Abstract
Recent studies have demonstrated that multiple co-occurring global changes can alter the abundance, diversity, and productivity of plant communities. Belowground processes, often mediated by soil microorganisms, are central to the response of these communities to global change. Very little is known, however, about the effects of multiple global changes on microbial communities. We examined the response of ammonia-oxidizing bacteria (AOB), microorganisms that mediate the transformation of ammonium into nitrite, to simultaneous increases in atmospheric CO2, precipitation, temperature, and nitrogen deposition, manipulated on the ecosystem level in a California grassland. Both the community structure and abundance of AOB responded to these simulated global changes. Increased nitrogen deposition significantly altered the structure of the ammonia-oxidizing community, consistently shifting the community toward dominance by bacteria most closely related to Nitrosospira sp. 2. This shift was most pronounced when temperature and precipitation were not increased. Total abundance of AOB significantly decreased in response to increased atmospheric CO2. This decrease was most pronounced when precipitation was also increased. Shifts in community composition were associated with increases in nitrification, but changes in abundance were not. These results demonstrate that microbial communities can be consistently altered by global changes and that these changes can have implications for ecosystem function.
Human activity is profoundly altering ecological systems. These alterations include increases in atmospheric CO2 due to fossil-fuel use and land-use change, with subsequent changes in air temperature, precipitation, and the deposition of nitrogen-containing compounds (1, 2). Studies of the ecological responses to these global changes have suggested that belowground processes, often mediated by soil microorganisms, are central to the response of ecological systems to global change (3, 4). Below-ground microbially mediated processes can both immobilize and release nutrients that limit primary production and can influence the long-term response of ecosystems to global change.
Although it is clear that multiple global changes can interact to alter communities of macroorganisms (1), it is less clear how microbial communities respond to such changes. There is some evidence that the aggregate properties of microbial communities (e.g., total microbial biomass, rates of microbial respiration, or biogeochemical transformations) can be altered by individual global changes (5). Some studies have also documented shifts in the abundance of microbial symbionts of plants in response to increased CO2 (6–8). The abundance and diversity of some free-living soil microorganisms have been shown to shift in response to agricultural fertilization, large changes in soil moisture, and extremes of soil temperature (9, 10). It is not clear, however, whether the abundance or diversity of soil microorganisms can be altered by the relatively subtle effects of realistic multifactorial global changes.
We investigated the response of soil bacteria to simulated multifactorial global change as part of the Jasper Ridge Global Change Experiment (JRGCE). The JRGCE is located on the Jasper Ridge Biological Preserve in the eastern foothills of the Santa Cruz Mountains in central California. The climate, grassland vegetation, and soil parameters, as well as the experimental design, are described in detail in refs. 1 and 8. The value of the multifactorial approach used in the JRGCE is evident from a number of studies (1, 11–13). For example, Shaw and colleagues observed that plant biomass in the JRGCE responded positively to increased precipitation, temperature, and nitrogen only when atmospheric CO2 was at ambient concentrations; these responses were dampened under elevated CO2 (1). Such antagonistic interactions between CO2 and other global-change factors had not previously been suspected.
We chose to focus on chemolitho-autotrophic ammonia-oxidizing bacteria (AOB) from the β subclass of the Proteobacteria as a model free-living bacterial group. This group of bacteria is an ideal model group for microbial ecology studies for a number of reasons (14). First, the bacteria are ubiquitous; they are found in nearly all soil, freshwater, and marine environments. Second, they are of great environmental importance; ammonia oxidation is the rate-limiting step of nitrification and is thus central to the global nitrogen cycle. Third, nitrification enzyme activity can be altered by environmental changes, including simulated global changes (15, 16). Finally, all AOB carry the gene amoA, which codes for the α-subunit of the ammonia monooxygenase enzyme, the protein that catalyzes the rate-limiting step in ammonia oxidation. The ubiquity of this gene among AOB has facilitated the development of amoA as a molecular marker, allowing the detection and enumeration of AOB without requiring laboratory culture. Because the vast majority of soil bacteria cannot be cultured in the laboratory, the accurate assessment of abundance and diversity requires the use of such molecular markers.
We report here the response of AOB community structure and abundance to simultaneous increases in atmospheric CO2, precipitation, atmospheric temperature, and nitrogen deposition.
Methods
JRGCE. The JRGCE was established in a grassland ecosystem dominated by annual grasses (Avena barbata and Bromus hordeaceus) and forbs (Geranium dissectum and Erodium botrys) growing in a sandstone-derived soil. The experiment was initiated in the fall of 1998 and includes four global-change manipulations with two levels: (i) ambient and elevated atmospheric CO2 (with a target concentration of 700 ppm in the elevated treatment) manipulated by means of a free air (FACE) system, (ii) ambient and elevated temperature (with a mean increase in the plant canopy of 0.8–1.0°C in the elevated treatment) manipulated with heat lamps, (iii) ambient and elevated nitrogen deposition (7 g·m–2·yr–1 in the elevated treatment) manipulated by application of Ca(NO3)2, and (iv) ambient and increased precipitation (with the elevated treatment 50% over ambient) manipulated by means of a spray irrigation system. The experiment was of a randomized block split-plot design, with atmospheric CO2 and temperature manipulated at the plot level (circles of grassland 2 m in diameter) and nitrogen deposition and precipitation manipulated at the subplot level (in 0.78-m2 wedge-shaped quadrants). Each of the quadrants was separated by aboveground (mesh) and belowground (solid) partitions. Each of the 16 possible treatment combinations was replicated eight times.
Soil Sampling. Four soil cores from each subplot were removed aseptically to a depth of 15 cm with a 2.2-cm-diameter corer. The cores were taken in late April 2000, at the end of the second growing season of the experiment. The soil was stored frozen until analysis. We used the UltraSoil DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA) to extract total community DNA from a 0.5-g subsample of the consolidated cores.
PCR Amplification and Sequencing of amoA. We initially assessed the different types of AOB in the JRGCE by PCR-amplifying amoA genes extracted from soil sampled from a subset of the plots (four ambient plots and four four-factor plots). In total, 77 clones were selected for sequencing. Because of the low abundance of AOB in our soils, amplification of amoA genes was performed by using a nested PCR approach, with aliquots of the first-round PCR products used as templates in the second round of PCR (see Supporting Text, which is published as supporting information on the PNAS web site). Nonradioactive sequencing of cloned amoA fragments was carried out as described in ref. 17. The identities of the amoA-gene sequences were confirmed by searching the international sequence databases with the blast program (www.ncbi.nlm.nih.gov/blast).
Phylogenetic Analysis. We used the maximum-likelihood algorithm of the arb software package (www.mikro.biologie.tumuenchen.de/pub/ARB) for phylogenetic inference. The robustness of the tree topology was verified by comparing with a neighbor-joining tree (18), constructed with arb software by using the Jukes–Cantor correction (19). The influence of the third codon position on the tree topology was tested by constructing a maximum-likelihood tree in which all nucleotides were specified according to their respective positions in the codons with paup 4.0 b10 software (Sinauer Associates, Sunderland, MA). Bootstrap analysis was performed (with 1,000 iterations) with arb software. The partial amoA gene sequences obtained in this study have been deposited in the European Molecular Biology Laboratory, GenBank, and DNA Data Base in Japan nucleotide sequence databases (accession nos. AY369266–AY369342).
Terminal Restriction Fragment Length Polymorphism (T-RFLP) Analysis of amoA. As a rapid screen for the relative abundance of different AOB groups, T-RFLP analysis of amoA fragments was performed as described by Horz et al. (20). The restriction enzymes TaqI, RsaI, and Bsh12362I were used, with reactions performed according to the protocols of the suppliers [New England Biolabs and Fermentas Life Sciences (Hanover, MD)]. Electrophoresis and determination of the relative abundance of the individual terminal restriction fragments were performed at the Genomics Technology Support Facility of Michigan State University (East Lansing; http://genomics.msu.edu). T-RFLP analysis with TaqI was used to determine whether Nitrosomonas species were likely to be present in our soil samples. The enzyme RsaI was used to distinguish clade JR1 (which produced a unique 210-bp terminal restriction fragment when cut with this enzyme) from other AOB present in the JRGCE. The enzyme Bsh12362I was used on a randomly chosen subset of 25 soil samples to verify the specificity of the RsaI assays. (JR1 also produces a unique terminal restriction fragment when cut with Bsh12362I.)
Quantitative PCR. Quantitative PCR (“real-time PCR”) of amoA was used to estimate the abundance of AOB (21). The quantification was based on the fluorescent dye SYBR-Green I (Molecular Probes), which binds to double-stranded DNA during PCR amplification. The quantitative PCR protocol was identical to the PCR performed for T-RFLP analysis, except that the reaction mix contained 0.25× the concentration of SYBR-Green I and primers without fluoresceine labels. For the quantitative PCR, a reoptimization of the thermal profile was necessary, which led to changes in cycle number and elongation time relative to those of T-RFLP PCR (see Table 1, which is published as supporting information on the PNAS web site). The amount of initial template DNA was estimated by determining the threshold cycle, the number of PCR cycles required for the fluorescence to exceed a threshold value higher than the background fluorescence. All PCRs were performed in triplicate. Compounds that reduce PCR efficiency are commonly present in soil, and differences in PCR efficiency among samples can reduce the accuracy of DNA quantification with quantitative PCR. Therefore, we estimated the PCR efficiency of each of our soil samples and corrected for differences (see Supporting Text).
Statistical Analysis. The structure and abundance data were analyzed by using a split-plot analysis of variance. The analysis was performed by using both the general linear model (GLM) and mixed procedures in the SAS statistical package (SAS Institute, Cary, NC). The two approaches gave qualitatively identical results. The results of the mixed procedure are presented here; the results of the GLM procedure are available in Tables 2 and 3, which are published as supporting information on the PNAS web site. Before analysis, the structure data were arcsin-transformed and the abundance was log-transformed (after coding by multiplying by 10,000 to eliminate negative logarithms).
Potential Nitrification Experiment. We measured potential nitrification rates by following the protocol of Hart et al. (22). In brief, 7 g of soil was incubated in 70 ml of buffer solution containing 27 μg of NH4 per ml, and NO3 accumulation was measured over time. The assay was replicated eight times for each soil sample. The samples were divided into two groups according to the proportion of JR1 present (high or low) or the abundance of amoA (high or low). The means of the two groups were compared with Student's t test.
Results and Discussion
Community Structure. We initially assessed the different types of AOB in the JRGCE by PCR-amplifying amoA genes extracted from soil, cloning the amplicons, and sequencing 77 of these clones. All amoA genotypes grouped within the phylogenetic radiation of Nitrosospira, the genus previously observed to be dominant in terrestrial ecosystems (14). Our T-RFLP analysis with TaqI was consistent with our clone library results; we detected only the terminal restriction fragment primarily characteristic of Nitrosospira.
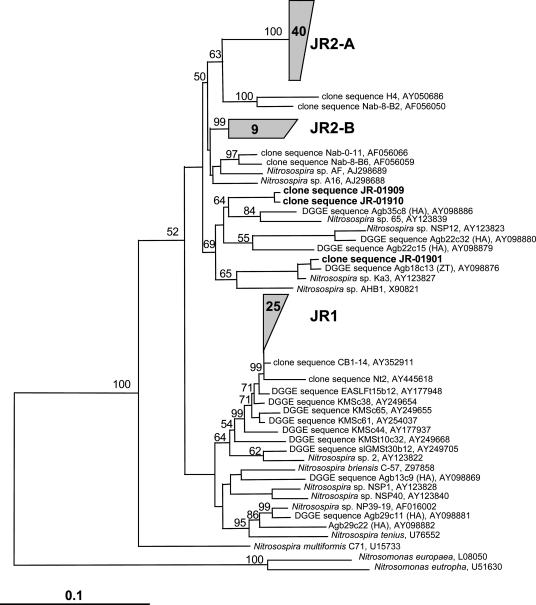
The sequences in our clone library clustered into three distinct clades: one (which we designated JR1) that was most closely related to Nitrosospira sp. 2 and two (which we designated JR2-A and JR2-B) that clustered outside of this group and were most closely related to Nitrosospira sp. isolate AF and isolate A16 (Fig. 1). The sequences in clade JR1 are nearly identical to sequence CB1-14 (accession no. AY352911), an amoA sequence cloned by Francis et al. (23). We developed a rapid screen for members of the JR1 clade based on T-RFLP of the amoA gene (20), which allowed us to determine the abundance of members of this clade relative to the total AOB abundance. This provided us a measure of the structure of the ammonia-oxidizing functional group, which could then be compared across the multifactorial global-change treatments.
Fig. 1.
The phylogenetic relationships among amoA gene types. Sequences obtained in this study are shown in bold with the prefix “JR”; amoA gene types available in public databases are shown for comparison. The majority of our sequences cluster together and are shown as boxes (JR1, JR-2A, and JR-2B). The numbers within the boxes indicate the number of sequenced clones. The tree was constructed by using the neighbor-joining approach with arb software. (Scale bar, 0.1 substitution per nucleotide.) The results of bootstrap analysis are indicated on the tree. DGGE, denaturing gradient gel electrophoresis.
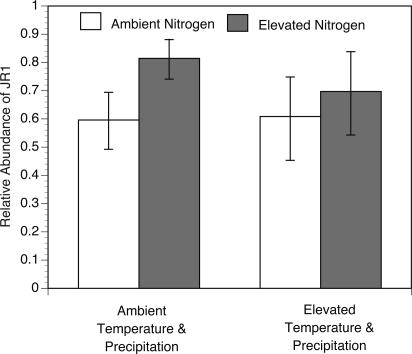
By using this approach, we observed that the structure of AOB responded significantly to simulated multifactorial global change. The structure of AOB shifted in response to nitrogen deposition, with an increased relative proportion of group JR1 with elevated nitrogen deposition (F1,26 = 10.83, P = 0.0029). There were also, however, significant interactions among the nitrogen, warming, and precipitation treatments (F1,26 = 6.91, P = 0.0142) such that the increase in the proportion of JR1 in response to increased nitrogen was greater when atmospheric temperature and precipitation were at ambient levels (Fig. 2).
Fig. 2.
The effect of nitrogen deposition, atmospheric temperature, and precipitation on the abundance of amoA gene type JR1 relative to the total AOB group. The mean relative abundance of JR1 is depicted for all samples, grouped by nitrogen, temperature, and precipitation treatments. For example, the first bar depicts the mean relative abundance of JR1 from all experimental plots under ambient nitrogen, ambient temperature, and ambient precipitation in the background of ambient and elevated CO2 treatments (n =16). Error bars, 95% confidence limits.
Could the shift in community structure that we observed be due to the presence of an AOB type not sampled by our clone library? It is possible that an AOB type unrelated to JR1 could generate the same terminal restriction fragment when cut with RsaI as JR1 and thus be misidentified by our rapid screen as a member of JR1. However, comparison of the T-RFLP results generated with different restriction enzymes suggests that this is unlikely. The relative abundance of JR1 estimated by T-RFLP analysis with RsaI was highly correlated with that estimated with Bsh12362I (r = 0.997, P < 0.0001, n = 25). It is highly unlikely that an organism unrelated to JR1/CB1-14 would have the same restriction sites for both of these enzymes.
The shift that we observed in community structure is consistent with other studies of AOB. Previous studies have reported that species from Nitrosospira 16S cluster 3 are often dominant under high nitrogen conditions (9, 24, 25), although not always (10). Clade JR1 is most closely related to species from cluster 3, whereas clades JR2-A and JR2-B are not; thus, our observation of an increase in the relative abundance of JR1 in response to nitrogen deposition is consistent with the majority of these previous studies. This finding is remarkable, given that nitrogen (as ammonium, rather than as nitrate) was applied at much higher rates in the previous studies than in our current study. It is plausible that clade JR1 may represent a “high-nitrogen” adapted clade. First, we observed that this clade was dominant under high-nitrogen conditions in our study. Second, Francis et al. (23) originally isolated JR1's twin, clone CB1-14, from a sediment sample taken from the upper reaches of an estuary with high levels of agricultural runoff and, consequently, very high concentrations of nitrate and ammonium.
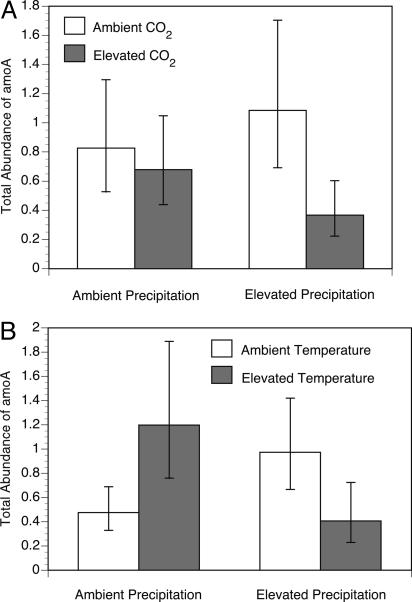
Community Abundance. In contrast to our results for community structure, nitrogen deposition had no effect on the total abundance of AOB. However, total abundance did respond to increased CO2, with the abundance of AOB significantly lower under elevated CO2 (F1,26 = 4.79, P = 0.0377; Fig. 3A). There was also a significant synergistic interaction between the CO2 and precipitation treatments (F1,26 = 4.85, P = 0.0368) such that the decrease in abundance under elevated CO2 was more pronounced when CO2 and precipitation were increased together (Fig. 3A). In addition, a significant antagonistic interaction between the temperature and precipitation treatments was observed (F1,26 = 19.09, P = 0.0002; Fig. 3B).
Fig. 3.
The effect of CO2 and precipitation (A) and atmospheric temperature and precipitation (B) on the abundance of the amoA gene. Abundance is standardized to a reference soil sample. Error bars, 95% confidence limits. The mean abundance of amoA is depicted for all samples, grouped by CO2 and precipitation treatments (A) and atmospheric temperature and precipitation (B). For example, the first bar in A depicts the mean total abundance of AOB from all experimental plots under ambient CO2 and ambient precipitation in the background of ambient and elevated nitrogen and temperature treatments (n = 32).
Could the difference in abundance we observed be due to shifts in structure rather than shifts in abundance? If an AOB species with a different amoA copy number changed in relative abundance, it is possible that this could appear as a change in total abundance of AOB even if total abundance did not change. However, the copy number of the amoA gene is believed to be constant (three copies per cell) for all Nitrosospira species except Nitrosospira tenuis, which has two copies (21, 26). None of the JRGCE sequences we cloned have N. tenuis as a close relative. Furthermore, the change in abundance we observed is too large to be explained as a change in amoA copy number from three to two alone. It is also likely that a shift in community structure would have been detected by our T-RFLP analysis, and we did not observe significant shifts in structure in response to increased CO2.
A previous study of culturable AOB in grassland soil reported no effect of elevated atmospheric CO2 on abundance (6). This is consistent with our observations; we observed a statistically significant effect only when both CO2 and water were elevated. Several studies have also reported significant increases in AOB abundance in response to nitrogen fertilization (21, 27, 28), an effect we did not observe in our system. However, those studies were designed to mimic agricultural fertilization, with nitrogen applied at much higher rates than in our study. In addition, manipulation in these studies occurred by means of a mixture of nitrogen sources, rather than by nitrate alone.
Possible Mechanisms Underlying These Responses. Our CO2 and warming treatments had only modest effects on the CO2 concentration and temperature of the soil. Thus, the change in abundance of AOB that we observed in response to these factors was probably a result of indirect effects, most likely mediated by the plant community. Indeed, AOB abundance was most strongly correlated with plant biomass (r = 0.18506, P = 0.0380, n = 126) and soil moisture (r =–0.19272, P = 0.0365, n = 118), environmental variables that exhibit plant-mediated changes in response to increased temperature and CO2 (1). Elevated soil moisture has been reported to alter AOB abundance both positively and negatively. Relatively moderate elevation of soil moisture can increase abundance by reducing water stress (29, 30); larger increases in soil moisture can depress abundance by decreasing the diffusion of oxygen into the soil (29). The optimal soil moisture for AOB therefore reflects the balance between these two effects. This balance may underlie the interaction we observed between elevated atmospheric temperature and precipitation (Fig. 3B). Elevated atmospheric temperature increases soil moisture at our site late in the growing season (the time of year at which we sampled) because of changes in the plant community that reduce transpiration (31). It is plausible that either elevated temperature or increased precipitation has a positive effect on AOB abundance because of a reduction in water stress. When both are elevated, however, the increase in soil moisture could exceed the optimum, leading to a net negative effect on AOB abundance because of a reduction in oxygen availability.
What mechanism might underlie the effect of CO2 on AOB abundance? It is possible, although not likely, that the effects of CO2 also are mediated through soil moisture. CO2 increases soil moisture at our site through changes in transpiration (31). The effect of elevated CO2 on soil moisture is, however, comparable to that of elevated temperature (31), and we observed a very different interaction between CO2 and precipitation (Fig. 3A) than between temperature and precipitation (Fig. 3B). Another possibility is that elevated CO2 alters the intensity of resource competition between AOB and heterotrophic microbes. Elevated CO2 increases soil heterotroph abundance and activity at our site (32), most likely because of increased root carbon exudation (33). AOB are inferior competitors for some resources (e.g., oxygen) (29) relative to heterotrophic microbes, and an increase in resource competition could result in a decrease in AOB abundance with elevated CO2. This decline could be exacerbated by an increase in precipitation if this also reduced the concentration of the limiting resource, either through leaching (for a mobile nutrient such as calcium) or by reducing diffusion (in the case of oxygen).
The change in AOB community structure that we observed in response to nitrogen deposition may have resulted from the direct effects of nitrate application to the soil. However, AOB structure was most strongly correlated with soil ammonium concentration (r = 0.23649, P = 0.0082, n = 124), rather than nitrate, suggesting that indirect effects may be the more likely cause of this response. The ammonium concentration was significantly higher in the N-deposition (i.e., nitrate) treatments (F1,27 = 9.61625, P = 0.00448), possibly because of increases in plant and microbial productivity and the subsequent mineralization of nitrogen, increased dissimilatory nitrate reduction to ammonium, and/or a shift in plant uptake to nitrate and a subsequent decrease in plant uptake of ammonium. This increase in ammonium may explain why we observed shifts in community structure in response to nitrate addition that were similar to those observed in previous studies of ammonium addition. Elevated precipitation can reduce available nitrogen at the JRGCE (J. Dukes and C.B.F., unpublished data). Elevated temperature increases soil moisture. It is possible that elevated temperature acts in concert with increased precipitation to increase the loss of available nitrogen. These effects may underlie the observed interactions among nitrogen deposition, temperature, and precipitation.
Implications for Ecosystem Functions. Our observations are important not only because free-living prokaryotes represent most of the earth's biodiversity (34) but also because they are key mediators of ecosystem functions. Changes in bacterial abundance and diversity could result in altered rates and/or controls of such functions. This is particularly likely for microbial groups involved in nitrogen transformations (e.g., nitrogen-fixers, denitrifiers, and nitrifiers), because these groups do not have a high degree of functional redundancy (35). Community structure is related to function in denitrifying bacteria, and there is some evidence for such a relationship for AOB (9, 36, 37). In the JRGCE, we measured the nitrification potentials of soil from six plots that varied in AOB abundance and structure. Although total abundance of AOB had no effect on nitrification potential, there was a significant effect of structure. Nitrification potential was significantly higher in those soils with a higher proportion of JR1 (df = 22, t = 2.5838, P = 0.0169). Nitrogen was added in abundance during these assays to eliminate the direct effect of nitrogen availability. Thus, the effect we observed is most likely due to the effects of AOB structure, further supporting the hypothesis that JR1 is a high-nitrogen-adapted clade. Not only does JR1 increase in relative abundance in response to increased nitrogen (Fig. 2) but also it is better able to metabolize ammonium at high concentrations.
Conclusions
This study demonstrates significant, previously unrecognized changes in the community structure and abundance of free-living soil bacteria in response to multifactorial global change. We observed that the response of AOB to multifactorial change was in some respects similar to that of plants: Community structure and abundance were altered by nitrogen and elevated CO2. Further, both interacted strongly with precipitation. This result is consistent with a number of studies that have shown that effects of global change on ecological communities are often driven by changes in the water budget, especially in grassland ecosystems (38). Some of the responses of AOB to the relatively subtle perturbations of simulated global change could be predicted by their responses to much more severe perturbations. For example, the response of AOB community structure to nitrogen deposition was similar to previously documented responses to agricultural fertilization. Finally, some of the responses we observed have the potential to alter ecosystem function. The enrichment of AOB clade JR1 in response to nitrogen deposition is associated with a higher potential for nitrification. This demonstrates the potential importance of feedbacks between the microbial and plant communities that could alter the overall ecosystem response to global change. Our study demonstrates that a complete understanding of the ecological impacts of global change will require understanding the responses of microorganisms, as well as macroorganisms, and that a multifactorial approach can play an important role in developing this understanding.
Supplementary Material
Acknowledgments
We appreciate the helpful comments of K. Carney, C. Davis, S. Forde, M. C. Horner-Devine, C. Jessup, G. Krukonis, L. Reddy, V. Rich, P. Vitousek, and an anonymous reviewer on previous drafts of the manuscript. We are very grateful for the advice and assistance of Karen Carney and the support of Pamela Matson for the nitrification potential assays. MJ Research (Cambridge, MA) generously provided the equipment used for the quantitative PCR assays, and we are grateful to Richard Kurtz for advice on its operation. This work was supported by awards from the Mellon Foundation, the David and Lucile Packard Foundation, and the National Science Foundation (DEB-0221838 and DEB-0108556).
Author contributions: H.-P.H., C.B.F., and B.J.M.B. designed research; H.-P.H., A.B., and B.J.M.B. performed research; H.-P.H., A.B., and B.J.M.B. analyzed data; and H.-P.H., C.B.F., and B.J.M.B. wrote the paper.
Abbreviations: AOB, ammonia-oxidizing bacteria; JRGCE, Jasper Ridge Global Change Experiment; T-RFLP, terminal restriction fragment length polymorphism.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY369266–AY369342).
References
- 1.Shaw, M. R., Zavaleta, E. S., Chiariello, N. R., Cleland, E. E., Mooney, H. A. & Field, C. B. (2002) Science 298, 1987–1990. [DOI] [PubMed] [Google Scholar]
- 2.Vitousek, P., Mooney, H., Lubchenco, J. & Melillo, J. (1997) Science 277, 494–499. [Google Scholar]
- 3.Hu, S., Firestone, M. & Chapin, F. (1999) Trends Ecol. Evol. 14, 433–437. [DOI] [PubMed] [Google Scholar]
- 4.Morgan, J. (2002) Science 298, 1903–1904. [DOI] [PubMed] [Google Scholar]
- 5.Zak, D., Pregitzer, K., King, J. & Holmes, W. (2000) New Phytol. 147, 201–222. [Google Scholar]
- 6.Schortemeyer, M., Hartwig, U., Hendrey, G. & Sadowsky, M. (1996) Soil Biol. Biochem. 28, 1717–1724. [Google Scholar]
- 7.Montealegre, C. M., Van Kessel, C., Blumenthal, J. M., Hur, H.-G., Hartwig, U. A. & Sadowsky, M. J. (2000) Global Change Biol. 6, 475–482. [Google Scholar]
- 8.Rillig, M., Wright, S., Shaw, M. & Field, C. (2002) Oikos 97, 52–58. [Google Scholar]
- 9.Bruns, M., Stephen, J., Kowalchuk, G., Prosser, J. & Paul, E. (1999) Appl. Environ. Microbiol. 65, 2994–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avrahami, S. & Conrad, R. (2003) Appl. Environ. Microbiol. 69, 6152–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oechel, W. C., Hastings, S. J., Vourlitis, G., Jenkins, M., Riechers, G. & Grulke, N. (1993) Nature 361, 520–523. [Google Scholar]
- 12.Chapin, F. S., III, Shaver, G. R., Giblin, A. E., Nadelhoffer, K. J. & Laundre, J. A. (1995) Ecology 76, 694–711. [Google Scholar]
- 13.Reich, P. B., Knops, J., Tilman, D., Craine, J., Ellsworth, D., Tjoelker, M., Lee, T., Wedin, D., Naeem, S., Bahauddin, D., et al. (2001) Nature 410, 809–812. [DOI] [PubMed] [Google Scholar]
- 14.Kowalchuk, G. & Stephen, J. (2001) Annu. Rev. Microbiol. 55, 485–529. [DOI] [PubMed] [Google Scholar]
- 15.Niklaus, P. A., Kandeler, E., Leadley, P. W., Schmid, B., Tscherko, D. & Körner, C. (2001) Oecologia 127, 540–548. [DOI] [PubMed] [Google Scholar]
- 16.Zak, D. R., Pregitzer, K. S., Curtis, P. S. & Holmes, W. E. (2000) Ecol. Appl. 10, 47–59. [Google Scholar]
- 17.Horz, H., Yimga, M. & Liesack, W. (2001) Appl. Environ. Microbiol. 67, 4177–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 19.Jukes, T. H. & Cantor, C. R. (1969) Evolution of Protein Molecules (Academic, New York).
- 20.Horz, H., Rotthauwe, J., Lukow, T. & Liesack, W. (2000) J. Microbiol. Methods 39, 197–204. [DOI] [PubMed] [Google Scholar]
- 21.Okano, Y., Hristova, K. R., Leutenegger, C. M., Jackson, L. E., Denison, R. F., Gebreyesus, B., Lebauer, D. & Scow, K. M. (2004) Appl. Environ. Microbiol. 70, 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart, S. C., Stark, J. M., Davidson, E. A. & Firestone, M. K. (1994) in Methods of Soil Analysis: Part 2—Microbiological and Biochemical Properties, ed. Bezdiecek, D. (Soil Science Society of America, Madison, WI), pp. 985–1018.
- 23.Francis, C. A., O'Mullan, G. D. & Ward, B. B. (2003) Geobiology 1, 129–140. [Google Scholar]
- 24.Kowalchuk, G., Stienstra, A., Heilig, G., Stephen, J. & Woldendorp, J. (2000) Environ. Microbiol. 2, 99–110. [DOI] [PubMed] [Google Scholar]
- 25.Kowalchuk, G., Stienstra, A., Heilig, G., Stephen, J. & Woldendorp, J. (2000) FEMS Microbiol. Ecol. 31, 207–215. [DOI] [PubMed] [Google Scholar]
- 26.Norton, J. M., Alzerreca, J. J., Suwa, Y. & Klotz, M. G. (2002) Arch. Microbiol. 177, 139–149. [DOI] [PubMed] [Google Scholar]
- 27.Mendum, T., Sockett, R. & Hirsch, P. (1999) Appl. Environ. Microbiol. 65, 4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermansson, A. & Lindgren, P.-E. (2001) Appl. Environ. Microbiol. 67, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belser, L. W. (1979) Annu. Rev. Microbiol. 33, 309–333. [DOI] [PubMed] [Google Scholar]
- 30.Hastings, R., Butler, C., Singleton, I., Saunders, J. & McCarthy, A. (2000) Lett. Appl. Microbiol. 30, 14–18. [DOI] [PubMed] [Google Scholar]
- 31.Zavaleta, E. S., Thomas, B. D., Chiariello, N. R., Asner, G. P., Shaw, M. R. & Field, C. B. (2003) Proc. Natl. Acad. Sci. USA 100, 9892–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hungate, B. A., Jaeger, C. H., III, Gamara, G., Chapin, F. S., III, & Field, C. B. (2000) Oecologia 124, 589–598. [DOI] [PubMed] [Google Scholar]
- 33.Luo, Y., Jackson, R. B., Field, C. B. & Mooney, H. A. (1996) Oecologia 108, 130–137. [DOI] [PubMed] [Google Scholar]
- 34.Whitman, W., Coleman, D. & Wiebe, W. (1998) Proc. Natl. Acad. Sci. USA 95, 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolters, V., Silver, W., Bignell, D., Coleman, D., Lavelle, P., Van der Putten, W., De Ruiter, P., Rusek, J., Wall, D., Wardle, D., et al. (2000) Bioscience 50, 1089–1098. [Google Scholar]
- 36.Phillips, C., Harris, D., Dollhopf, S., Gross, K., Prosser, J. & Paul, E. (2000) Appl. Environ. Microbiol. 66, 5410–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavigelli, M. & Robertson, G. (2001) Soil Biol. Biochem. 33, 297–310. [Google Scholar]
- 38.Volk, M., Niklaus, P. A. & Korner, C. (2000) Oecologia 125, 380–388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.