Abstract
The rapid and sensitive determination of pathogenic bacteria is extremely important in biotechnology, medical diagnosis, and the current fight against bioterrorism. Current methods either lack ultrasensitivity or take a long time for analysis. Here, we report a bioconjugated nanoparticle-based bioassay for in situ pathogen quantification down to single bacterium within 20 min. The bioconjugated nanoparticle provides an extremely high fluorescent signal for bioanalysis and can be easily incorporated with biorecognition molecules, such as antibody. The antibody-conjugated nanoparticles can readily and specifically identify a variety of bacterium, such as Escherichia coli O157:H7, through antibody–antigen interaction and recognition. The single-bacterium-detection capability within 20 min has been confirmed by the plate-counting method and realized by using two independent optical techniques. The two detection methods correlated extremely well. Furthermore, we were able to detect multiple bacterial samples with high throughput by using a 384-well microplate format. To show the usefulness of this assay, we have accurately detected 1–400 E. coli O157 bacterial cells in spiked ground beef samples. Our results demonstrate the potential for a broad application of bioconjugated nanoparticles in practical biotechnological and medical applications in various biodetection systems. The ultimate power of integrating bionanotechnology into complex biological systems will emerge as a revolutionary tool for ultrasensitive detection of disease markers and infectious agents.
We have developed a bioassay for the accurate determination of a single bacterial cell within 20 min by using bioconjugated nanoparticles in a fluorescence-based immunoassay. The analysis of bacteria is vital for food safety, clinical diagnosis and therapies, portable water, and prevention strategies to combat bioterrorism agents. Escherichia coli O157:H7 is one of the most dangerous agents of food-borne diseases (1). Several of the reported outbreaks of E. coli O157:H7 have led to death, especially in cases involving children and the elderly (2–5). Given the low infectious dose of E. coli O157:H7 (≈10–100 cells), the presence of even a single bacterium in food may pose a serious health risk (1, 6). Therefore, a simple, rapid, and sensitive detection of trace amounts of E. coli O157:H7 and other bacterial pathogens is critical for minimizing or eliminating potential infections. Traditional methods for the detection of trace amounts of bacteria require amplification or enrichment of the target bacteria in the sample (1, 7, 8). These methods tend to be laborious and time-consuming because of the complicated assay procedures (9–18). Recently, many attempts have been made to improve the sensitivity of bacteria detection without the need for target amplification and enrichment (19–21). However, rapid bacteria detection at the single-cell level, in a given sample, has been quite challenging.
The two major challenges for the rapid detection of a single bacterium are the achievement of (i) short to real-time detection and (ii) ultrasensitivity in bioanalysis. To reduce the time required for target detection, a minimal amount of sample manipulation is essential. The sensitivity of the detection method has to be high enough to eliminate the need for target amplification and enrichment steps and also allow for the accurate identification of a single bacterium in a short period. Recently, many novel techniques have been developed to amplify analytical signals from biorecognition events to improve the sensitivity of various bioassays for bacteria detection (22–24).
Using fluorescent-bioconjugated silica nanoparticles, we have developed ultrasensitive methods for bioassays (25–27). Each nanoparticle encapsulates thousands of fluorescent dye molecules in a protective silica matrix, providing a highly amplified and reproducible signal for fluorescence-based bioanalysis. Compared with conventional immunoassays, where only one or a few dye molecules are linked to an antibody molecule and then used to signal an antibody–antigen binding event, the bioconjugated nanoparticles enable significant amplification of the analytical signal because of the many dye molecules inside each nanoparticle, which is attached to the antibody molecule. For a bacterium, there are many surface antigens available for specific recognition by using antibody-conjugated nanoparticles. Therefore, thousands of nanoparticles can bind to each bacterium, thereby producing a greatly amplified signal. Using these nanoparticles, we have developed an assay tool, enabling the detection of one bacterium cell per given sample in <20 min with a spectrofluorometer. In addition, we have designed a simple flow cytometry device to detect antibody-conjugated nanoparticles bound to single bacterial cells. The two detection methods correlated extremely well. Furthermore, we were able to detect multiple samples with high throughput by using a 384-well microplate format. To show the usefulness of this assay, we have accurately detected 1–400 E. coli O157 bacterial cells in spiked ground beef samples.
Experimental Protocol
Materials. Tetraethylorthosilicate, Triton X-100, Tris(2,2′-bipyridyl) dichlororuthenium(II) hexahydrate (RuBpy), succinic anhydride, Z-morpholinoethanesulfonic acid (Mes), BSA, 1-ethyl-3–3(3-dimethylaminopropyl) carbodiimide hydrochloride, and N-hydroxy-succinimide were purchased from Sigma-Aldrich. Trimethoxysilyl-propyldiethylenetriamine was purchased from United Chemical Technologies (Bristol, PA), and N-(trimethoxysilylpropyl)-ethylenediamine was purchased from Gelest (Morrisville, PA). Ammonium hydroxide (28–30 wt%), N,N-dimethylformamide, and all other chemicals of analytical reagent grade were obtained from Fisher Scientific. mAbs against E. coli O157:H7 were purchased from Biodesign International (Kennebunkport, ME). E. coli O157:H7 and E. coli DH5α were obtained from the American Type Culture Collection. Distilled, deionized water (EasyPure LF, Barnstead) was used in the preparation of all aqueous solutions.
Instrumentation. Spectrofluorometric analysis was done with a Tecan (Maennedorf, Switzerland) Spectrofluor Plus plate reader with magellan software. Dye-doped silica nanoparticle size and uniformity were measured with an H-7000 transmission electron microscope (data not shown) and an FE S-4000 scanning electron microscope (Hitachi, Tokyo). Fluorescence images were obtained with an inverted fluorescence microscope (IX70-S8F, (Olympus, Melville, NY) assembled with a charge-coupled device (CCD) camera (Pixera, Los Gatos, CA) and xenon lamp (Olympus) for excitation. The CCD camera was controlled with imageview and studio software (Pixeria). The laboratory-made flow cytometer used an Ar+ laser (model series 532, Omnichrome, Chino, CA) as the excitation light source. The sample flow channel was a glass capillary (i.d. 50 μm) purchased from Polymicro Technologies (Phoenix). The photomultiplier tube signal was sent to a computer interfaced with a data-acquisition card (NI DAQPad-6020E, National Instruments, Austin, TX) for data collection. The acquisition board was controlled with the National Instruments labview program, and data were analyzed with custom-made software.
The optical detection system in the flow cytometer is a homemade set-up, comprising a micrometer-sized capillary channel to flow the sample at a steady flow rate. An Ar+ Laser (Omnichrome), at 488 nm, is tightly focused to the central region of the channel to probe the bacteria species with bioconjugated nanoparticles. An ultrasensitive optical detection scheme was designed to detect the fluorescence signal as each bacterium passed through the probing volume. Fluorescence events produced at the probing region were collected by using a microscope objective (×40), followed by an optical beam splitter and filter system. Subsequently, the fluorescence signals caused by a single bacterial cell were detected with a highly sensitive photomultiplier tube (Hammamatsu, Middlesex, NJ), which has a built-in amplifier. The bursts of fluorescence from each bacterial species were recorded through a data-acquisition system (NI DAQPad-6020E) interfaced to a computer and analyzed with custom-built software (labview).
Chemical Modification of the Nanoparticle Surface. Before immobilizing mAbs against E. coli O157 onto the nanoparticles, the surfaces of the RuBpy-doped silica nanoparticles were chemically modified. To form the amine-functionalized group on the nanoparticle surfaces, 32 mg of silica nanoparticles was reacted with 20 ml of 1% trimethoxysilyl-propyldiethylenetriamine in 1 mM acetic acid for 30 min at room temperature, with continuous stirring. The amine-functionalized nanoparticles were obtained. These nanoparticles were thoroughly washed three times in distilled, deionized water. After washing with N,N-dimethylformamide, the nanoparticles were reacted with 10% succinic anhydride in N,N-dimethylformamide solution under N2 gas for 6 h with continuous stirring. By doing so, carboxyl groups were formed onto the silica nanoparticle surface for conjugation of antibodies. In an alternative nanoparticle synthesis method, carboxylated nanoparticles would be directly produced by adding a carboxylated siliane, N-(trimethoxysilylpropyl)-ethylenediamine, during the postcoating of the silica nanoparticles. With storage at 4°C, the chemically modified RuBpy-doped, silica-coated nanoparticles were viable for several months. After a thorough water wash, the carboxylated nanoparticles were activated by using 5 ml of 100 mg/ml 1-ethyl-3–3(3-dimethylaminopropyl) carbodiimide hydrochloride and 5 ml of 100 mg/ml of N-hydroxy-succinimide in a Z-morpholinoethanesulfonic acid (Mes) buffer (pH 6.8), for 25 min at room temperature with continuous stirring. Water-washed nanoparticles were dispersed in 10 ml of 0.1 M PBS (pH 7.3). To covalently immobilize mAbs against E. coli O157 onto the nanoparticle surface, 5 ml of 0.1 mg/ml nanoparticles was reacted with 2 ml of 5 μg/ml antibody for E. coli O157 for 2–4 h at room temperature with continuous stirring to form the resultant antibody-conjugated nanoparticles, followed by washing with a PBS buffer. To reduce the effects of nonspecific binding in the subsequent immunoassay, the antibody-conjugated nanoparticles were reacted with 1% BSA and washed in 0.1 M PBS (pH 7.3) before use.
Preparation of Single-Bacterium Samples. A 500-μl bacterial sample, which contained 25 bacteria based on plate-counting results, was dispersed into 1.0 ml of 0.1 mg/ml antibody-conjugated nanoparticles in a 0.1 M PBS buffer (pH 7.3) for 10 min. To remove the free antibody-conjugated nanoparticles that did not bind to the bacteria, the samples were centrifuged at 20,817 × g for 30 s, and then the supernatant was removed. The samples were washed again to remove all unbound antibody-conjugated nanoparticles, and 1.5 ml of PBS buffer was added to the samples. The sample was divided into 100 aliquots and transferred to individual cuvettes or a 384-well plate, and 85 μl of the PBS buffer was added to each to provide 100 μl of detectable sample volume. A multipipettor was used for sample preparation. The fluorescence intensity in each sample was detected with 430-nm excitation and 595-nm emission by using a Tecan Spectrofluor Plus fluorometer. Control samples were obtained by using the same experimental procedures but without the addition of bacteria. Average fluorescence intensity of the controls was considered background. Signals above background plus three times the SD were considered to be positive signals. To obtain reliable statistical results, >20 samples in each test were prepared and analyzed at the same time. The total time spent on each determination was <20 min. Other bacterial samples, such as E. coli O157, Salmonella, and Bacillus cereus, were prepared with a similar procedure and analyzed with the same strategy. Specific antibodies were used for the recognition of the individual bacterial samples.
Preparation of Ground Beef Samples. Fresh ground beef was purchased from a local grocery store and ground further with a blender into a paste-like consistency. Several 25-g ground beef samples were divided into 25 1-g samples and stored in sterile 15-ml conical tubes. Freshly cultured bacteria [≈109 colony-forming units (CFU)/ml] were serially diluted 10-fold until there was 1 cell per ml (as determined by CFU count) in a 0.1 M PBS buffer (pH 7.3). The spiked samples were then prepared by adding 1 ml of different concentrations of E. coli O157:H7 to the 1-g ground beef samples. The samples were mixed by vortexing. Then, 8 ml of buffer E (0.05% Tween 20 and 0.5% bovine albumin in the 0.1 M PBS buffer, pH 7.3) was added to each ground beef sample. The entire slurry of ground beef, bacteria, and buffer was mixed by using a vortex for 1 min, followed by centrifugation at 500 × g for 5 min. At this point, there were three layers in the tubes: (i) a bottom layer of ground beef, (ii) a middle layer of buffer containing the E. coli O157:H7, and (iii) a top layer of fat. The middle layer was removed for testing; half of the sample was detected with the spectrofluorometer-based method, and the other half was detected with the conventional plating method as described above. Sterile PBS buffer was used in place of the E. coli O157:H7 solution (i.e., middle layer of the sample mixture) in negative controls. For positive controls, the ground beef was replaced with the PBS buffer. Five parallel samples were prepared for each concentration.
Results
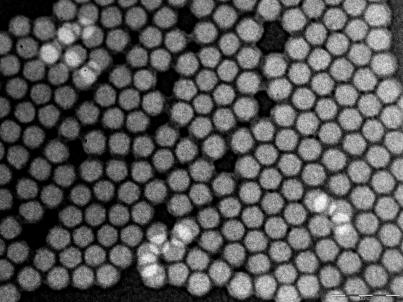
Highly Photostable Dye-Doped Silica Nanoparticles for Antibody Conjugation. We synthesized RuBpy-doped silica nanoparticles (17) and several other dye-doped silica nanoparticles (28) with a water-in-oil microemulsion method. The dye molecules were trapped inside a silica matrix to form the dye-doped nanoparticles. The size of the nanoparticles was uniform, with a diameter of 60 ± 4 nm (Fig. 1). The size of the nanoparticles could be manipulated, as needed, by changing the water-to-surfactant molar ratio (W0 value, for 60 nm, W0 = 10) (25, 26). Because of the protective function of the silica matrix, the nanoparticles were highly photostable. To verify the photostability of the nanoparticles, we compared pure RuBpy dye molecules with RuBpy-doped silica nanoparticles. Both of them were continuously irradiated with 450 nm of light for 1,000 s. The fluorescence intensity of the pure dye molecules was reduced by 81%, whereas that of the nanoparticles remained constant (data not shown). This high photostability of the nanoparticles provides a foundation for precise and reproducible bioanalytical measurements. The silica matrix not only provides the high photostability of the dye molecules inside the nanoparticle, but it also enables the combination of biology and nanotechnology (i.e., the conjugation of various biomolecules to the nanoparticles). After biochemical modification of the nanoparticle surface, mAbs against the O-antigen of E. coli O157:H7 were covalently immobilized onto the nanoparticles, which were then used in the immunoassay. When stored at 4°C, the antibody-nanoparticle conjugates are viable for antigen recognition for up to 4 weeks, whereas the nanoparticles are stable for several months when stored at –20°C.
Fig. 1.
Fluorescent nanoparticles. Transmission electron microscope image of RuBpy-doped silica nanoparticles before bioconjugation. Each of these nanoparticles contains tens of thousands of dye molecules inside, emits strong fluorescence signal, has excellent photostability, and can be used for easy and effective biomolecule conjugation for biorecognition.
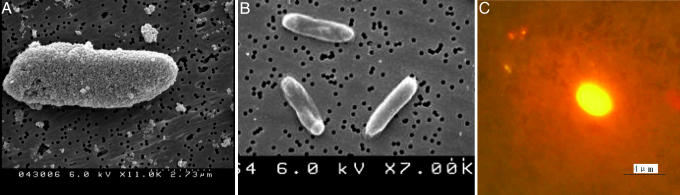
Significant Signal Amplification of Antibody-Bioconjugated Nanoparticles. Because of the thousands of dye molecules encapsulated within each nanoparticle, high signal amplification was achieved when the antibody-conjugated nanoparticles bound to antigens on the surface of the bacteria. The mAb was highly selective for E. coli O157:H7 in the immunoassay because the antibody-conjugated nanoparticles specifically associated with E. coli O157:H7 cell surfaces (Fig. 2A) but not with E. coli DH5α, which lacks the surface O157:H7 antigen (Fig. 2B). The scanning electron microscope image of the E. coli O157:H7 cell after incubation with the nanoparticles shows that there were thousands of antibody-conjugated nanoparticles bound to a single bacterium, providing significant fluorescent signal amplification as compared with a single dye molecule. The nanoparticle-based signal amplification can be easily seen in a fluorescent image, as shown in Fig. 2C. After 20 min of continuous excitation, the fluorescence intensity remained constant. In solution-based experiments for bacteria detection, we have demonstrated signal amplification by the antibody-conjugated nanoparticles that is >1,000 times greater than that produced with dye molecule-labeled antibody. In the comparison experiment, an organic fluorophore, tetramethylrhodamine, was chosen to label E. coli O157:H7, and the conjugated dye-labeled antibody was used for bacterium imaging. The signal from the bacterium cell was weak. The high fluorescence signal enhancement by the nanoparticle-based antibody provides the foundation for the rapid detection of a single bacterium in solution samples.
Fig. 2.
Images of bacterial cells. (A) Scanning electron microscope image of E. coli O157:H7 cell incubated with antibody-conjugated nanoparticles. (B) Scanning electron microscope image of E. coli DH5α cell (negative control) incubated with nanoparticles conjugated with antibody for E. coli O157:H7. (C) Fluorescence image of E. coli O157:H7 after incubation with antibody-conjugated nanoparticles. The fluorescence intensity is strong, enabling single-bacterium cell identification in aqueous solution.
Detection of a Single Bacterial Cell Within 20 Min. To achieve fast single-bacterium detection, we used a spectrofluorometer to detect the fluorescence signals of single bacterial cell samples in solution. The spectrofluorometer-based method needed only minimum sample preparation, as specified in Experimental Protocols. The assay was rapid, taking <20 min to complete sample preparation, instrumentation preparation, and sample determination. To confirm that we were able to detect a single bacterium, a sampling method was designed to ensure reproducibility and accuracy of detecting single bacterial cells. The single-bacterium counting method was based on a technique used in single-molecule studies (29), where the sample was diluted to a concentration in which there was only a 25% chance that a specific volume of the sample would have a bacterium cell. Based on OD600 data and further verification by plate counting, a golden standard method in bacterium counting in microbiology and cell biology (30), E. coli O157:H7 was accurately diluted into 10 cells per sample. Then, each sample was divided into 40 aliquots. By using the conventional plating method (30), each aliquot was plated and grown on an agar plate for 16–18 h in a 37°C incubator. Each plate either had none or only one CFU, confirming that our sample-preparation method enabled us to obtain samples with single bacterial cells.
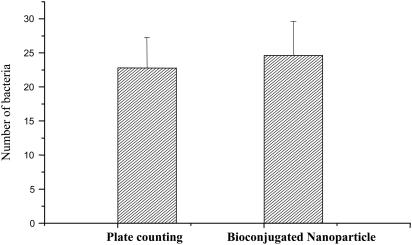
To obtain an accurate bacteria count, we first prepared 21 samples with ≈50 bacteria in each sample. Each sample was then divided into two parts. The first half of each sample was grown on agar plates to obtain an accurate number of bacterium by CFU counts as described above. The result showed that the average bacterial count was 22.8 ± 4.4 (mean ± SD), as shown in Fig. 3. The second half of each sample was used for single bacterial cell determination by using a spectrofluorometer. After incubating with the antibody-conjugated nanoparticles and washing, as described in Experimental Protocol, the second part of the 21 samples was used for fluorescence measurements. A bacterial cell was confirmed only when the fluorescence intensity was above the background plus three times the SD of the controls. Control samples were obtained by using the same experimental procedures, but without the addition of bacteria or nontarget bacteria of E. coli DH5α. For the 21 tested samples, the average number of bacteria was 24.6 ± 5.0, as shown in Fig. 3. The fluorescence-based bacterial detection results highly correlated with that of the plating method, confirming the validity of the spectrofluorometer method for single bacterial cell detection.
Fig. 3.
Single-bacterium quantitation. Comparison of single bacterium detection with the plate-counting method [a golden standard for bacteria counting (22)] vs. the spectrofluorometer method with antibody-conjugated nanoparticles. In both experiments, 21 samples of 25 bacteria were used. The variation in counted number of bacteria is caused by the sampling nature of bacteria-containing solutions, not the detection methods.
High-Throughput Determination of Single Bacterium. The high-throughput determination of multiple bacterial samples is critical in toxicology screening, the detection of bioterrorism agents, and medical diagnosis. The single-bacterium assay can be adapted for multiple-sample determination, as shown in the previous section, when many aliquots of samples were tested simultaneously. With the plate reader fluorometer, we were able to detect >300 samples at one time with a single-bacterium detection limit. The identification of a bacterium was based on the fluorescence intensities measured in each well of a 384-well plate. By using the same procedure as described in the previous section, each aliquot of the sample, with a 25% probability of having a single bacterium, was dispensed into the 384-well plates. For a given plate, control samples without bacteria were added into 20 wells of the plate for the determination of background signal and SD. The total number of bacteria in each sample was counted based on the same statistical method detailed in Experimental Protocol. An example of fluorescence intensity signals obtained from one bacteria sample is shown in Fig. 6, which is published as supporting information on the PNAS web site. Using this method, we were able to determine the existence of a single bacterium with 99.99% accuracy when compared with the golden standard for CFU count on agar plates (22).
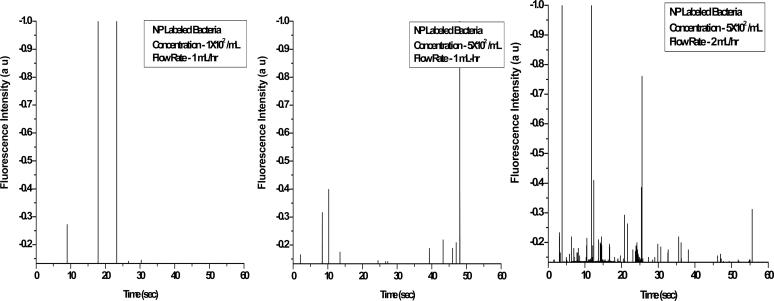
Single Bacterial Cell Detection with a Simple Optical Flow Cytometer. To further confirm the above results, we also detected the antibody-conjugated nanoparticles bound to bacteria samples by using a laboratory-made flow cytometer, which can precisely detect a single bacterial cell by giving a fluorescent spike when the cell flows through the detection zone (a channel in this case). Laser excitation and an optical design for the collection of the fluorescence emission in the orthogonal direction of the forward scattered light beam made the cytometric analysis more efficient and accurate. In the current scheme, a micrometer-sized capillary flow cell and the narrow focusing of the excitation light beam reduces the probing volume of the sample to a few picoliters. Moreover, this design decreases the chance of detection of two or multiple events simultaneously. The total time for the sample detection and analysis with the present system was 60 s, which minimized the duration of the bacterial assay even further. Detailed information about the set-up is described in Experimental Protocol. Fig. 4 shows the measurement of typical fluorescence bursts as different concentrations of bacteria flow through the homemade cytometry. Each spike, which was higher than the background plus 3 σ, represented one bacterial cell. The height of the spikes was not uniform, which might be caused partly by the rod-shaped bacterial rotation as the signal beams are collected by the detector.
Fig. 4.
A laboratory-made flow cytometer was used to detect single bacterium. Detection of different concentrations of bacteria after incubation with antibody-conjugated nanoparticles (NP) was done with a laboratory-made flow cytometer. The trace was recorded under different experimental conditions as those described in the keys.
Multiple-Pathogen Quantitation. We have also used highly fluorescent-bioconjugated nanoparticles for in situ rapid, simultaneous multiple pathogen quantification in water samples with the ability to detect pathogens with one bacterium sensitivity. Using the same principle and the same strategies, we tested other bacteria and spores such as Salmonella typhimurium and B. cereus. The nanoparticles with antibodies specific to the target pathogens immobilized at the nanoparticle surface can quantitate the presence of pathogens in our artificial aqueous samples. We have demonstrated the use of this method for simultaneous quantification of model pathogens, E. coli O157, S. typhimurium, and B. cereus spores. We were able to count the individual target cells. This result clearly shows that our bioassay, using a simple principle and an easily implementable experimental strategy, can be widely useful for rapid and ultrasensitive detection of multiple target cells with high specificity.
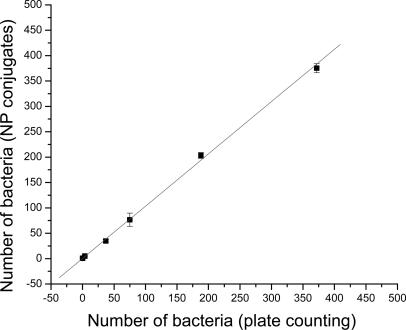
Single-Bacterium Determination with Beef Samples. To test the usefulness of our bioassay for bacteria detection in real samples, we determined the number of E. coli O157:H7 in several spiked ground beef samples. Following a reported sample preparation method (21), the recovery rate of the spiked bacteria from the ground beef increased from 50% to 90% as the number of spiked bacteria increased from 2 to 400. The recovered samples were equally divided into two portions as described above. One portion was used for the CFU count on LB agar plates, whereas the other portion was subjected to fluorescence detection with the antibody-conjugated nanoparticles. It should be noted that the colony morphology of the E. coli O157:H7 on LB agar was easily distinguishable from other bacteria derived from the ground beef. As shown in Fig. 5, the number of bacterial cells determined by the two methods highly correlated, with a correlation factor of 0.99. This result clearly demonstrates that our bacterium assay based on bioconjugated nanoparticles can be used to effectively detect a single bacterium in solution recovered from a ground beef sample within 20 min. We conducted both positive and negative control experiments to confirm that the effects of potential interference, such as fat in the ground beef, were negligible.
Fig. 5.
Single-bacterium detection with beef sample. Detection of E. coli O157:H7 in spiked ground beef was done with the plate-counting method and the antibody-conjugated, nanoparticle (NP)-based method. Bacteria in the range of 1–400 cells per sample were detected. The two methods had linear correlation, with an R value of 0.99. Total detection time for the beef sample was ≈20 min for the antibody-conjugated, nanoparticle-based method, and that for the plate-counting method was >1 day.
Conclusion and Discussion
In summary, we have developed a fast and ultrasensitive immunological method for bacterial detection that uses antibody-conjugated nanoparticles. A single bacterium can be detected quickly and accurately without any amplification or enrichment. Antibody-bioconjugated nanoparticles provide significant signal amplification for the bioanalysis and enumeration of bacteria. This bioassay is rapid (<20 min from bacterium binding to detection and analysis and could be further shortened), convenient, and highly selective. Furthermore, because multiple samples can be analyzed simultaneously, this assay is adaptable to high-throughput bioanalysis for multiple pathogens. In addition, the accurate and reliable detection of trace amounts of E. coli O157:H7 bacteria in spiked ground beef samples demonstrates the practical usefulness of this assay system. This study clearly exhibits the excellent properties of bioconjugated nanomaterials in applications in bioanalysis and biodetection. This bionanotechnology could be adapted in studies using antibodies specific for various bacterial pathogens for the detection of a wide variety of bacterial pathogens used as bioterrorism agents in food, clinical samples, and environmental samples. Our results demonstrate the potential for a broad application of this type of bionanotechnology in practical biotechnological applications in various biodetection systems.
Recently, nanomaterials have demonstrated their unique advantages when they are combined with biomolecules for bioanalysis and biotechnology applications. The demand for highly sensitive nonisotopic bioanalysis systems for biotechnology applications, such as in clinical diagnostics, food quality control, drug delivery, etc., has driven nanomaterials more toward biomedical fields and biotechnology. Each of the nanoparticles described in this article can emit an extremely strong fluorescent signal, enabling us to achieve enormous signal amplification for ultrasensitive target detection and for monitoring rare events that would be otherwise undetectable with existing labeling technologies. Furthermore, the nanoscale size of the particles minimized physical interference with the biological recognition events, whereas the nature of silica particles enables us to easily modify the surface for conjugation with various biomolecules for a wide range of applications in the bioassay systems. Moreover, the potential to prepare the nanoparticles with any existing fluorophores provides the diversity of nanoparticles for various applications. By integrating nanotechnology into complex biological systems, we can achieve the detection and prevention of disease at the earliest stages of its development. Nanotechnology promises scientific and commercial opportunities that are virtually unimaginable at this time. The ultimate power of the bioconjugated nanoparticles will emerge as a revolutionary tool for ultrasensitive detection of disease markers as well as infectious agents. Indeed, by using the dye-doped nanoparticles as fluorescent markers, highly sensitive target detection has been achieved (27), opening the possibility for the fabrication of truly smart bioprobes and biosensors for rapid and ultrasensitive determination of bacterium samples.
Supplementary Material
Acknowledgments
W.T. was supported by grants from the National Science Foundation, the Packard Foundation, and the National Institutes of Health, and S.J. was supported by grants from the National Institutes of Health and the Cystic Fibrosis Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RuBpy, Tris(2,2′-bipyridyl) dichlororuthenium(II) hexahydrate; CFU, colonyforming units.
References
- 1.Phillips, C. A. (1999) J. Sci. Food Agric. 79, 1367–1381. [Google Scholar]
- 2.Groseclose, S. L., Brathwaite, W. S., Hall, P. A., Conner, F. J., Sharp, P., Anderson, W. J., Fagan, R. F., Aponte, J. J., Jones, G. F., Nitschke, D. A., et al. (2004) Morbid. Mortal. Wkly. Rep. 51, 1–84. [PubMed] [Google Scholar]
- 3.Varma, J. K., Greene, K. D., Reller, M. E., DeLong, S. M., Trottier, J., Nowicki, S. F., DiOrio, M., Koch, E. M., Bannerman, T. L., York, S. T., et al. (2003) J. Am. Med. Assoc. 290, 2709–2712. [DOI] [PubMed] [Google Scholar]
- 4.Bruce, M.G., Curtis, M. B., Payne, M. M., Gautom, R. K., Thompson, E. C., Bennett, A. L. & Kobayashi, J. I. (2003) Arch. Pediatr. Adolescent Med. 157, 1016–1021. [DOI] [PubMed] [Google Scholar]
- 5.Mohle-Boetani, J. C., Farrar, J. A., Werner, S. B., Minassian, D., Bryant, R., Abbott, S., Slutsker, L. & Vugia, D. J. (2001) Ann. Intern. Med. 135, 239–247. [DOI] [PubMed] [Google Scholar]
- 6.Nataro, J. P. & Kaper, J. B. (1998) Clin. Microbiol. Rev. 11, 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deisingh, A. K. & Thompson, M. (2004) J. Appl. Microbiol. 96, 419–429. [DOI] [PubMed] [Google Scholar]
- 8.Iqbal, S. S., Mayo, W. M., Bruno, J. G., Bronk, B. V., Batt, C. A. & Chambers, J. P. (2000) Biosens. Bioelectron. 15, 549–578. [DOI] [PubMed] [Google Scholar]
- 9.Alexandre, M. & Prado, V. (2003) Exp. Rev. Mol. Diagn. 3, 105–115. [DOI] [PubMed] [Google Scholar]
- 10.Bettelheim, K. A. & Beutin, L. (2003) J. Appl. Microbiol. 95, 205–217. [DOI] [PubMed] [Google Scholar]
- 11.Ibekwe, A. M. & Grieve, C. M. (2003) J. Appl. Microbiol. 94, 421–431. [DOI] [PubMed] [Google Scholar]
- 12.Kourkine, I. V., Ristic-Petrovic, M., Davis, E., Ruffolo, C. G., Kapsalis, A. & Barron, A. (2003) Electrophoresis 24, 655–661. [DOI] [PubMed] [Google Scholar]
- 13.Feldsine, P. T., Kerr, D. E., Leung, S. C., Lienau, A. H., Miller, S. M., Mui, L. A., Anderson, G., Beasley, M., Dillon, J., Dombroski, P., et al. (2002) J. Assoc. Off. Anal. Chem. Int. 85, 1037–1044. [Google Scholar]
- 14.Tu, S.-I., Golden, M., Andreotti, P. & Irwin, P. (2002) J. Rapid Methods Automat. Microbiol. 10, 37–48. [Google Scholar]
- 15.Yu, L. S. L., Reed, S. A & Golden, M. H. (2002) J. Microbiol. Methods 49, 63–68. [DOI] [PubMed] [Google Scholar]
- 16.Henry, Y. M., Natrajan, N. & Lauer, W. F. (2001) J. Assoc. Off. Anal. Chem. Int. 84, 752–760. [PubMed] [Google Scholar]
- 17.Edwards, R., ed. (1996) Immunoassays: Essential Data (Wiley, New York), pp. 84–87.
- 18.Delves, P. J., ed. (1995) Antibody Applications: Essential Techniques (Wiley, New York), pp. 43–51.
- 19.Song, J. M. & Vo-Dinh, T. (2004) Anal. Chim. Acta 507, 115–121. [Google Scholar]
- 20.Muhammad-Tahir, Z. & Alocilja, E. C. (2003) IEEE Sens. J. 3, 345–351. [Google Scholar]
- 21.Weimer, B. C., Walsh, M. K., Beer, C., Koka, R. & Wang, X. (2001) Appl. Environ. Microbiol. 67, 1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, J. A. & Hsu, H-W. (2003) Anal. Chem. 75, 4330–4334. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M., Oh, S. & Durst, R. A. (2003) J. Microbiol. Biotechnol. 13, 509–516. [Google Scholar]
- 24.Wu, C.-F., Valdes, J. J., Bentley, W. E. & Sekowski, J. W. (2003) Biosens. Bioelectron. 19, 1–8. [DOI] [PubMed] [Google Scholar]
- 25.Santra, S., Zhang, P., Wang, K., Tapec, R. & Tan, W. (2001) Anal. Chem. 73, 4988–4993. [DOI] [PubMed] [Google Scholar]
- 26.Zhao, X., Tapec-Dytioco, R., Wang, K. & Tan, W. (2003) Anal. Chem. 75, 3476–3483. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, X., Tapec-Dytioco, R. & Tan, W. (2003) J. Am. Chem. Soc. 125, 11474–11475. [DOI] [PubMed] [Google Scholar]
- 28.Zhao, X., Bagwe, R. & Tan, W. (2004) Adv. Mater. 16, 173–176. [Google Scholar]
- 29.Nie, S & Zare, R. N. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 567–596. [DOI] [PubMed] [Google Scholar]
- 30.Boyd, R. F. (1988) General Microbiology (Times Mirror/Mosby, St. Louis), 2nd Ed., pp. 401–404.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.