Abstract
The function of upstream binding factor (UBF), an essential component of the RNA polymerase (pol) I preinitiation complex, is unclear. Recently, UBF was found distributed throughout ribosomal gene repeats rather than being restricted to promoter regions. This observation has led to the speculation that one role of UBF binding may be to induce chromatin remodeling. To directly evaluate the impact of UBF on chromatin structure, we used an in vivo assay in which UBF is targeted via a lac repressor fusion protein to a heterochromatic, amplified chromosome region containing lac operator repeats. We show that the association of UBF with this locus induces large-scale chromatin decondensation. This process does not appear to involve common remodeling complexes, including SWI/SNF and histone acetyltransferases, and is independent of histone H3 lysine 9 acetylation. However, UBF recruits the pol I-specific, TATA box-binding protein containing complex SL1 and pol I subunits. Our results suggest a working hypothesis in which the dynamic association of UBF with ribosomal DNA clusters recruits the pol I transcription machinery and maintains these loci in a transcriptionally competent configuration. These studies also provide an in vivo model simulating ribosomal DNA transactivation outside the nucleolus, allowing temporal and spatial analyses of chromatin remodeling and assembly of the pol I transcription machinery.
Ribosomal RNA (rRNA) is encoded by tandem arrays of rDNA genes that are organized in human cells into nucleolar organizing regions (NORs) located on five chromosome pairs. A specific set of transcription factors is dedicated to transcription of rDNA into pre-rRNA that is subsequently processed into 28S, 18S, and 5.8S rRNAs. The rRNAs are packaged with ribosomal proteins to form the large and small subunits of ribosomes (1). Transcription of rDNA is highly specific and extensively regulated, involving a large number of proteins (1–3). Studies based mostly on in vitro assays suggest that transcriptional activation of rDNA involves association of the preinitiation complex with the promoter region followed by recruitment of other factors and RNA polymerase (pol) I subunits. The preinitiation complex has been shown to contain upstream binding factor (UBF) (4) and the TATA box-binding protein (TBP) containing complex SL1 (5).
UBF is highly conserved in vertebrate cells from Xenopus to human. It contains five high mobility group boxes, an N terminus necessary for nuclear translocation, and a highly acidic C terminus essential for nucleolar localization (4, 6, 7). Although UBF is involved in pol I transcriptional activation, the mechanism by which it acts remains unclear. The consensus derived mostly from in vitro and some in vivo correlative studies places UBF at the early steps of rDNA transactivation (1, 2, 8). Analyses using in vitro transcription assays indicate that UBF initially binds DNA and is necessary for SL1 binding to form the preinitiation complex (9). Recently, UBF was discovered to literally coat DNA throughout entire NORs (10), suggesting that UBF might play a role in determining NOR structure. In support of a role in chromatin structure, in vitro analyses have demonstrated that UBF homodimers can bend and loop DNA in solution (11–14). However, there is not a direct in vivo experimental system that can single out the function of UBF in mammalian cells.
To set up such a system to directly analyze the role of UBF in chromatin structure and pol I transactivation in vivo, we took advantage of a lac operator–repressor-based system that allows visualization of the effect of transcriptional activators on large-scale chromatin remodeling in mammalian cells (15). The A03_1 Chinese hamster ovary DG44 cell line contains an ≈90-Mbp amplified, heterochromatic region consisting of multiple copy vector repeats, ≈400 kbp in size, separated by large regions of coamplified genomic DNA (16). Throughout most of the cell cycle, this amplified chromosome region is condensed into a compact mass. Targeting certain transcriptional activators to this heterochromatic locus via a lac repressor fusion protein leads to large-scale decondensation of this region (17–27).
There are concerns regarding the physiological relevance of the effects observed in this system because of the highly repetitive nature of the chromatin template and the large number of binding sites for the transcription factor–lac repressor fusion protein. However, there are several reasons that it represents a physiologically relevant model for analyzing the role of UBF in chromatin remodeling of the rDNA locus. First, the lac operator array is structurally analogous to NORs because both are highly repetitive and the core repeats are flanked by intergenic sequences. Second, UBF binds throughout entire rDNA clusters without apparent sequence specificity, implying that the binding is either mediated by a higher-order DNA structural recognition or by chaperones. The targeting of UBF to the array through repressor binding mimics a chaperone-mediated association. Lastly, binding of repressor–UBF to the closely spaced lac operator repeats would resemble the coating properties of UBF throughout NORs. Therefore, this system provides a DNA template that is structurally similar to NORs but is located outside of nucleoli, allowing evaluation of the primary role of UBF in modulating chromatin structure.
To analyze the effect of UBF on the heterochromatic locus, we used UBF–lac repressor fusion proteins to target UBF to the lac operator repeats contained within this locus. Our results demonstrate that targeting UBF to the heterochromatic lac operator array is sufficient to induce a large-scale chromatin decondensation and to initiate the assembly of the pol I transcription apparatus.
Materials and Methods
Cell Culture. A03_1 Chinese hamster ovary DG44 cells carry a gene-amplified chromosome region containing lac operator repeats (16). These cells were cultured at 37°C with 5% CO2 in Ham's F-12 media without hypoxanthine and thymidine, with 50 units/ml penicillin, 50 μg/ml streptomycin, and 0.3 μM methotrexate, without phenol red and with 10% dialyzed FBS (HyClone Labs) treated with charcoal-dextran. Phenol-red-free trypsin was used to passage cells. HeLa (cervix epithelial carcinoma) cells were grown in DMEM supplemented with 10% FBS (GIBCO, Invitrogen), 50 units/ml penicillin, and 50 μg/ml streptomycin in a humidified 37°C incubator with 5% CO2.
Plasmid Constructs. The p3′SS-EGFP-dimer lac repressor-VP16 AAD construct has been described (22). Plasmids p3′SS-EGFP-dimer lac repressor and p3′SS-EYFP-dimer lac repressor, which express the GFP- and yellow fluorescent protein (YFP)-dimer lac repressor–simian virus 40 nuclear localization signal fusion protein under control of the F9–1 promoter, respectively (22), were used in these studies. UBF1 was amplified by PCR using primers that contain AscI sites. The primers that amplified wild-type UBF1 are T TGGCGCGCCAGATGA ACGGAGAAGCCGACTGC for the N terminus and TTGGCGCGCCAGGTTGGAGTCAGAGTCTGAGGA for the C terminus. The PCR products were ligated into the AscI-digested vectors to create in-frame fusions. The pHcRed1-C1-UBF1 plasmid was generated by ligating a KpnI–BamHI fragment containing wild-type human UBF1 into the pHcRed1-C1 vector (BD Biosciences Clontech). All fusion constructs were sequenced and shown to be faithful copies of UBF1.
Transfection. Constructs were transiently transfected into A03_1 Chinese hamster ovary DG44 cells and HeLa cells by electroporation (28). Cells were subsequently seeded onto glass coverslips that were mounted on the bottom of 35-mm Petri dishes and grown for 24 h.
Immunolabeling. Cells were immediately fixed in 2% paraformaldehyde in 1× PBS for 10 min, washed in PBS three times for 5 min, and blocked in 0.5% BSA for 20 min. Washing cells in PBS three times after a 1-h primary antibody incubation was followed by staining for 1 h with secondary antibodies. The following primary antibodies and titers were used: 1:100 mouse anti-heterochromatin protein-1α (HP1α) (Chemicon); 1:50 rabbit anti-Brg1 (Santa Cruz Biotechnology); 1:50 goat anti-Brm (Santa Cruz Biotechnology); 1:50 rabbit anti-Mi2 CHD3/4 (Upstate Biotechnology, Lake Placid, NY); 1:50 rabbit anti-hGCN5 (Shelley L. Berger, The Wistar Institute, Philadelphia); 1:50 rabbit anti-acetylated histone H3 (K9) (Upstate Biotechnology), 1:500 rabbit anti-acetylated histone H3 (David Allis, The Rockfeller University, New York); 1:50 anti-phospho-Histone H1 (Upstate Biotechnology); 1:50 mouse anti-UBF (Santa Cruz Biotechnology); 1:100 rabbit anti-TBP (Santa Cruz Biotechnology); 1:100 rabbit anti-TFIIB (Nouria Hernandez, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY); 1:50 goat anti-TAFI p95/110 (Santa Cruz Biotechnology); 1:50 goat anti-TAFII p32 (Santa Cruz Biotechnology); 1:50 goat anti-TFIIIB (Santa Cruz Biotechnology); 1:50 rabbit anti-RPA39 (Angus Lamond, University of Dundee, Dundee, Scotland); 1:15 human anti-fibrillarin (ANA-N, Sigma). The immunolabeling signals were subsequently detected by incubating cells with Cy5 or Texas red-conjugated secondary antibodies (Jackson ImmunoResearch). Coverslips were washed in PBS and stained with 0.2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) before mounting. Fluorescent images were collected on a Nikon Eclipse E800 microscope or a Zeiss 510 confocal laser scanning microscope equipped with an argon–krypton laser.
Results
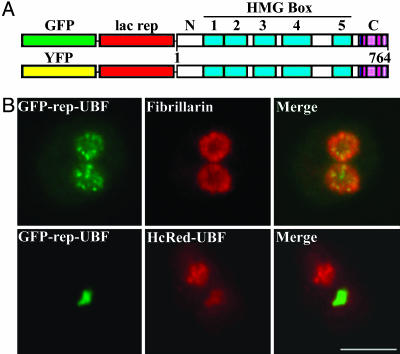
GFP- or YFP-Repressor-UBF Behaves Similarly to Its Endogenous Counterparts. To evaluate the effect of UBF on the lac-operator arrays of A03_1 cells, we constructed a GFP- or YFP-lac repressor-UBF fusion protein (Fig. 1A). To verify that the fusion protein behaved similarly to endogenous UBF, its localization and protein–protein interactions were compared to wild-type UBF. When HeLa cells that did not contain the lac–operator arrays were transfected with GFP-repressor-UBF, the fusion protein localized to punctate dots within the nucleoli, most likely representing the fibrillar structures observed by electron microscopy where the synthesis and processing of pre-rRNA take place (29) (Fig. 1B). This punctate localization pattern is identical to that of the endogenous UBF (data not shown), and similar to the labeling of fibrillarin, a protein involved in pre-rRNA processing and shown to colocalize with UBF (Fig. 1B), as has been demonstrated (30).
Fig. 1.
GFP- or YFP-repressor-UBF behaves similarly to its endogenous counterparts. (A) A diagram of the expression constructs for fusion proteins that contain GFP or YFP, lac repressor, and UBF. (B) The fusion protein GFP-repressor-UBF behaves similarly to endogenous UBF with regards to its subnucleolar localization pattern in HeLa cells (Upper) and its dimerization with cotransfected HcRed-UBF in A03_1 cells (Lower). (Scale bar, 10 μm.)
UBF forms homodimers (31). If the fusion proteins remain functionally similar to the endogenous UBF, we expected that they should form homodimers with wild-type UBF. Two approaches were used to evaluate the dimerization capacity of the fusion proteins. First, A03_1 cells were either singly transfected with HcRed-UBF1 or cotransfected with HcRed-UBF and GFP-repressor-UBF. Although HcRed-UBF by itself does not localize to the lac operator array because of the lack of repressor (data not shown), the overlay of the cotransfected cells demonstrates that GFP-repressor-UBF and HcRed-UBF colocalize at the amplified chromosome region (Fig. 1B), with HcRed-UBF also localizing to nucleoli. These results are consistent with HcRed-UBF being recruited to the amplified chromosome region through binding to GFP-repressor-UBF, most likely through dimerization. Second, we asked whether GFP or YFP-repressor-UBF could coimmunoprecipitate endogenous UBF. HeLa and A03_1 cells were transfected with either GFP- or YFP-repressor-UBF, and cell lysates were immunoprecipitated with anti-GFP antibody. The precipitates were then Western blotted with anti-UBF antibody. The results show that endogenous UBF can be coimmunoprecipitated with GFP or YFP-repressor-UBF, demonstrating that the fusion proteins form complexes with endogenous UBF, most likely through dimerization (data not shown). In addition, the dynamics of GFP- or YFP-repressor-UBF in HeLa nucleoli were compared with GFP-UBF (28, 32) by using fluorescence recovery after photo-bleaching analyses. The results demonstrate similar paces of fluorescence recovery among all these fusion proteins in HeLa nucleoli (data not shown). Together, these studies show that both GFP- and YFP-repressor-UBF behave similarly to endogenous UBF in vivo.
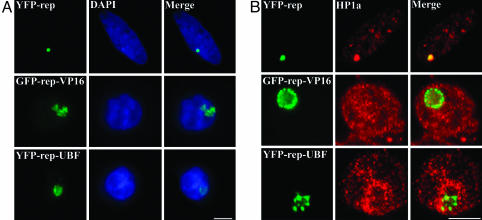
Targeting GFP- or YFP-Repressor-UBF to the Heterochromatic Lac Operator Locus Induces Large-Scale Chromatin Decondensation. To evaluate the role of UBF in chromatin structure, GFP- or YFP-repressor-UBF expression constructs were transiently transfected into A03_1 cells. Binding of GFP- or YFP-repressor to the lac operator array did not change the dense nature of the heterochromatic locus, as it still remained a round dot that colocalized with a similarly shaped DNA structure labeled with DAPI (Fig. 2A). In comparison, the association of GFP- or YFP-repressor-UBF with the lac operator array transformed the appearance of the array from condensed round dots to a larger, irregularly shaped diffuse pattern (Fig. 2 A). DAPI staining of the same locus showed a lower DNA density than that observed in the absence of UBF, suggesting that targeting of UBF to this chromosomal locus induced decondensation of the heterochromatic lac operator array. Around one third (34 ± 6%) of cells expressing YFP-repressor-UBF exhibited significant decondensation at the lac operator array, as compared to only 1% of cells expressing repressor alone. This assay has been used to show that several pol II transcription factors, including the viral VP16 acidic activation domain (19, 33), can also induce decondensation of the locus when fused to the lac repressor. We compared the effect of GFP-repressor-VP16 to that of the YFP-repressor-UBF, and found that the VP16 fusion protein induced greater decondensation of the lac operator locus (judging by the size of the “puff”) than did the UBF fusions (Fig. 2 A).
Fig. 2.
Targeting YFP-repressor-UBF to the lac operator array induces chromatin decondensation. (A) The decondensation induced by UBF (A Bottom) is often to a lesser extent than the decondensation induced by VP-16 (A Middle). A03_1 cells were transfected with YFP-repressor (Top), YFP-repressor-VP16 (Middle), and YFP-repressor-UBF (Bottom). (Left) The localization of the lac operator array visualized by its binding to YFP-repressor and fusion proteins. (Center) The same cells stained with DAPI. (Right) Overlay images. (B) The lac operator array loses characteristic HP1α binding of heterochromatin upon UBF binding. (Left) The expression of GFP or YFP-repressor and their fusion proteins. (Center) Immunolabeling of the same cells with either anti-HP1α antibody. (Right) The overlay images. (Scale bar, 10 μm.)
The Repressor-UBF Fusion Protein Induces Chromatin Decondensation in the Absence of Extensive Histone H3-K9 Acetylation. To understand the mechanisms by which repressor-UBF induces chromatin decondensation at the lac-operator array, we analyzed changes at this region upon UBF association. HP1 is a signature of the heterochromatic state of chromatin and has a nuclear localization pattern that mirrors that of heterochromatin (34). In A03_1 cells transfected with YFP-repressor, the concentrated localization of HP1α overlapped with the condensed locus of the lac operator array, as seen in the merged image with the YFP-repressor (Fig. 2B Top, yellow overlay). This finding demonstrates the heterochromatic nature of the locus in the absence of transcription factor mediated chromatin remodeling. When the lac operator locus was remodeled by association with VP16, a significant reduction of HP1α labeling was observed (Fig. 2B Middle), as has been described (27). A similar reduction in HP1α labeling was also observed upon YFP-repressor-UBF binding (Fig. 2B Bottom), suggesting that UBF association induces alterations of the chromatin structure that significantly reduced HP1α binding.
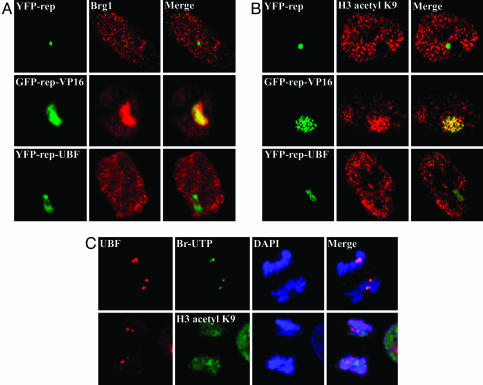
A well characterized chromatin remodeling pathway involves the recruitment of SWI/SNF and HAT components, followed by demethylation and acetylation of lysine residues located on the N-terminal tails of histone H3 (35). Because earlier studies demonstrated that VP16 mediated decondensation of lac operator array in A03_1 cells follows this pathway (22, 33), we evaluated whether UBF induced chromatin decondensation undergoes similar processes. Antibodies specific to Brg1 (a component of the SWI/SNF complex), GCN5 (HATs), trimethylated lysine 9 (K9) of H3, acetylated K9 of H3, or overall acetylated histone H3 were used to immunostain A03_1 cells expressing YFP-repressor, GFP-repressor-VP16, or YFP-repressor-UBF. As expected, Brg1 was not found at the condensed lac operator array in cells expressing repressor alone (Fig. 3A Top), but was heavily recruited to the VP16 associated and decondensed locus (Fig. 3A Middle). Interestingly, Brg1 was not extensively recruited to the UBF-associated lac operator array (Fig. 3A Bottom). Correspondingly, little acetylated histone labeling was observed at the condensed lac operator array in GFP-repressor-expressing cells (Fig. 3B Top), contrasting the massive amount of labeling at the decondensed locus in cells expressing GFP-repressor-VP16 (Fig. 3B Middle) that occurs in a time frame coincident with the large-scale chromatin decondensation, as previously shown (22, 33). In comparison, the decondensation of lac-operator repeats induced by UBF targeting did not show significant increases in the immunolabeling of the locus by anti-acetylated histone H3, H3-K9 acetylation (Fig. 3B Bottom), or GCN5 (data not shown), although demethylation at K9 of H3 was detected in the decondensed locus (data not shown).
Fig. 3.
The decondensed chromatin induced by UBF association does not recruit components of SWI/SNF complex (Brg1) (A) and does not show massive histone acetylations. (B)(Left) The expression of GFP or YFP-repressor and their fusion proteins. (Center) Immunolabeling of the same cells with either anti-Brg1 antibody (A) or anti-K9 actylated histone H3 (B). (Right) Overlay images. (C) Double labeling BrUrd incorporation for 5 min and UBF at anaphase–telophase tranisition (Upper) and double labeling of acetylated K9 of histone H3 and UBF at the same stage of mitosis (Lower, arrowheads). (Scale bar, 10 μm.)
To evaluate whether the deficiency in H3 K9 acetylation during UBF-mediated chromatin decondensation reflects the transactivation of rDNA clusters, we examined the immunolabeling of the acetylated K9 of H3 during rDNA activation at the end of mitosis in HeLa cells. BrdUrd incorporation coupled with UBF immunolabeling demonstrated that transcriptional activation of rDNA clusters takes place at the anaphase–telophase transition (Fig. 3C). Colabeling of either GCN5 or acetylated H3 K9 with UBF at the rDNA clusters during this transition showed little enrichment of either the enzyme (data not shown) or the acetylated H3 at the activated rDNA clusters (Fig. 3C, arrowheads). These findings suggest that the chromatin remodeling induced by UBF and rDNA transactivation may involve chromatin remodeling pathways distinct from that mediated by SWI/SNF and HATs.
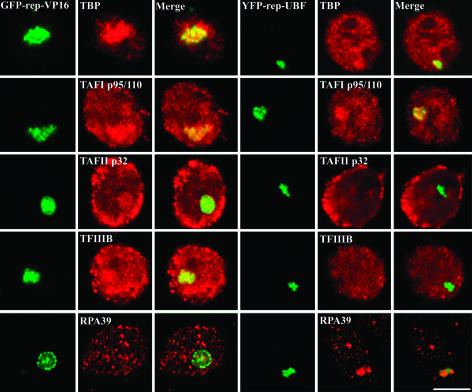
The UBF Fusion Protein Recruits Pol I-Specific Transcription Factors to the Lac Operator Array. To analyze the pathways or players that might be involved in UBF-induced chromatin remodeling, we examined the localization of a battery of other known chromatin remodeling and transcription factors in A03_1 cells expressing either YFP-repressor, GFP-repressor-VP16, or YFP-repressor-UBF. Additional factors examined included members of the CHRAC complex, the NuRD complex, and general transcription factors. Although immunolabeling of the other remodeling proteins did not show enriched labeling at the decondensed array mediated by UBF targeting (data not shown), some of the pol I-specific transcription factors were localized there. TBP is an essential component of three complexes, SL1, TFIID, and TFIIIB, that are specific for pol I, -II (TFIID), and -III (TFIIIB) polymerase function, respectively (36). Immunolabeling using anti-TBP antibody demonstrated that TBP was enriched at the decondensed lac-operator array when the decondensation was induced by either VP16 or UBF (Fig. 4), but was absent from the condensed locus associated with repressor alone (data not shown). This finding suggests that, like VP16, UBF is able to recruit basal transcription factors. Because TBP is part of three complexes that serve different polymerases, we were interested in determining whether the TBP-containing complex recruited by UBF was specific to any or all of these three classes of transcription. To do so, antibodies specific to TAF I (p95/110) for SL1, TAFII (p32) for TFIID, or TFIIIB were used to immunolabel transfected A03_1 cells. The binding of repressor alone did not recruit any of the factors, whereas VP16 recruited all three classes of TBP associated proteins (Fig. 4). In comparison, UBF detectably recruited p95/100, but not p32 or TFIIIB (Fig. 4). These results demonstrate that UBF selectively recruits the pol I-specific TBP-containing complex SL1. To further evaluate whether other pol I transcription factors are also recruited to the UBF-associated lac operator array, a specific antibody against pol I subunit RPA 39 was also used to immunolabel the transfected A03_1 cells. The results showed that UBF, but not VP16, recruited the pol I subunit (Fig. 4). The mass amount of recruitment of pol I factors to the lac operator arrays significantly decreased the localization of these factors to the endogenous nucleoli (Fig. 4, arrowhead). These findings demonstrate that UBF is sufficient to recruit SL1 and the pol I subunit to the lac operator array.
Fig. 4.
The association of UBF with the lac operator array recruits the pol I-specific TBP-containing complex, SL1, and pol I subunit RPA39. This finding is in contrast to the association of VP-16 with the lac operator array, which indiscriminately recruits all TBP-associated complexes and does not recruit RPA39. Arrowheads indicate the labeling of RPA39 in the endogenous nucleolus. Specifics are labeled in the images. (Scale bar, 10 μm.)
Discussion
Although many of the factors involved in rDNA transcription have been identified and characterized (1–3, 37) as discussed in the Introduction, the specific mechanisms that remodel the rDNA chromatin and initiate pol I transcription remain unclear. Studies primarily in vitro and some correlative ones in vivo over the past 20 years have led to a general model that the preinitiation complex, which contains UBF and SL1, binds the promoter elements and facilitates the assembly of the complete transcription apparatus (1). UBF is believed to stabilize SL1, interact with the nucleosomal DNA (13, 14), replace H1 (38, 39), and recruit pol I to the promoter (1).
Interestingly, more recent studies showed that UBF binds rDNA throughout entire NORs (10). In addition, when a human acrocentric chromosome that contains rDNA was introduced into mouse nuclei, where they remained transcriptionally inactive, they were associated with both UBF and nucleoli (40). These studies generated a rather puzzling picture regarding the role of UBF as a specific component of the preinitiation complex. One possible explanation is that UBF is a special chromatin protein for the NORs. This idea is consistent with the finding that UBF homodimers bend rDNA into 140-bp loops (11–14). In addition, studies of NORs in Physarum polycephalum demonstrated that transcriptionally active NOR chromosome regions have a unique, unfolded chromatin architecture (41). A logical speculation based on these studies was that binding of UBF throughout the NORs could induce chromatin remodeling to assist their conversion to a transcriptionally active state. However, no direct in vivo experimental evidence addressed this hypothesis.
Here we have begun to directly test this model by tethering UBF to a heterochromatic chromosome region that contains a large array of repetitive sequences, resembling the highly repetitive rDNA locus. However, the lac operator DNA is unlikely to recruit other pol I factors that might specifically recognize rDNA sequences, thus allowing isolated analyses of the primary role of UBF on the chromatin structure and pol I transactivation outside of nucleoli. Our experiments show that targeting UBF across the lac operator array induces large-scale chromatin decondensation characterized by the loss of heterochromatic features of the locus. In contrast to the chromatin decondensation induced by targeting the VP16 pol II acidic activation domain, decondensation induced by UBF occurs in the absence of SWI/SNF complex, HAT recruitment, and histone H3-K9 acetylation. However, UBF association does recruit pol I-specific transcription factors. These findings demonstrate that UBF binding is sufficient to induce chromatin remodeling in vivo and to recruit pol I-specific transcription factors in a sequence-independent manner, because the lac operator does not share any sequence similarity with rDNA. The lack of extensive histone H3 tail acetylation is consistent with the overall acetylated H3 nuclear distribution patterns, where nucleoli often appear deficient in labeling (Fig. 3), and with the activation of endogenous rDNA clusters at the anaphase–telophase transition, where little acetylated K9 of H3 (Fig. 3) or GCN5 were detected at the activation loci marked by UBF labeling. In addition to UBF, BRCA1 (18) or estrogen receptor (17) mediated chromatin decondensation of the lac operator array is also independent of extensive histone acetylation. However, the lack of abundant histone H3 acetylation in UBF-mediated chromosome decondensation is not entirely consistent with recent studies in plants that demonstrated the involvement of K9 deacetylation and methylation of H3 in the epigenetic silencing switch of rDNA clusters (42, 43), although positive chromatin immunoprecipitation of H3-K9 acetylation was only observed in a deacetylase null mutant (42). These inconsistencies could be due partly to the large evolutionary distance between animal and plant cells, because plants do not have UBF. In addition, another study using an antibody recognizing multiple acetylated histones showed that histone acetylation associates with nucleosome-disrupted rDNA in cells treated with deacetylase inhibitor (44). Taken together, histone acetylation might be involved in rDNA activation, although the classic SWI/SNF pathway may not be a main player in this process. Further studies are needed to clarify this possibility.
The question now is how UBF contributes to NOR chromatin remodeling and/or maintenance of an open chromatin configuration. There are several possibilities that are not mutually exclusive. Dimeric UBF could act to remodel chromatin by binding rDNA and bending the DNA such that its association with nucleosomes is disrupted. Second, UBF could recruit chromatin remodeling proteins, unrelated to those investigated, that cooperatively change the NOR structure. Lastly, UBF could recruit components of the pol I transcription apparatus, which might themselves possess chromatin remodeling capacities or recruit proteins with those capacities. These possibilities need to be investigated.
The association of UBF with lac operator arrays recruits not only SL1, but also pol I subunits, suggesting that the pol I transcription complex can be assembled upon the binding of UBF to DNA in a sequence-independent manner. This finding is consistent with the low consensus found between vertebrate rDNA promoter regions, which contrast with the highly conserved nature of the pol I transcription factors, including UBF. The recruitment of pol I transcription factors suggests that chromatin remodeling is linked to transcriptional activation. Examples of chromatin remodeling coupled with transactivation have been elegantly illustrated in some classic studies, including those that observed puffs on Drosophila polytene chromosomes (45–47). The findings that UBF binding results in chromosome remodeling and recruitment of transcription factors not only support the model that UBF acts at the early stages of pol I transcription (1), but also indicate that UBF association could be the first step in the process in vivo. In addition, these findings validate usefulness of the lac operator array system as a model for analyzing the function of UBF.
The lac array system also uncouples the assembly of pol I transcription complex from transcription itself, because in situ incorporation of BrdUrd showed little labeling at the decondensed locus induced by UBF (Fig. 5, which is published as supporting information on the PNAS web site). This finding is in contrast to the targeting of VP16 to the same locus that stimulates RNA synthesis, as assayed by BrdUrd incorporation (22). It is conceivable that the association of the pol I transcription machinery may help maintain the open configuration of the NORs without necessarily being active in transcription. This idea is supported by findings that showed the association of pol I transcription factors with mitotic NORs when rDNA transcription is silent (2, 9). Transactivation could be regulated through signaling pathways that facilitate transcription through modifications of the factors, including phosphorylation, acetylation, methylation, and ubiqutination (1, 37). Elongation could depend on the recognition of an initiation site, mediated through sequence specific interacting factors.
In summary, our studies provide in vivo evidence that UBF acts at the earliest steps in chromatin remodeling and assembly of the pol I transcription apparatus. The integration of our findings and those of others leads us to propose a working model in which a dynamic association of UBF to NORs maintains rDNA in a transcriptionally competent form at all times, including the period of transcriptional silencing during mitosis. Transcription may take place when the activated initiation complex recruits the rest of the pol I transcriptional machinery, facilitating elongation through site-specific recognition. Future studies are necessary to test this working model.
Supplementary Material
Acknowledgments
We are grateful for constructive discussions and manuscript readings by our colleagues, Drs. Steve Adam, Eric Flitney, Yosef Gruenbaum, Daniel Leary, and Tom Volpe. We are also thankful to the generous gifts of antibodies from Drs. David Allis, Shelley Berger, Edward Chan (The Scripps Research Institute, La Jolla, CA), Nouria Hernandez, Angus Lamond, and Larry Rothblum. This study was supported by National Cancer Institute Grant R01 CA77560 (to S.H.) and National Institutes of Health Grant R01 GM58460 (to A.B.).
Abbreviations: NOR, nucleolar organizing region; pol, RNA polymerase; UBF, upstream binding factor; TBP, TATA box-binding protein; YFP, yellow fluorescent protein; HP1α, heterochromatin protein 1α; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Grummt, I. (2003) Genes Dev. 17, 1691–1702. [DOI] [PubMed] [Google Scholar]
- 2.Grummt, I. (1999) Prog. Nucleic Acid Res. Mol. Biol. 62, 109–154. [DOI] [PubMed] [Google Scholar]
- 3.Moss, T. & Stefanovsky, V. Y. (2002) Cell 109, 545–548. [DOI] [PubMed] [Google Scholar]
- 4.Jantzen, H. M., Admon, A., Bell, S. P. & Tjian, R. (1990) Nature 344, 830–836. [DOI] [PubMed] [Google Scholar]
- 5.Learned, R. M., Cordes, S. & Tjian, R. (1985) Mol. Cell. Biol. 5, 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maeda, Y., Hisatake, K., Kondo, T., Hanada, K., Song, C. Z., Nishimura, T. & Muramatsu, M. (1992) EMBO J. 11, 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Mahony, D. J. & Rothblum, L. I. (1991) Proc. Natl. Acad. Sci. USA 88, 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannan, K. M., Hannan, R. D. & Rothblum, L. I. (1998) Front. Biosci. 3, d376–398. [DOI] [PubMed] [Google Scholar]
- 9.Bell, S. P., Learned, R. M., Jantzen, H. M. & Tjian, R. (1988) Science 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- 10.O'Sullivan, A. C., Sullivan, G. J. & McStay, B. (2002) Mol. Cell. Biol. 22, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanovsky, V. Y., Pelletier, G., Bazett-Jones, D. P., Crane-Robinson, C. & Moss, T. (2001) Nucleic Acids Res. 29, 3241–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefanovsky, V. Y., Bazett-Jones, D. P., Pelletier, G. & Moss, T. (1996) Nucleic Acids Res. 24, 3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copenhaver, G. P., Putnam, C. D., Denton, M. L. & Pikaard, C. S. (1994) Nucleic Acids Res. 22, 2651–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bazett-Jones, D. P., Leblanc, B., Herfort, M. & Moss, T. (1994) Science 264, 1134–1137. [DOI] [PubMed] [Google Scholar]
- 15.Carpenter, A. E. & Belmont, A. S. (2004) Methods Enzymol. 375, 366–381. [DOI] [PubMed] [Google Scholar]
- 16.Li, G., Sudlow, G. & Belmont, A. S. (1998) J. Cell Biol. 140, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nye, A. C., Rajendran, R. R., Stenoien, D. L., Mancini, M. A., Katzenellenbogen, B. S. & Belmont, A. S. (2002) Mol. Cell. Biol. 22, 3437–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye, Q., Hu, Y. F., Zhong, H., Nye, A. C., Belmont, A. S. & Li, R. (2001) J. Cell Biol. 155, 911–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tumbar, T. & Belmont, A. S. (2001) Nat. Cell Biol. 3, 134–139. [DOI] [PubMed] [Google Scholar]
- 20.Tsukamoto, T., Hashiguchi, N., Janicki, S. M., Tumbar, T., Belmont, A. S. & Spector, D. L. (2000) Nat. Cell Biol. 2, 871–878. [DOI] [PubMed] [Google Scholar]
- 21.Stenoien, D. L., Nye, A. C., Mancini, M. G., Patel, K., Dutertre, M., O'Malley, B. W., Smith, C. L., Belmont, A. S. & Mancini, M. A. (2001) Mol. Cell. Biol. 21, 4404–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tumbar, T., Sudlow, G. & Belmont, A. S. (1999) J. Cell Biol. 145, 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belmont, A. S., Li, G., Sudlow, G. & Robinett, C. (1999) Methods Cell Biol. 58, 203–222. [DOI] [PubMed] [Google Scholar]
- 24.Belmont, A. S. & Straight, A. F. (1998) Trends Cell Biol. 8, 121–124. [DOI] [PubMed] [Google Scholar]
- 25.Vazquez, J., Belmont, A. S. & Sedat, J. W. (2001) Curr. Biol. 11, 1227–1239. [DOI] [PubMed] [Google Scholar]
- 26.Vazquez, J., Belmont, A. S. & Sedat, J. W. (2002) Curr. Biol. 12, 1473–1483. [DOI] [PubMed] [Google Scholar]
- 27.Janicki, S. M., Tsukamoto, T., Salghetti, S. E., Tansey, W. P., Sachidanandam, R., Prasanth, K. V., Ried, T., Shav-Tal, Y., Bertrand, E., Singer, R. H., et al. (2004) Cell 116, 683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen, D. & Huang, S. (2001) J. Cell Biol. 153, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raska, I. (2003) Trends Cell Biol. 13, 517–525. [DOI] [PubMed] [Google Scholar]
- 30.Hernandez-Verdun, D., Roussel, P. & Gebrane-Younes, J. (2002) J. Cell Sci. 115, 2265–2270. [DOI] [PubMed] [Google Scholar]
- 31.McStay, B., Frazier, M. W. & Reeder, R. H. (1991) Genes Dev. 5, 1957–1968. [DOI] [PubMed] [Google Scholar]
- 32.Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L. I., Phair, R. D. & Misteli, T. (2002) Science 298, 1623–1626. [DOI] [PubMed] [Google Scholar]
- 33.Memedula, S. & Belmont, A. S. (2003) Curr. Biol. 13, 241–246. [DOI] [PubMed] [Google Scholar]
- 34.Kamakaka, R. T. (2003) Curr. Biol. 13, R317–R319. [DOI] [PubMed] [Google Scholar]
- 35.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez, N. (1993) Genes. Dev. 7, 1291–1308. [DOI] [PubMed] [Google Scholar]
- 37.Grummt, I. & Pikaard, C. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 641–649. [DOI] [PubMed] [Google Scholar]
- 38.Kuhn, A. & Grummt, I. (1992) Proc. Natl. Acad. Sci. USA 89, 7340–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn, A., Stefanovsky, V. & Grummt, I. (1993) Nucleic Acids Res. 21, 2057–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan, G. J., Bridger, J. M., Cuthbert, A. P., Newbold, R. F., Bickmore, W. A. & McStay, B. (2001) EMBO J. 20, 2867–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prior, C. P., Cantor, C. R., Johnson, E. M., Littau, V. C. & Allfrey, V. G. (1983) Cell 34, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 42.Lawrence, R. J., Earley, K., Pontes, O., Silva, M., Chen, Z. J., Neves, N., Viegas, W. & Pikaard, C. S. (2004) Mol. Cell 13, 599–609. [DOI] [PubMed] [Google Scholar]
- 43.Probst, A. V., Fagard, M., Proux, F., Mourrain, P., Boutet, S., Earley, K., Lawrence, R. J., Pikaard, C. S., Murfett, J., Furner, I., et al. (2004) Plant Cell 16, 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutskov, V. J., Russanova, V. R., Dimitrov, S. I. & Pashev, I. G. (1996) J. Biol. Chem. 271, 11852–11857. [DOI] [PubMed] [Google Scholar]
- 45.Ashburner, M. (1970) Chromosoma 31, 356–376. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner, M. (1972) Chromosoma 38, 255–281. [DOI] [PubMed] [Google Scholar]
- 47.Nowak, S. J. & Corces, V. G. (2000) Genes Dev. 14, 3003–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.