Abstract
The Arabidopsis CBF cold response pathway has a central role in cold acclimation, the process whereby plants increase in freezing tolerance in response to low nonfreezing temperatures. Here we examined the changes that occur in the Arabidopsis metabolome in response to low temperature and assessed the role of the CBF cold response pathway in bringing about these modifications. Of 434 metabolites monitored by GC-time-of-flight MS, 325 (75%) were found to increase in Arabidopsis Wassilewskija-2 (Ws-2) plants in response to low temperature. Of these 325 metabolites, 256 (79%) also increased in nonacclimated Ws-2 plants in response to overexpression of C-repeat/dehydration responsive element-binding factor (CBF)3. Extensive cold-induced changes also occurred in the metabolome of Arabidopsis Cape Verde Islands-1 (Cvi-1) plants, which were found to be less freezing tolerant than Ws-2 plants. However, low-temperature-induced expression of CBF1, CBF2, CBF3, and CBF-targeted genes was much lower in Cvi-1 than in Ws-2 plants, and the low-temperature metabolome of Cvi-1 plants was depleted in metabolites affected by CBF3 overexpression. Taken together, the results indicate that the metabolome of Arabidopsis is extensively reconfigured in response to low temperature, and that the CBF cold response pathway has a prominent role in this process.
Many plants have the ability to sense low temperature and respond by activating mechanisms that lead to an increase in freezing tolerance, an adaptive process known as cold acclimation (1, 2). At present, the best-understood genetic system with a role in cold acclimation is the Arabidopsis CBF cold response pathway (3). Exposing Arabidopsis plants to low temperature results in rapid induction of a small family of genes encoding transcriptional activators known either as C-repeat (CRT)/dehydration responsive element (DRE)-binding factor (CBF)1, -2, and -3 (4-6) or DREB1b, -c, and -a, respectively (7). These genes encode transcription factors that belong to the AP2/ERF domain family of DNA-binding proteins (8). The CBF1-3 proteins recognize a cis-acting regulatory element known as the CRT/DRE present in the promoters of many cold-inducible genes, including COR15a and COR78/RD29a (4, 9, 10). Expression of the >100 genes that comprise the CBF regulon, i.e., those cold-regulated genes that are responsive to CBF expression (refs. 11-13; J. Vogel and M.F.T., unpublished data), then brings about an increase in freezing tolerance. This increase in freezing tolerance involves the action of multiple mechanisms, including the production of cryoprotective polypeptides, such as COR15a (14), and synthesis of low molecular-weight cryoprotectants, including proline and raffinose (15, 16).
Genetic studies suggest that the CBF cold response pathway is not the only system that contributes to freezing tolerance. The eskimo1 mutant of Arabidopsis (17) is constitutively more freezing tolerant than wild-type plants, yet the COR genes are not expressed, indicating that the mutation affects a pathway outside the CBF cold response pathway. Similarly, ada2 mutants of Arabidopsis (ADA2 encodes a transcriptional adaptor protein) (18) are constitutively more freezing tolerant than wild-type plants, but again, COR genes are not constitutively expressed (19). These results suggest that in wild-type plants grown at warm temperature, the ADA2 protein is involved in inhibiting expression of a freezing tolerance pathway that is distinct from the CBF cold response pathway.
Transcript profiling experiments using DNA microarrays have shown that extensive changes occur in the transcriptome of Arabidopsis in response to low temperature (11-13, 20, 21). Further, the results indicate that the CBF cold response pathway has a significant role in bringing about these changes. Fowler and Thomashow (12) surveyed expression of ≈8,000 Arabidopsis genes and found that transcripts for ≈4% (306) of the genes were responsive to low temperature, with 3% (218) being up-regulated and 1% (88) being down-regulated. Of the cold-responsive genes, 12% could be assigned to the CBF regulon. However, at least 28% of the cold-responsive genes were not affected by expression of the CBF transcription factors, including 15 encoding known or putative transcription factors. It was therefore concluded that cold acclimation is associated with the activation of multiple low-temperature regulatory pathways. Similar conclusions have been reached by others also studying the Arabidopsis low-temperature transcriptome (11-13, 20).
Here, our objectives were to explore on a large scale the changes that occur in the metabolome of Arabidopsis in response to low temperature and to determine the extent to which these changes involve action of the CBF cold response pathway. The experiments included a comparison of the metabolite profiles of two Arabidopsis ecotypes, Wassilewskija-2 (Ws-2) and Cape Verde Islands-1 (Cvi-1); the former was found to be more freezing tolerant than the latter. Taken together, the results indicate that cold acclimation is associated with extensive changes in the metabolome, and that these changes can be largely mimicked by constitutive overexpression of CBF3. In addition, the results indicate that, upon exposure to low temperature, CBF1, -2, and -3 and CBF-targeted genes are expressed at higher levels in Arabidopsis Ws-2 plants, as compared with Cvi-1 plants, and that the low-temperature metabolome of Cvi-1 plants is depleted in metabolites controlled by the CBF regulon. We conclude that the metabolome of Arabidopsis is extensively reconfigured in response to low temperature, and that the CBF cold response pathway has a prominent role in this process.
Materials and Methods
Plant Materials and Growth. Arabidopsis thaliana ecotypes Ws-2 and Cvi-1, collected from Russia and the Cape Verde Islands, respectively (obtained from the Arabidopsis Biological Resource Center at Ohio State University, Columbus), and transgenic plants constitutively overexpressing CBF3 in the Ws-2 background (A28 and A30) (16) were used in this study. For the metabolome experiments, plants were gown in potted soil (seeds were stratified at 4°C for 4 days before sowing) in controlled environment chambers with illumination of 150 μmol·m-2·s-1 from cool white lights. Plants were grown either under a short day (8-h light, 20°C/16-h dark, 16°C) or long day (16-h light, 20°C/8-h dark, 16°C) photoperiod. Plants were cold-acclimated at 4°C for 14 days with illumination of 60-80 μmol·m-2·s-1 and photoperiod corresponding to that used for warm conditions. For the gene expression experiments, plants were grown on Gamborg's B-5 medium, pH 5.7, solidified with 0.8% phytagar (Invitrogen). Plates were incubated at 24°C under continuous illumination of 100 μmol·m-2·s-1.
Metabolite Extraction and Detection. Tissue samples were prepared by homogenizing individual plants in liquid nitrogen with a Retsch mixer mill MM200 (Haan, Germany). Metabolites were extracted from 25-30 mg of each tissue sample with 1 ml of chloroform/methanol/water solution (2:5:2) for 5 min at 4°C. After this, 500 μl of water was added to partition against the chloroform, and the samples were centrifuged for 2 min at 14,000 × g. An aliquot of the aqueous phase was lyophilized for later analysis. Replicate samples were used in lieu of an internal standard for metabolite recovery. For nonacclimated and cold-acclimated wild-type plant samples, 24 individual plants for each condition were analyzed. For the CBF3-overexpressing lines and the corresponding wild-type control (nonacclimated) samples, 12 individual plants for each were analyzed. Metabolites were detected by GC-time-of-flight MS, as described (22, 23).
Data Analysis. Metabolite levels were normalized to milligrams of fresh weight of plant tissue. A given metabolite was judged to increase in response to cold acclimation if the ratio of the average peak areas for the metabolite in samples produced from cold-acclimated and nonacclimated plants (harvested at both dawn and dusk) was >1 with a t test P value of <0.001. A given metabolite was judged to increase in response to CBF3 overexpression if the ratio between the average peak areas for the metabolite in samples produced from the CBF3-overexpressing transgenic plants and nonacclimated Ws-2 plants (harvested at dusk) was >1 with a t test P value of <0.001. Microsoft excel Version 5.0 and access were used for statistical analysis and other calculations.
Whole-Plant Freeze Tests. Nonacclimated Arabidopsis Ws-2 and Cvi-1 plants were grown for 45 days at 20°C. Cold-acclimated plants were grown for 41 days at 20°C followed by 14 days cold acclimation at 4°C. Plants were frozen at -5°C in the dark for 3 h, after which ice chips were added to promote ice formation. Plants were incubated an additional 3 days at -5°C and then were allowed to thaw at 4°C for 12 h and recover for 5 days in the original growth conditions. Three independent experiments were conducted by testing ≈100 plants for each ecotype.
RNA Gel Blot Hybridization Analysis. Total RNA was extracted, and Northern transfers were prepared, hybridized, and washed at high stringency as described (4). Gene-specific probes for CBF1-3 (5) and PAL2 (24) were as described. A cDNA clone for AtGolS3 (15) was obtained from the Arabidopsis Biological Resource Center at Ohio State University. Probes for CHS (25), 25S rRNA (26), COR78, COR15a, P5CS, and SuSy (16) were as described. Probes were labeled with 32P by using the Random Primers DNA Labeling System (Invitrogen) as directed by the manufacturer. Membranes were exposed either to x-ray film or to a PhosphorImager (Molecular Dynamics) plate that was then visualized by scanning the plate in a Fluor-S MultiImager (Bio-Rad). Quantification was performed by using quantity one software, Version 4.2.2 (Bio-Rad).
Results
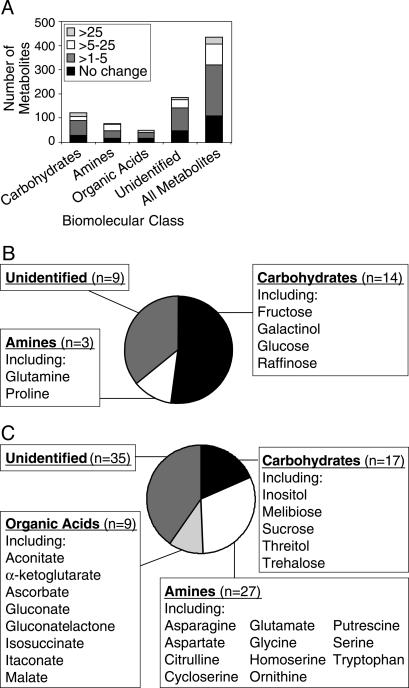
Cold Acclimation in Arabidopsis Ws-2 Plants Is Associated with Extensive Changes in the Metabolome. Changes in metabolite levels associated with cold acclimation in Arabidopsis Ws-2 plants were assessed on a large scale by using GC-time-of-flight MS. In the analysis, a total of 434 low-molecular-weight carbohydrates, amines, organic acids, and other polar molecules were monitored. When using a rigorous statistical cutoff (P < 0.001), the results indicated that the metabolome of Ws-2 plants was extensively altered in response to low temperature. Of the 434 metabolites monitored, 325 (75%) were found to increase in cold-acclimated plants (Fig. 1A and Table 1, which is published as supporting information on the PNAS web site). The increases observed varied from <2- to >25-fold, with 114 (35%) increasing at least 5-fold (Fig. 1 B and C).
Fig. 1.
Effect of low temperature on the metabolome of Arabidopsis Ws-2 plants. (A) Total metabolites that increased in response to low temperature. (B) Metabolites that increased >25-fold (n = 26). (C) Metabolites that increased between 5- and 25-fold (n = 88).
Of the metabolites that increased with cold acclimation, 75 were identified; the others were categorized according to class (i.e., carbohydrate, organic acid, etc.) or were unknown (Table 1). Several of the identified metabolites had previously been shown to increase in Arabidopsis plants upon exposure to low temperature, including the amino acid proline and the sugars glucose, fructose, inositol, galactinol, raffinose, and sucrose (15, 16, 27, 28). To our knowledge, however, the majority of the identified metabolites had not previously been shown to increase in Arabidopsis. This includes trehalose, putrescine, and ascorbate, which have potential roles in cold tolerance (see Discussion). In addition, the increases observed in ornithine and citrulline, precursors to polyamine biosynthesis, suggest up-regulation of the urea cycle. Furthermore, increases in α-ketoglutarate, fumarate, malate, and citrate, precursors to amino acid biosynthesis, suggest up-regulation of the citric acid cycle.
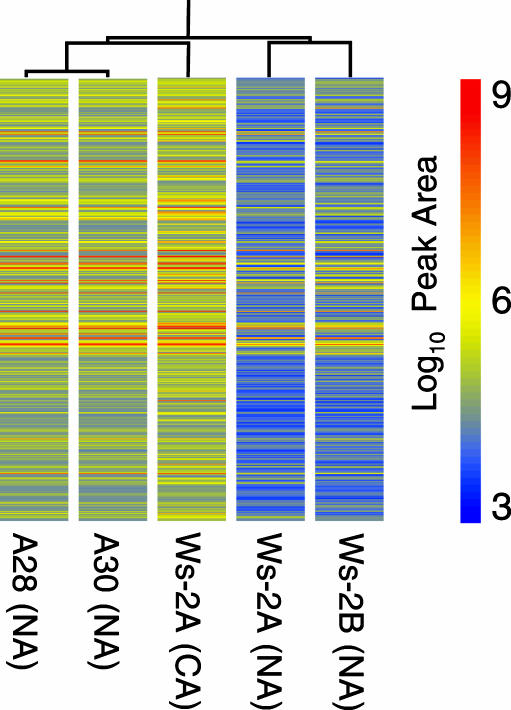
Nonacclimated Plants Expressing the CBF Regulon Have a Metabolome Resembling That of Cold-Acclimated Plants. To explore the extent to which the low-temperature metabolome of Arabidopsis is conditioned by the CBF cold response pathway, we compared the metabolomes of nonacclimated and cold-acclimated wild-type Arabidopsis Ws-2 plants with the metabolome of nonacclimated transgenic Arabidopsis Ws-2 plants overexpressing CBF3 (transgenic lines A28 and A30). The results indicated that expression of the CBF regulon brought about extensive changes in the metabolome similar to those that occurred in response to low temperature. Of the 325 metabolites that were identified as increasing in response to low temperature, 256 (79%) were also found to increase in response to overexpression of CBF3 (P < 0.001) (Table 1). Moreover, of the 114 metabolites that increased >5-fold in response to low temperature, 102 (90%) also increased in response to CBF3 overexpression (Table 1). These results indicated that the metabolome of nonacclimated CBF3- overexpressing plants was similar to that of cold-acclimated plants. To test this further, the levels of the 434 metabolites in the nonacclimated CBF3-overexpressing plants were compared with those in nonacclimated and cold-acclimated Ws-2 plants, and the results were subjected to hierarchical clustering (Fig. 2). The analysis indicated that the metabolome of the nonacclimated CBF3-overexpressing plants more closely resembled the metabolome of cold-acclimated wild-type plants than the metabolome of nonacclimated wild-type plants.
Fig. 2.
The metabolome of nonacclimated CBF3-overexpressing plants resembles that of cold-acclimated wild-type Arabidopsis plants. Plants were grown at 20°C (NA) and, where indicated, were cold-acclimated at 4°C for 14 days (CA). Ws-2A plants were grown under a short-day photoperiod; plants from Ws-2B- and CBF3-overexpressing transgenic lines A28 and A30 were grown under a long-day photoperiod. Plant material was harvested just before the end of the photoperiod. The mean log-transformed values of the peak area of each metabolite from the individual samples were calculated and used to generate a hierarchical cluster of the various treatments by using a Pearson correlation included in the genespring software package (Silicon Genetics, Redwood City, CA).
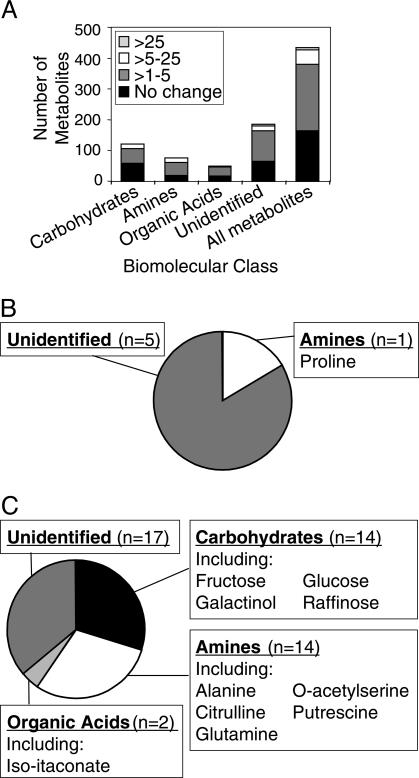
Arabidopsis Cvi-1 Plants Are Less Freezing Tolerant Than Ws-2 Plants and Have a Metabolome Depleted in Metabolites Regulated by the CBF Cold Response Pathway. The Arabidopsis Cvi-1 ecotype is less freezing tolerant than the Landsberg ecotype (J. Salinas, personal communication). Likewise, we found that Cvi-1 plants were less freezing tolerant than Ws-2 plants. Whereas cold-acclimated Ws-2 plants were able to survive freezing at -5°C for 3 days, Cvi-1 plants treated in an equivalent manner were not (Fig. 7, which is published as supporting information on the PNAS web site). This finding prompted us to determine whether the metabolomes of Ws-2 and Cvi-1 plants were configured differently in response to low temperature. To test this, the metabolite profiles of nonacclimated and cold-acclimated Cvi-1 plants were determined and compared with those of Ws-2 plants. The results indicated that, as with Ws-2 plants, extensive changes occurred in the metabolome of Cvi-1 plants upon exposure to low temperature. Of the 434 metabolites that were monitored, 269 (62%) were found to increase in cold-treated Cvi-1 plants (Fig. 3A and Table 2, which is published as supporting information on the PNAS web site), with 6 increasing at least 25-fold and 47 increasing at least 5-fold (Fig. 3 B and C). A comparison of these results with those for Ws-2 plants (Fig. 1 and Table 1) indicated there was considerable overlap in the changes that occurred with cold acclimation in the Cvi-1 and Ws-2 plants. Of the 269 metabolites that increased in response to low temperature in the Cvi-1 plants, 244 (91%) also increased in cold-treated Ws-2 plants.
Fig. 3.
Effect of low temperature on the metabolome of Arabidopsis Cvi-1 plants. (A) Total metabolites that increased in response to low temperature. (B) Metabolites that increased >25-fold (n = 6). (C) Metabolites that increased between 5- and 25-fold (n = 47).
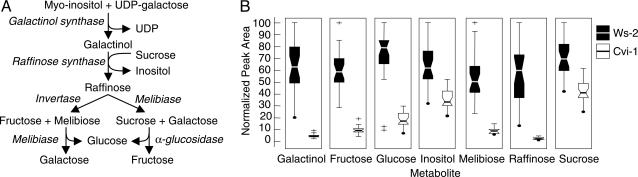
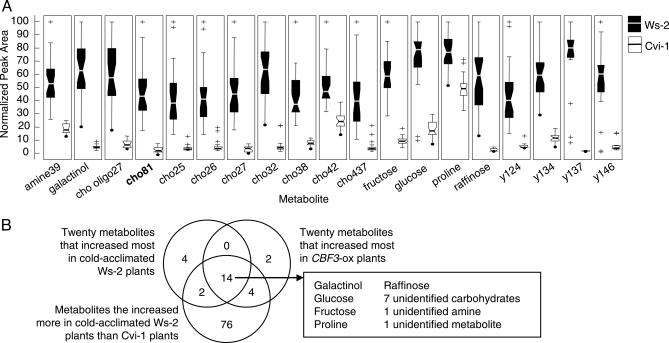
Although it was true that most of the metabolites that increased in response to low temperature in Ws-2 plants also increased in Cvi-1 plants, it was also true that there were considerable quantitative differences in the low-temperature metabolomes of these two ecotypes. Of the 325 metabolites that increased in Ws-2 plants upon cold treatment, 96 (30%) accumulated to lower levels in Cvi-1 plants (Table 3, which is published as supporting information on the PNAS web site). Moreover, many of the metabolites that accumulated in response to CBF3 overexpression accumulated to lower levels in the Cvi-1 plants. Galactinol and raffinose, sugars that are synthesized through the action of the CBF-targeted gene AtGolS3, as well as melibiose, fructose, glucose, inositol, and sucrose, sugars associated with the raffinose pathway, were all at lower levels in Cvi-1 plants as compared with Ws-2 plants (Fig. 4). In addition, proline levels, which are determined in part by expression of the CBF-targeted P5CS2 gene, were twice as high in Ws-2 plants compared with Cvi-1 plants (Table 3). More broadly, of the 96 metabolites that increased less in Cvi-1 plants than in Ws-2 plants, 80 (83%) increased in response to CBF3 overexpression. Moreover, of the 26 metabolites that increased 25-fold or more in response to low temperature in Ws-2 plants, 19 accumulated to a greater level in Ws-2 than in Cvi-1 plants (Fig. 5A), and of these, all but one, compound cho81, increased in response to CBF3 overexpression. Similarly, a comparison of the 20 metabolites that increased the most in Ws-2 plants in response to low temperature with the 20 metabolites that increased the most in response to CBF3 overexpression indicated that 14 metabolites were common to the two treatments, all of which increased less in Cvi-1 than in Ws-2 plants in response to low temperature (Fig. 5B).
Fig. 4.
Cold-acclimated Arabidopsis Cvi-1 plants are deficient in metabolites associated with the raffinose biosynthetic pathway. (A) The raffinose biosynthetic pathway (www.genome.ad.jp/kegg/pathway.html). Substrates and products are in plain text, and enzymes are in italics. (B) Box and Whisker plots showing accumulation of the raffinose pathway metabolites in cold-acclimated Ws-2 and Cvi-1 plants. The top and bottom of each box represent the 25th and 75th percentiles, the center line indicates the median, and the extents of the whiskers show the extent of the data (except for outliers). Outliers are indicated by crosses and are defined as data points that are distant from either the 25th or 75th percentiles by >1.5 times the interquartile range. The black dot indicates absence of outliers. For each metabolite, the maximal measured peak area was normalized to a value of 100.
Fig. 5.
Cold-acclimated Arabidopsis Cvi-1 plants are deficient in metabolites that increase markedly in cold-acclimated Ws-2 plants and accumulate in nonacclimated transgenic Arabidopsis Ws-2 plants overexpressing CBF3. (A) Box and Whisker plots showing accumulation in cold-acclimated Ws-2 and Cvi-1 plants of 19 metabolites that increased >25-fold in cold-acclimated Ws-2 plants. Plot specifics are as described in the legend to Fig. 5. The bold text indicates that the metabolite was not increased by CBF3 overexpression. (B) Venn diagram showing commonalities in metabolites that increased most in cold-acclimated Ws-2 plants, increased most in nonacclimated CBF3-overexpressing Ws-2 plants, and increased more in cold-acclimated Ws-2 plants relative to Cvi-1 plants.
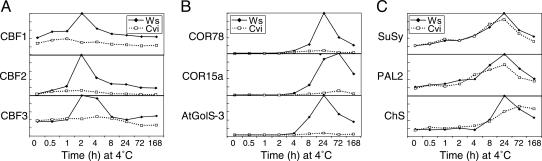
Arabidopsis Cvi-1 Plants Have a Weak CBF Locus. The results presented above suggested there might be differences in the CBF cold response pathways of Cvi-1 and Ws-2 plants. To test this, we asked whether the CBF1-3 genes were expressed differently in Cvi-1 plants as compared with Ws-2 plants. Northern analysis indicated they were. Transcripts for the CBF1, -2, and -3 genes were found to accumulate to much lower levels in cold-treated Cvi-1 plants than in cold-treated Ws-2 plants (Fig. 6A). In addition, transcripts for the known CBF-targeted genes COR15a, COR78, and AtGolS3 (12, 15, 29, 30) accumulated to much lower levels in cold-treated Cvi-1 than in cold-treated Ws-2 plants (Fig. 6B). In contrast, the transcript levels for three cold-regulated genes that are not CBF targets, those encoding sucrose synthase (SuSy), phenylalanine ammonia lyase (PAL2), and chalcone synthase (ChS) (11, 12), were similar in cold-treated Cvi-1 and Ws-2 plants, indicating that not all cold-regulated gene expression was at low levels in the Cvi-1 ecotype (Fig. 6C). Taken together, these results indicate that the CBF1-3 loci were weakly expressed in Cvi-1 plants, resulting in corresponding low-level expression of CBF-targeted genes.
Fig. 6.
Reduced cold-induced expression of CBF cold-response pathway genes in Arabidopsis Cvi-1 plants relative to Ws-2 plants. Transcript levels of CBF genes (A), CBF target genes (B), and CBF independent cold-regulated genes (C).
Discussion
It has long been known that biochemical changes occur with cold acclimation (31), including the accumulation of certain sugars and amino acids, of which some, such as sucrose and proline, have cryoprotective activities (32-34). However, the degree to which the metabolome is altered in response to low temperature has been a matter for speculation. This is due primarily to the limitations in methodologies that have been available for metabolite analysis. The recent development of highly sensitive metabolite profiling technologies now makes such comprehensive analysis possible (35, 36).
Here we used GC-time-of-flight MS to detail the changes that occur in the metabolome of Arabidopsis in response to low temperature and to assess the extent to which the low-temperature metabolome is configured by the action of the CBF cold response pathway. The results reveal in unparalleled fashion that cold acclimation is associated with extensive changes in the plant metabolite profile. Of the 434 metabolites that were monitored, 325 (75%) increased in level in response to low temperature in Arabidopsis Ws-2 plants, 114 (35%) of which increased at least 5-fold. Furthermore, the results indicate that the CBF cold response pathway has a prominent role in determining the composition of the low-temperature metabolome. Of the 325 metabolites that increased in response to low temperature, 256 (79%) also increased in transgenic Arabidopsis plants overexpressing CBF3. Moreover, when the metabolite profiles for nonacclimated and cold-acclimated wild-type plants were compared with nonacclimated CBF3-overexpressing plants and the data subjected to hierarchical clustering, the results indicated that the metabolome of nonacclimated CBF3-overexpressing plants was more similar to that of cold-acclimated than to that of nonacclimated wild-type plants. Similar results would likely be obtained with plants overexpressing CBF1 or CBF2, because a survey of transcript levels for ≈8,000 Arabidopsis genes revealed no obvious differences between plants overexpressing CBF1, -2, or -3 (37).
A comparison of the metabolomes of Ws-2 and Cvi-1 plants, the former of which was found to be more freezing tolerant than the latter, indicated there was considerable overlap in the metabolite changes that occurred in these two ecotypes in response to low temperature. However, quantitative differences were evident indicating natural variation for this “trait” among Arabidopsis ecotypes. In addition, the results provided further evidence for the CBF cold response pathway having an important role in shaping the low-temperature metabolome. This follows from the findings that CBF1, -2, and -3 and representative CBF-target genes were expressed at much lower levels in cold-treated Cvi-1 than in cold-treated Ws-2 plants, and that the low-temperature metabolome of the Cvi-1 plants had reduced levels of many of the metabolites that fall under the control of the CBF cold response pathway. Although it remains to be tested, it seems likely that the “weak” expression of the CBF cold response pathway and reduced levels of metabolites affected by CBF3 expression observed in the Cvi-1 plants, as compared with the Ws-2 plants, contributes to the differences in freezing tolerance observed between the two ecotypes.
The metabolome profiling experiments resulted in the identification of a large number of metabolites (>50) that increased with cold acclimation that, to our knowledge, had not been shown to do so before in Arabidopsis. It seems likely that at least some of these, including trehalose, putrescine, and ascorbate, contribute to cold tolerance. Trehalose is known to be an important stress protectant in a variety of organisms, including bacteria, yeast, and invertebrates (38, 39). In vivo studies suggest that trehalose is able to protect membranes and proteins from deleterious alterations that can occur due to dehydration or oxidative stress (32, 40). Putrescine is found in many organisms, including plants, where its levels have been shown to increase in response to multiple abiotic stresses, including low temperature (41, 42). A number of observations indicate a role for putrescine in plant stress tolerance, such as its exogenous application decreasing membrane leakage caused by low temperature in tomato (43) and mutants of Arabidopsis (ADC2) that are deficient in endogenous putrescine being more sensitive to abiotic stresses (42). Finally, ascorbate is an abundant antioxidant in plants, providing protection against reactive oxygen species that are produced in a variety of ways, including exposure of plants to low temperature (44).
At present, only a few of the changes that occur in the Arabidopsis metabolome in response to low temperature can be ascribed to the activities of specific genes and enzymes. The increases in free proline and the levels of sugars associated with the raffinose pathway involve increased expression of the CBF-targeted genes P5CS2 and AtGolS3, respectively (15, 16). Presumably other changes that occur involve the >100 identified cold-inducible genes that encode enzymes involved in amino acid, carbohydrate, lipid, nucleotide, and secondary metabolism (11-13; J.V. and M.F.T., unpublished data). Integrating the results of previous transcriptome analyses with the metabolome results presented here, however, is substantially limited by the fact that only ≈22% of the metabolites monitored in this study were identified; most could be assigned only to a chemical category (carbohydrate, amine, etc.) or were completely unknown. This situation does not impact the general conclusions that we draw here about the low-temperature metabolome and the role of the CBF cold response pathway in configuring it. However, it limits the assigning of metabolites to specific metabolic pathways. Indeed, 84 of the 114 metabolites that increased >5-fold in response to low temperature in Ws-2 plants fell into the unidentifiable class, 20 of which increased >25-fold. Future identification of these metabolites will increase our understanding of how metabolism is reprogrammed in Arabidopsis at low temperature and may lead to the identification of novel metabolites with roles in stress tolerance.
Supplementary Material
Acknowledgments
We thank Anne Eckardt for help with the GC-time-of-flight MS analysis, Rebecca Grumet and Gareth Catchpole for help with statistical analysis, and Julio Salinas for information on the freezing tolerance of Cvi-1 plants. D.C. was a recipient of the Alfred Toepfer Award from the von Humboldt Foundation, which supported the research that he conducted at the Max Planck Institute of Molecular Plant Physiology. This research was also supported by National Science Foundation Plant Genome Project Grant DBI 0110124 (to M.F.T.), Department of Energy Grant DEFG0291ER20021 (to M.F.T.), the Michigan Agricultural Experiment Station (M.F.T.), and the Max Planck Institute of Molecular Plant Physiology (O.F.).
Author contributions: M.F.T., D.C., and O.F. designed the research; D.C. and O.F. performed the research; D.C., S.F., O.F., and M.F.T. analyzed the data; and D.C., S.F., O.F., and M.F.T. wrote the paper.
Abbreviations: CRT, C-repeat; DRE, dehydration responsive element; CBF, CRT/DRE-binding factor; Cvi-1, Cape Verde Islands-1; Ws-2, Wassilewskija-2.
See Commentary on page 14996.
References
- 1.Smallwood, M. & Bowles, D. J. (2002) Philos. Trans. R. Soc. London B 357, 831-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomashow, M. F. (1998) Plant Physiol. 118, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomashow, M. F. (2001) Plant Physiol. 125, 89-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockinger, E. J., Gilmour, S. J. & Thomashow, M. F. (1997) Proc. Natl. Acad. Sci. USA 94, 1035-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M. & Thomashow, M. F. (1998) Plant J. 16, 433-442. [DOI] [PubMed] [Google Scholar]
- 6.Medina, J., Bargues, M., Terol, J., Perez-Alonso, M. & Salinas, J. (1999) Plant Physiol. 119, 463-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1998) Plant Cell 10, 1391-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riechmann, J. L. & Meyerowitz, E. M. (1998) Biol. Chem. 379, 633-646. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi-Shinozaki, K. & Shinozaki, K. (1994) Plant Cell 6, 251-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker, S. S., Wilhelm, K. S. & Thomashow, M. F. (1994) Plant Mol. Biol. 24, 701-713. [DOI] [PubMed] [Google Scholar]
- 11.Seki, M., Narusaka, M., Abe, H., Kasuga, M., Yamaguchi-Shinozaki, K., Carninci, P., Hayashizaki, Y. & Shinozaki, K. (2001) Plant Cell 13, 61-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler, S. & Thomashow, M. F. (2002) Plant Cell 14, 1675-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., Shimada, Y., Yoshida, S., Shinozaki, K. & Yamaguchi-Shinozaki, K. (2004) Plant J. 38, 982-993. [DOI] [PubMed] [Google Scholar]
- 14.Steponkus, P. L., Uemura, M., Joseph, R. A., Gilmour, S. J. & Thomashow, M. F. (1998) Proc. Natl. Acad. Sci. USA 95, 14570-14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taji, T., Ohsumi, C., Iuchi, S., Seki, M., Kasuga, M., Kobayashi, M., Yamaguchi-Shinozaki, K. & Shinozaki, K. (2002) Plant J. 29, 417-426. [DOI] [PubMed] [Google Scholar]
- 16.Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D. & Thomashow, M. F. (2000) Plant Physiol. 124, 1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin, Z. & Browse, J. (1998) Proc. Natl. Acad. Sci. USA 95, 7799-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stockinger, E. J., Mao, Y., Regier, M. K., Triezenberg, S. J. & Thomashow, M. F. (2001) Nucleic Acids Res. 29, 1524-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachonasios, K. E., Thomashow, M. F. & Triezenberg, S. J. (2003) Plant Cell 15, 626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreps, J. A., Wu, Y., Chang, H. S., Zhu, T., Wang, X. & Harper, J. F. (2002) Plant Physiol. 130, 2129-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki, M., Narusaka, M., Ishida, J., Nanjo, T., Fujita, M., Oono, Y., Kamiya, A., Nakajima, M., Enju, A., Sakurai, T., et al. (2002) Plant J. 31, 279-292. [DOI] [PubMed] [Google Scholar]
- 22.Weckwerth, W., Loureiro, M. E., Wenzel, K. & Fiehn, O. (2004) Proc. Natl. Acad. Sci. USA 101, 7809-7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weckwerth, W., Wenzel, K. & Fiehn, O. (2004) Proteomics 4, 78-83. [DOI] [PubMed] [Google Scholar]
- 24.Wanner, L. A., Li, G., Ware, D., Somssich, I. E. & Davis, K. R. (1995) Plant Mol. Biol. 27, 327-338. [DOI] [PubMed] [Google Scholar]
- 25.Pelletier, M. K. & Shirley, B. W. (1996) Plant Physiol. 111, 339-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delseny, M., Cooke, R. & Penon, P. (1983) Plant Sci. Lett. 30, 107-119. [Google Scholar]
- 27.McKown, R., Kuroki, G. & Warren, G. (1996) J. Exp. Bot. 47, 1919-1925. [Google Scholar]
- 28.Wanner, L. A. & Junttila, O. (1999) Plant Physiol. 120, 391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O. & Thomashow, M. F. (1998) Science 280, 104-106. [DOI] [PubMed] [Google Scholar]
- 30.Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1999) Nat. Biotechnol. 17, 287-291. [DOI] [PubMed] [Google Scholar]
- 31.Levitt, J. (1980) Responses of Plants to Environmental Stresses (Academic, New York).
- 32.Rudolph, A. S. & Crowe, J. H. (1985) Cryobiology 22, 367-377. [DOI] [PubMed] [Google Scholar]
- 33.Strauss, G. & Hauser, H. (1986) Proc. Natl. Acad. Sci. USA 83, 2422-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter, J. F. & Crowe, J. H. (1988) Cryobiology 25, 244-255. [DOI] [PubMed] [Google Scholar]
- 35.Fiehn, O. (2002) Plant Mol. Biol. 48, 155-171. [PubMed] [Google Scholar]
- 36.Tolstikov, V. V. & Fiehn, O. (2002) Anal. Biochem. 301, 298-307. [DOI] [PubMed] [Google Scholar]
- 37.Gilmour, S. J., Fowler, S. G. & Thomashow, M. F. (2004) Plant Mol. Biol. 54, 767-781. [DOI] [PubMed] [Google Scholar]
- 38.Arguelles, J. C. (2000) Arch. Microbiol. 174, 217-224. [DOI] [PubMed] [Google Scholar]
- 39.Chen, Q., Ma, E., Behar, K. L., Xu, T. & Haddad, G. G. (2002) J. Biol. Chem. 277, 3274-3279. [DOI] [PubMed] [Google Scholar]
- 40.Colaco, C., Sen, S., Thangavelu, M., Pinder, S. & Roser, B. (1992) Biotechnology 10, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 41.Bouchereau, A., Aziz, A., Larher, F. & Martin-Tanguy, J. (1999) Plant Sci. 140, 103-125. [Google Scholar]
- 42.Urano, K., Yoshiba, Y., Nanjo, T., Ito, T., Yamaguchi-Shinozaki, K. & Shinozaki, K. (2004) Biochem. Biophys. Res. Commun. 313, 369-375. [DOI] [PubMed] [Google Scholar]
- 43.Kim, T. E., Kim, S. K., Han, T. J., Lee, J. S. & Chang, S. C. (2002) Physiol. Plant 115, 370-376. [DOI] [PubMed] [Google Scholar]
- 44.Conklin, P. L. (2001) Plant Cell Environ. 24, 383-394. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.