F., Nanni P., and Picci P. c-kit receptor expression in Ewing's sarcoma: lack of prognostic value but therapeutic targeting opportunities in appropriate conditions. J Clin Oncol 21, 1952, 2003 [DOI] [PubMed] [Google Scholar]
- 26.Romano P., Manniello A., Aresu O., Armento M., Cesaro M., and Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res 37, D925, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahlin R.L., Meretoja V.V., Ni M.W., Kasper F.K., and Mikos A.G. Design of a high-throughput flow perfusion bioreactor system for tissue engineering. Tissue Eng Part C Methods 18, 817, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jerome L., Shiry L., and Leyland-Jones B. Deregulation of the IGF axis in cancer: epidemiological evidence and potential therapeutic interventions. Endocr Relat Cancer 10, 561, 2003 [DOI] [PubMed] [Google Scholar]
- 29.Sasamoto A., Nagino M., Kobayashi S., Naruse K., Nimura Y., and Sokabe M. Mechanotransduction by integrin is essential for IL-6 secretion from endothelial cells in response to uniaxial continuous stretch. Am J Physiol Cell Physiol 288, C1012, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Antonov A.S., Antonova G.N., Munn D.H., Mivechi N., Lucas R., Catravas J.D., and Verin A.D. Alpha V beta 3 integrin regulates macrophage inflammatory eesponses via PI3 kinase/Akt-dependent NF-kappa B activation. J Cell Physiol 226, 469, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig J.A., Lamhamedi-Cherradi S.E., Lee H.Y., Naing A., and Benjamin R. Dual targeting of the insulin-like growth factor and collateral pathways in cancer: combating drug resistance. Cancers 3, 3029, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez-Lopez E., Flashner-Abramson E., Shalapour S., Zhong Z., Taniguchi K., Levitzki A., and Karin M. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene 35, 2634, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W., Zong C.S., Hermanto U., Lopez-Bergami P., Ronai Z., and Wang L.H. RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol Cell Biol 26, 413, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., and June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 3, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.