The NKD transcription factors regulate diverse and key pathways in developing maize endosperm, including cell fate, storage compound accumulation, and seed maturation.
Abstract
NAKED ENDOSPERM1 (NKD1) and NKD2 are duplicate INDETERMINATE DOMAIN (IDD) transcription factors important for maize (Zea mays) endosperm development. RNA-seq analysis of the nkd1 nkd2 mutant endosperm revealed that NKD1 and NKD2 influence 6.4% of the transcriptome in developing aleurone and 6.7% in starchy endosperm. Processes regulated by NKD1 and NKD2 include gene expression, epigenetic functions, cell growth and division, hormone pathways, and resource reserve deposition. The NKD1 and NKD2 proteins bind a consensus DNA sequence of TTGTCGT with slightly different properties. This motif was enriched in the promoters of gene transcripts differentially expressed (DE) in mutant endosperm. DE genes with a NKD binding motif in the 5′ promoter region were considered as likely direct targets of NKD1 and NKD2 regulation, and these putative direct target genes were notably enriched for storage proteins. Transcription assays demonstrate that NKD1 and NKD2 can directly regulate gene transcription, including activation of opaque2 and viviparous1 promoters. NKD2 functions as a negative regulator of nkd1 transcription, consistent with previously reported feedback regulation. NKD1 and NKD2 can homo- and heterodimerize through their ID domains. These analyses implicate NKD1 and NKD2 as central regulators of gene expression in developing maize endosperm.
INTRODUCTION
Cereal endosperm nourishes the developing embryo and germinating seedling, composes a major portion of human and livestock diets, and has important industrial applications. Maize (Zea mays) endosperm consists of seven cell types as defined by histology and gene expression patterns (Leroux et al., 2014; Zhan et al., 2015). In mature kernels, the major cell types include starchy endosperm (SE), basal endosperm transfer (BET) cells, and aleurone (AL) (Becraft and Gutierrez-Marcos, 2012). SE cells constitute the majority of the endosperm mass and function in nutrient reserve storage, primarily starch and protein. BET cells transport nutrients from maternal tissue into the developing endosperm. AL is important for digestion and remobilization of stored reserves during germination as well as mineral storage and pathogen defense (Stewart et al., 1988; Fath et al., 2000; Jerkovic et al., 2010).
Endosperm development begins with the formation of the coenocyte followed by cellularization. Subsequent cell differentiation involves the perception and response to positional cues that specify the different cell fates (Becraft and Asuncion-Crabb, 2000; Geisler-Lee and Gallie, 2005; Gruis et al., 2006). Maturation entails genomic endoreduplication and the accumulation of resource reserves in SE, acquisition of desiccation tolerance in AL, culminating in programmed cell death of the SE and BET cells, metabolic quiescence of the AL, and desiccation of the grain as a whole (Sabelli and Larkins, 2009; Becraft and Gutierrez-Marcos, 2012).
The duplicate genes naked endosperm1 (nkd1) and nkd2 encode INDETERMINATE DOMAIN (IDD) proteins (Yi et al., 2015), and the nkd1 nkd2 double mutant shows pleiotropic effects, including multiple layers of peripheral endosperm cells with compromised cell identity, decreased anthocyanin accumulation, opaque and floury endosperm texture, decreased carotenoid accumulation, decreased kernel dry weight, and occasional vivipary (Becraft and Asuncion-Crabb, 2000; Yi et al., 2015). These phenotypes indicate that nkd1 and nkd2 functions are required for cell patterning and differentiation, resource reserve deposition, and seed maturation. In the wild type, nkd1 and nkd2 transcripts accumulate in both AL and SE, consistent with the pleiotropic phenotype (Yi et al., 2015).
IDD proteins are a plant-specific family of transcription factors (TFs) whose members function in a broad range of developmental and signaling processes (Morita et al., 2006; Welch et al., 2007; Tanimoto et al., 2008; Hassan et al., 2010; Feurtado et al., 2011; Ogasawara et al., 2011; Seo et al., 2011b; Cui et al., 2013; Wu et al., 2013; Yoshida et al., 2014; Yoshida and Ueguchi-Tanaka, 2014; Jöst et al., 2016). The IDD is composed of a nuclear localization signal and four highly conserved tandem zinc fingers: one standard C2H2 zinc finger, one irregular C2H2 zinc finger, and two irregular CCHC zinc fingers. There are 17 IDD family members in maize, some with tissue-specific expression differences (Colasanti et al., 2006; Sekhon et al., 2013; Yi et al., 2015). The founding member of the IDD family, INDETERMINATE1 (ID1), controls flowering time in maize (Colasanti et al., 1998). ID1 localizes to the nucleus and binds an 11-bp DNA consensus sequence of TTTGTCGTTTT, which NKD1 (IDDveg9) can also bind, although with different specificity (Kozaki et al., 2004; Wong and Colasanti, 2007; Yi et al., 2015).
In this study, we analyze NKD1 and NKD2 function in developing maize endosperm. We identify the DNA binding specificities of NKD1 and NKD2, discern that NKD1 and NKD2 proteins can homo- and heterodimerize through their IDDs, and demonstrate they regulate transcription by binding DNA. RNA-sequencing (RNA-seq) analysis of nkd1 nkd2 versus wild-type endosperm revealed that NKD1 and NKD2 regulate widespread processes, including nutrient reserve deposition as well as cell growth and proliferation. Analyses of transcriptomic data lead to functional predictions that were validated by phenotypic analyses of nkd1 nkd2 mutants.
RESULTS
Identification of Endosperm Gene Transcripts Regulated by NKD1 and NKD2
To identify genes and biological processes directly or indirectly regulated by NKD1 and NKD2 (NKD1/2) in developing endosperm, a transcriptomic analysis was undertaken to identify gene transcripts differentially expressed between the wild type and nkd1 nkd2 mutant. Laser capture microdissection (LCM) coupled with RNA-seq was previously performed on AL and SE cells from the wild type (B73 inbred) versus nkd1 nkd2 mutant endosperms at 15 d after pollination (DAP) (Yi et al., 2015). These 50-base paired-end reads were realigned and mapped to the maize reference genome (B73 RefGen-V3, GSE61057). A total of 1.14 × 109 raw reads were generated, with a minimum of 256 × 108 per tissue, summed over three biological replicates (Supplemental Table 1). Reads that uniquely mapped to the genome and were properly paired were selected for further analysis, which ranged from 40.7 to 72.6% of the raw reads per tissue. Expression of 34,014 genes was detected in AL cells, whereas 32,629 genes were expressed in SE. Among these, 31,792 genes were expressed in all endosperm tissues sampled (wild type and nkd1 nkd2 mutant; AL and SE).
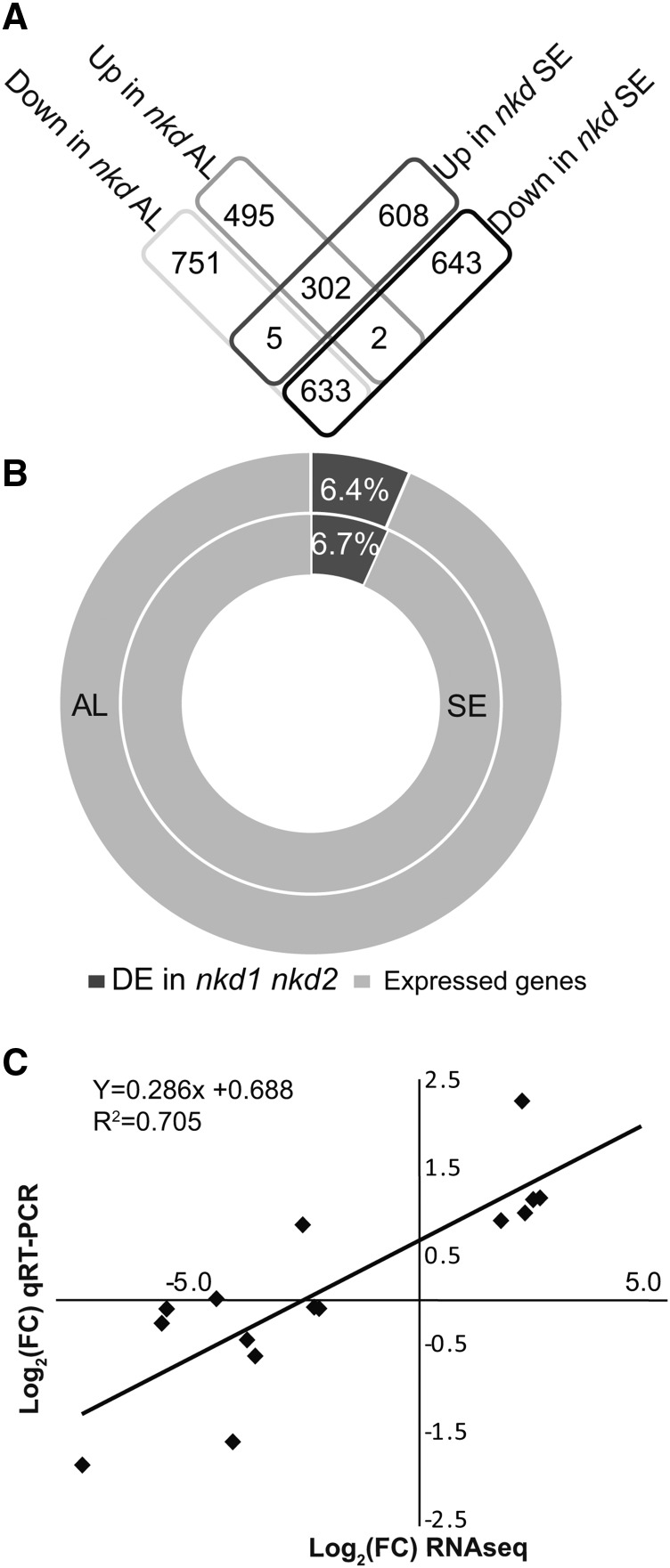
Differential expression (DE) analysis comparing nkd1 nkd2 mutant to the wild type identified 2188 DE genes in AL and 2193 in SE (adjusted P value ≤ 0.01) (Figure 1; Supplemental Table 2; Supplemental Data Sets 1 and 2). An MA plot of gene expression changes did not reveal any systemic bias in the data (Supplemental Figure 1A). Multidimensional scaling analysis of gene expression variance among samples revealed that AL exhibited lesser variance than SE (Supplemental Figure 1B). Furthermore, nkd1 nkd2 genotype had a greater overall impact on gene expression than did tissue type. Transcript abundance among the DE genes ranged from highs with mean normalized read counts up to 8 × 106 to lows of less than 10. Among DE genes, 63.48% had decreased transcript abundance in mutant AL and 58.28% had decreased transcript abundance in mutant SE. The AL and SE DE data sets showed statistically significant overlap and similar transcript fold change trends (P < 0.001; Z-score of 80.8). A total of 935 genes were DE between the wild type and mutant in both tissues (Figure 1A).
Figure 1.
Summary of Transcriptomic Analysis
(A) Shared and unique differentially expressed genes in nkd1-R nkd2-R mutant AL and SE.
(B) Proportion of genes with ≥1 read counts detected in RNA-seq (expressed) to number of DE genes in AL and SE.
(C) Scatterplot of RNA-seq transcript abundance fold change values in nkd1-R nkd2-R mutant AL relative to wild-type AL (on the x axis) against RT-PCR expression values (on y axis) for the15 RNA-seq DE genes in nkd1 nkd2 mutant AL tested by qRT-PCR.
To validate the RNA-Seq data, 15 DE genes were tested by qRT-PCR on independent, unamplified RNA from 15-DAP wild-type and nkd1 nkd2 AL. The qRT-PCR expression values showed a positive correlation (R2 = 0.7036) with the RNA sequencing log2 fold changes, indicating minimal bias in LCM-RNA sequencing results (Figure 1C). Eight of these transcripts exhibited statistically significant DE as expected based on the RNA-seq analysis (Supplemental Figure 1C). Verified transcripts showing increased expression in nkd1 nkd2 AL included cell cycle related genes (tubulin1, cell division cycle2-like, actin-1, and proliferating cell nuclear antigen2) and genes annotated as involved in epigenetic regulation (e.g., one encoding FASCIATA1-LIKE). Transcripts showing decreased mutant expression included the AL cell identity marker phospholipid transfer protein homolog1 (Gruis et al., 2006) and genes related to nutrient reserve accumulation (granule bound starch synthase and prolamin-box binding factor1).
nkd1 and nkd2 Regulate Genes Involved in Diverse Processes
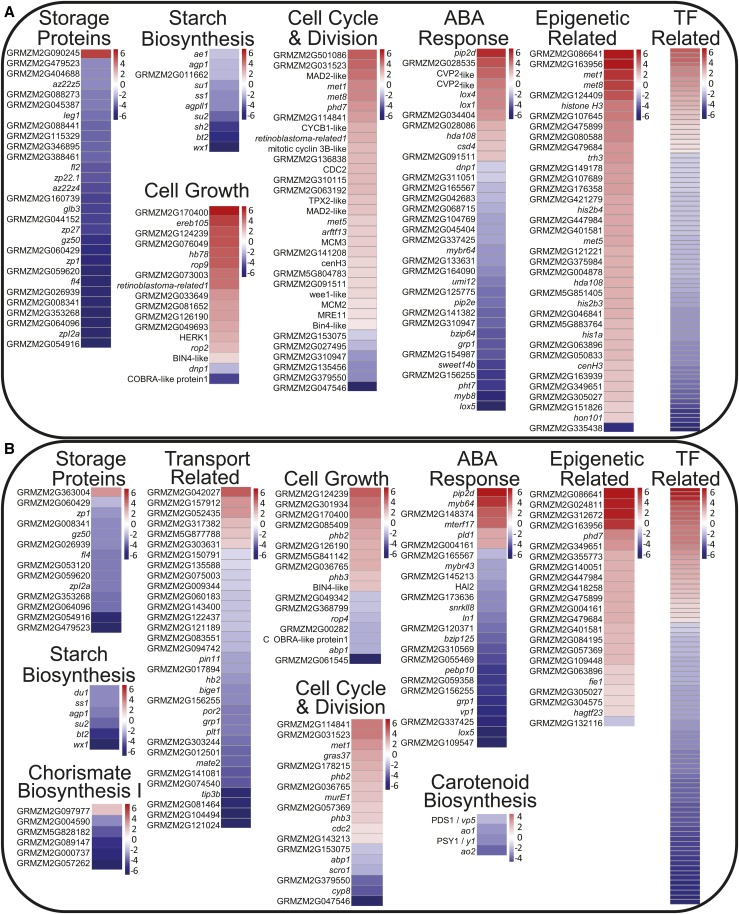
To determine biological processes regulated by nkd1 and nkd2, pathway analyses were performed on DE genes using Gene Ontology (GO), MaizeCyc, and CornCyc (Monaco et al., 2013). Complete lists of significantly enriched GO terms are shown in Tables 1 and 2, and individual genes within each pathway are reported in Supplemental Data Sets 1 and 2. There were notable effects on genes functioning in the regulation of gene expression, including 80 TFs with altered expression in mutant AL and 86 in SE (Figure 2). Epigenetic related factors were also affected, showing a general trend of increased expression in both mutant AL and SE. Thirty-seven epigenetic related factors were DE in AL and 23 in SE. Cell division and cell growth pathways trended to increased expression in the mutant AL. Thirty-three cell division-related factors were DE in AL and 18 in SE, while 17 cell growth factors were DE in AL and 17 in SE. The AL and SE each also showed wide ranging effects on hormone-related pathways including biosynthesis, signaling, and response in systems including auxin, abscisic acid (ABA), ethylene, jasmonate, brassinosteroids, cytokinins, and gibberellins (Figure 2; Supplemental Figures 2 and 3).
Table 1. Enriched GO Terms in nkd1 nkd2 Mutant Aleurone Differentially Expressed Genes.
| GO Term | Ontology | Description | DE ALa | AL Tran.b | P Valuec | FDRd |
|---|---|---|---|---|---|---|
| GO:0045735 | F | Nutrient reservoir activity | 28 | 56 | 3.30E-18 | 3.70E-15 |
| GO:0032993 | C | Protein-DNA complex | 28 | 106 | 4.60E-10 | 3.10E-07 |
| GO:0000786 | C | Nucleosome | 26 | 101 | 3.40E-09 | 1.20E-06 |
| GO:0044427 | C | Chromosomal part | 35 | 173 | 8.20E-09 | 1.90E-06 |
| GO:0031497 | P | Chromatin assembly | 27 | 109 | 4.30E-09 | 3.80E-06 |
| GO:0006334 | P | Nucleosome assembly | 27 | 107 | 2.80E-09 | 3.80E-06 |
| GO:0071103 | P | DNA conformation change | 33 | 152 | 3.40E-09 | 3.80E-06 |
| GO:0065004 | P | Protein-DNA complex assembly | 27 | 110 | 5.30E-09 | 3.80E-06 |
| GO:0034728 | P | Nucleosome organization | 27 | 107 | 2.80E-09 | 3.80E-06 |
| GO:0006333 | P | Chromatin assembly or disassembly | 27 | 111 | 6.50E-09 | 3.90E-06 |
| GO:0006323 | P | DNA packaging | 27 | 115 | 1.50E-08 | 7.50E-06 |
| GO:0000785 | C | Chromatin | 27 | 122 | 5.50E-08 | 9.40E-06 |
| GO:0006325 | P | Chromatin organization | 39 | 233 | 2.60E-07 | 0.00012 |
| GO:0005694 | C | Chromosome | 37 | 230 | 1.40E-06 | 0.00019 |
| GO:0051276 | P | Chromosome organization | 42 | 271 | 8.00E-07 | 0.00032 |
| GO:0009501 | C | Amyloplast | 5 | 7 | 2.80E-05 | 0.0032 |
| GO:0046982 | F | Protein heterodimerization activity | 26 | 148 | 9.40E-06 | 0.0052 |
| GO:0034622 | P | Cellular macromolecular complex assembly | 41 | 297 | 1.90E-05 | 0.0068 |
| GO:0016701 | F | Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen | 14 | 58 | 2.90E-05 | 0.011 |
| GO:0007017 | P | Microtubule-based process | 28 | 179 | 4.00E-05 | 0.013 |
| GO:0019252 | P | Starch biosynthetic process | 8 | 21 | 4.40E-05 | 0.013 |
| GO:0004866 | F | Endopeptidase inhibitor activity | 12 | 49 | 9.00E-05 | 0.02 |
| GO:0030414 | F | Peptidase inhibitor activity | 12 | 49 | 9.00E-05 | 0.02 |
| GO:0015630 | C | Microtubule cytoskeleton | 27 | 189 | 0.00025 | 0.025 |
| GO:0034621 | P | Cellular macromolecular complex subunit organization | 43 | 340 | 9.60E-05 | 0.027 |
| GO:0003777 | F | Microtubule motor activity | 15 | 79 | 0.00028 | 0.052 |
| GO:0009536 | C | Plastid | 188 | 2144 | 0.00068 | 0.058 |
| GO:0006112 | P | Energy reserve metabolic process | 5 | 10 | 0.00028 | 0.06 |
| GO:0005977 | P | Glycogen metabolic process | 5 | 10 | 0.00028 | 0.06 |
| GO:0005978 | P | Glycogen biosynthetic process | 5 | 10 | 0.00028 | 0.06 |
| GO:0007018 | P | Microtubule-based movement | 15 | 79 | 0.00028 | 0.06 |
| GO:0005975 | P | Carbohydrate metabolic process | 100 | 1023 | 0.00036 | 0.071 |
| GO:0009507 | C | Chloroplast | 178 | 2036 | 0.001 | 0.076 |
| GO:0003774 | F | Motor activity | 16 | 95 | 0.00072 | 0.078 |
| GO:0005506 | F | Iron ion binding | 57 | 534 | 0.00077 | 0.078 |
| GO:0016798 | F | Hydrolase activity, acting on glycosyl bonds | 46 | 407 | 0.00071 | 0.078 |
| GO:0004867 | F | Serine-type endopeptidase inhibitor activity | 9 | 36 | 0.00057 | 0.078 |
| GO:0051213 | F | Dioxygenase activity | 11 | 52 | 0.00068 | 0.078 |
| GO:0019843 | F | rRNA binding | 11 | 55 | 0.0011 | 0.097 |
| GO:0004553 | F | Hydrolase activity, hydrolyzing O-glycosyl compounds | 42 | 371 | 0.0011 | 0.097 |
| GO:0005875 | C | Microtubule associated complex | 15 | 92 | 0.0015 | 0.1 |
| GO:0065003 | P | Macromolecular complex assembly | 44 | 382 | 0.00062 | 0.12 |
| GO:0046983 | F | Protein dimerization activity | 58 | 566 | 0.0018 | 0.13 |
| GO:0008483 | F | Transaminase activity | 10 | 50 | 0.0018 | 0.13 |
| GO:0016769 | F | Transferase activity, transferring nitrogenous groups | 10 | 50 | 0.0018 | 0.13 |
| GO:0016702 | F | Oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | 10 | 51 | 0.0022 | 0.14 |
| GO:0051087 | F | Chaperone binding | 7 | 28 | 0.0023 | 0.14 |
| GO:0044430 | C | Cytoskeletal part | 28 | 229 | 0.0023 | 0.14 |
| GO:0044422 | C | Organelle part | 262 | 3190 | 0.0029 | 0.14 |
| GO:0044446 | C | Intracellular organelle part | 262 | 3187 | 0.0028 | 0.14 |
| GO:0005576 | C | Extracellular region | 49 | 474 | 0.0032 | 0.14 |
| GO:0031226 | C | Intrinsic to plasma membrane | 11 | 61 | 0.0027 | 0.14 |
| GO:0031225 | C | Anchored to membrane | 9 | 46 | 0.0036 | 0.14 |
| GO:0046658 | C | Anchored to plasma membrane | 9 | 46 | 0.0036 | 0.14 |
| GO:0005856 | C | Cytoskeleton | 31 | 272 | 0.0041 | 0.15 |
| GO:0043232 | C | Intracellular non-membrane-bounded organelle | 123 | 1405 | 0.0045 | 0.15 |
| GO:0043228 | C | Non-membrane-bounded organelle | 123 | 1405 | 0.0045 | 0.15 |
| GO:0051301 | P | Cell division | 22 | 155 | 0.00096 | 0.17 |
| GO:0048046 | C | Apoplast | 35 | 323 | 0.0054 | 0.17 |
| GO:0033293 | F | Monocarboxylic acid binding | 5 | 16 | 0.0035 | 0.2 |
Table 2. Enriched GO Terms in nkd1 nkd2 Mutant Starchy Endosperm Differentially Expressed Genes.
| GO Term | Ontology | Description | DE SEa | SE Tran.b | P Valuec | FDRd |
|---|---|---|---|---|---|---|
| GO:0006261 | P | DNA-dependent DNA replication | 16 | 56 | 1.00E-06 | 0.0037 |
| GO:0005730 | C | Nucleolus | 53 | 432 | 8.10E-05 | 0.057 |
| GO:0031974 | C | Membrane-enclosed lumen | 79 | 755 | 0.00042 | 0.1 |
| GO:0070013 | C | Intracellular organelle lumen | 77 | 747 | 0.00075 | 0.1 |
| GO:0048046 | C | Apoplast | 39 | 324 | 0.00089 | 0.1 |
| GO:0009536 | C | Plastid | 193 | 2128 | 0.00046 | 0.1 |
| GO:0043233 | C | Organelle lumen | 77 | 747 | 0.00075 | 0.1 |
| GO:0009507 | C | Chloroplast | 179 | 2023 | 0.0019 | 0.18 |
| GO:0005618 | C | Cell wall | 60 | 579 | 0.0022 | 0.18 |
| GO:0031981 | C | Nuclear lumen | 63 | 613 | 0.0022 | 0.18 |
| GO:0044422 | C | Organelle part | 265 | 3160 | 0.004 | 0.2 |
| GO:0030312 | C | External encapsulating structure | 60 | 596 | 0.0042 | 0.2 |
| GO:0044435 | C | Plastid part | 105 | 1128 | 0.0031 | 0.2 |
| GO:0044446 | C | Intracellular organelle part | 265 | 3157 | 0.0038 | 0.2 |
| GO:0005576 | C | Extracellular region | 49 | 467 | 0.0041 | 0.2 |
| GO:0032993 | C | Protein-DNA complex | 16 | 105 | 0.0028 | 0.2 |
Number of genes associated with each GO term that are differentially expressed between wild-type and nkd1 nkd2 starchy endosperm.
Total number of genes in each GO category expressed in the starchy endosperm transcriptome.
Fisher’s exact test for GO term enrichment.
False discovery rate.
Figure 2.
Pathway Analysis of Differentially Expressed Gene Transcripts
Log2 fold change heat maps of differentially expressed genes functioning in selected disrupted pathways in nkd1 nkd2 mutants. Aleurone (A) and starchy endosperm (B).
Pathways involved in the metabolism or accumulation of storage compounds generally showed decreased transcript levels in nkd1 nkd2 endosperm (Figure 2; Supplemental Figures 2 and 3). Notably, many pathways involved in carbohydrate metabolism were affected. The mutant SE and AL both exhibited decreased expression of genes functioning in starch/glycogen biosynthesis, sucrose biosynthesis, and sucrose, galactose, and glycerol degradation, while mutant AL also showed decreased expression of gluconeogenesis genes. Protein metabolism was also affected, with mutant SE and AL both showing decreased expression of genes for storage protein accumulation (nutrient reservoir activity) and amino acid biosynthesis, while mutant SE also showed decreased expression of chorismate biosynthesis genes. Mutant AL also displayed decreased expression of lipid metabolism pathways, including linoleate biosynthesis and triacylglycerol degradation (Supplemental Figure 2A). SE showed decreased expression of genes involved in nutrient transport pathways (Figure 2). Of final note, genes involved in defense response were altered in both AL and SE, with an overall decrease in defense response-related gene expression in the mutant (Supplemental Figures 2C and 3C). Functional characterization performed by evaluating GO overrepresentation produced results consistent with pathway analyses, with AL DE genes enriched for GO terms related to storage proteins (nutrient reservoir activity), sugar metabolism, epigenetics, microtubule-related processes, and cell division (Table 1). SE GO terms were enriched for DNA replication, nucleolus, plastid, and cell wall (Table 2).
Pathway analysis suggested that nkd1 and nkd2 may regulate cellular transport activities. BET cells are sites of intensive nutrient transport from the maternal plant tissue into the developing endosperm (Thompson et al., 2001) and decreased BET cell transport activity would be consistent with the decreased kernel weight and storage reserve accumulation in the mutant (Table 3). Because BET tissues were not included in the transcriptomic analysis, potential NKD1 NKD2 regulation of BET gene expression was tested using qRT-PCR on BET cell marker genes. RNA was isolated from the lower quarter of 12-DAP wild-type and nkd1 nkd2 endosperms. The expression of maternally expressed gene1 (meg1; P = 0.027) and basal endosperm transfer layer2/bap2 (betl2; P = 0.0195) was significantly increased in nkd1 nkd2 mutant, while betl1 (P = 0.482) and myb related protein1 (mrp1; P = 0.1445) were not significantly different (Supplemental Figure 4). These results suggest that nkd1 and nkd2 likely regulate transcription in BET cells but are not required for BET cell differentiation.
Table 3. Wild-Type and nkd1 nkd2 Mutant Seed Compositions.
| Seed Component | Wild Type (±se) | nkd1 nkd2 (±se) | P Valuea |
|---|---|---|---|
| Total seed weight (mg) | 220.16 (1.49) | 128.12 (4.52) | 1.01E-03 |
| Pericarp (mg) | 11.34 (0.7) | 9.61 (0.41) | 0.12 |
| Endosperm (mg) | 186.45 (1.39) | 91.67 (3.74) | 4.72E-04 |
| Embryo (mg) | 22.38 (0.52) | 26.84 (0.86) | 0.02 |
| Endosperm protein (mg) | 16.63 (1.05) | 7.96 (0.53) | 5.39E-03 |
| Endosperm nitrogen (%) | 1.58 (0.05) | 1.72 (0.12) | 0.34 |
| Endosperm starch (mg) | 108.19 (5.75) | 73.02 (2.98) | 2.97E-05 |
Student’s t test.
Effects of the nkd1 nkd2 Mutant on Endosperm Composition
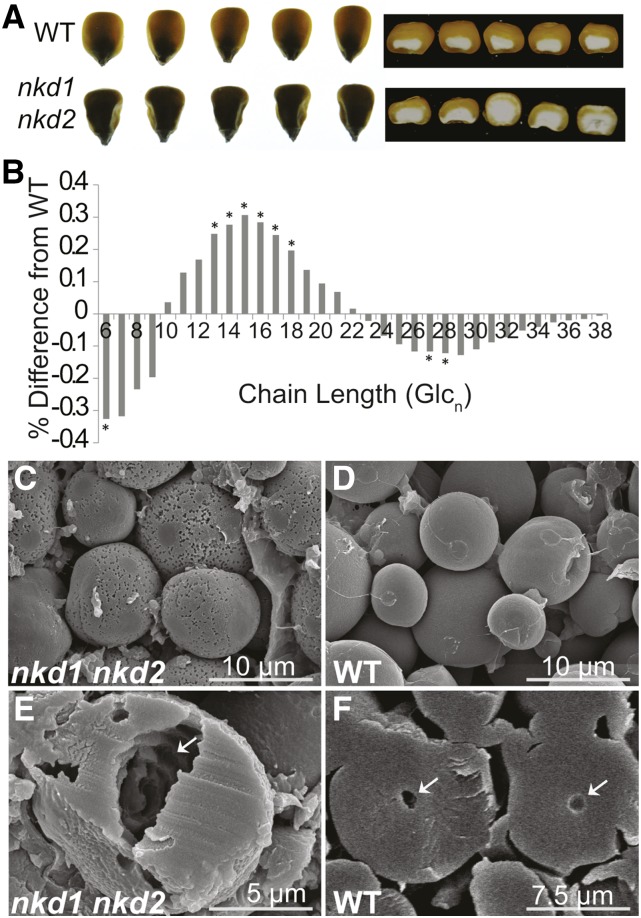
The nkd1 nkd2 mutant was previously reported to cause decreased total kernel weight and opaque endosperm texture (Yi et al., 2015), and transcriptomic analysis suggested alterations in starch and storage protein pathways, prompting examination of seed composition. Wild-type and nkd1-Ds nkd2-Ds0297 mutant kernels from the same F2 segregating ears were compared. Mutants contained less vitreous endosperm than the wild type (Figure 3A) and showed a 41.8% decrease of total kernel dry weight with a 50.8% reduction in endosperm (Table 3). The nkd1-Ds nkd2-Ds0297 mutant kernels showed a 32.5% reduction in total starch abundance as determined by amyloglucosidase digestion quantified by colorimetric glucose oxidase-peroxidase (GOPOD) assays (Table 3). Some of the DE gene transcripts were implicated in determining starch structure (Figure 2), of which glucan branch chain length distribution is a major component. Glucan branch chain length abundance was measured by fluorophore-assisted carbohydrate electrophoresis of debranched endosperm starch (O’Shea and Morell, 1996). Difference plots of nkd1 nkd2 mutant minus wild-type starch chain length abundance show the mutant starch had a decrease in the frequency of branch chains of 6 and 27 to 28 glucose units, while chains 13 to 18 glucose units in length were significantly increased in abundance (Figure 3B).
Figure 3.
Disrupted Nutrient Reserve Deposition in nkd1 nkd2 Mutant Endosperm
(A) Opaque endosperm phenotype in wild-type and nkd1-Ds nkd2-Ds0297 mutant kernels from an F2 segregating ear
(B) Difference plots of nkd1-Ds nkd2-Ds0297 mutant minus wild-type mean starch branch chain length abundance. Asterisk denotes statistically significant differences (P < 0.05) determined by Student’s t test.
(C) to (F) Scanning electron microscopy of starch grains of mature segregating nkd1-Ds nkd2-Ds0297 mutant ([C] and [E]) and wild type ([D] and [F]) kernels. Arrows indicate hollow core in bisected starch granules.
Starch grain morphology was examined with scanning electron microscopy. Whereas the wild-type starch granules had smooth surfaces and were nearly spherical in shape, mutant granules had distinctly pitted surfaces and were more irregularly shaped, sometimes appearing faceted (Figures 3C and 3D; Supplemental Figure 5). Furthermore, bisected mutant starch grains often showed irregular hollow cores with internal pitting, while wild-type grains were nearly solid with a small hollow center (Figures 3E and 3F).
Protein content in the wild type versus nkd1-Ds nkd2-Ds0297 mutant endosperm was estimated from total nitrogen content measured using Dumas N combustion (Schindler and Knighton, 1999). There was a 52.11% reduction in total protein content in endosperm of mutant segregants (P = 0.0054; Table 3).
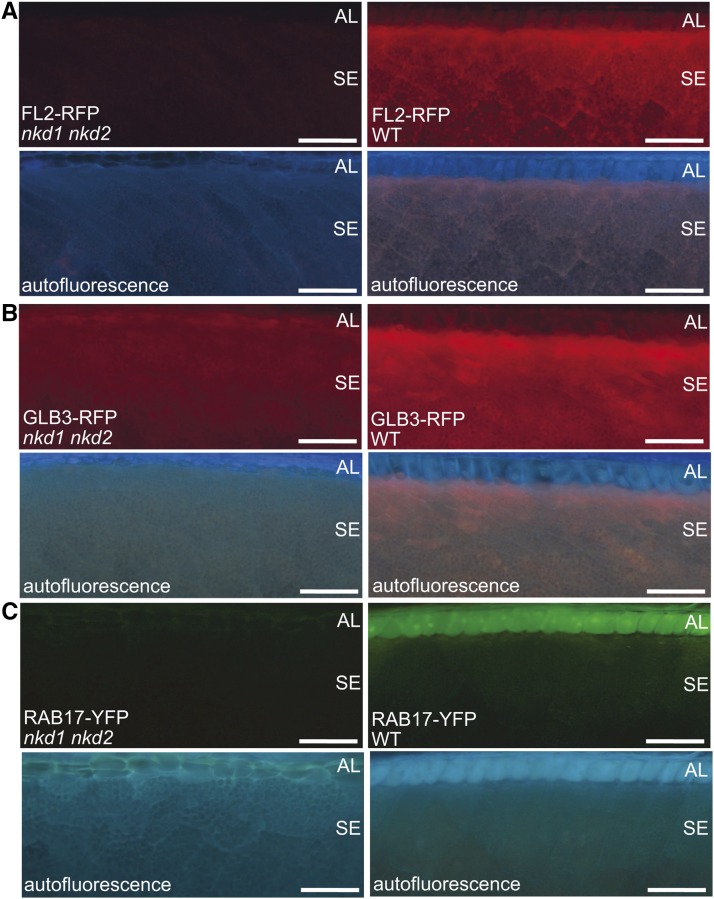
Fluorescently tagged transgenes were available for several genes DE in nkd1 nkd2 mutant endosperm, including two storage protein genes, floury2 (Coleman et al., 1997) and globulin3 (glb3) (Woo et al., 2001), and a putative ABA response gene, responsive to aba17 (rab17) (Kizis and Pagès, 2002). The nkd1-R nkd2-R mutant alleles were introduced into FL2-RFP, GLB3-RFP, and RAB17-YFP transgenic lines, which consist of fluorescent proteins translationally fused to the maize gene coding sequences controlled by their native regulatory elements (Mohanty et al., 2009). Mature kernels were sampled from segregating F2 ears and epifluorescence imaging demonstrated that each reporter showed decreased fluorescence intensity in mutant compared with the wild type (Figure 4). Thus, decreased expression levels of these transgenic reporter proteins were consistent with the RNA-seq analysis of native gene transcripts.
Figure 4.
Marker Transgene Expression in Wild-Type and nkd1 nkd2 Mutant Endosperm.
Florescence microscopy using narrow violet-broad range (NV) filter to visualize autofluorescence and an mCherry filter to visualize FL2-RFP (A) and GL3B-RFP (B) or a YFP florescence filter for RAB17-YFP (C). Bars = 100 μm.
Many variables can influence grain yield and composition but principal component analysis (PCA) clearly identified genotype as the primary contributor to the observed variance in these traits. Genotype explained 99.9% of the variance for endosperm weight and total protein, as well as for starch content and branch length, while the remaining 0.1% of variance was explained by independent cob, or independent kernel for protein and starch, respectively (Supplemental Figure 6).
Identification of NKD1 and NKD2 Target DNA Binding Sequences
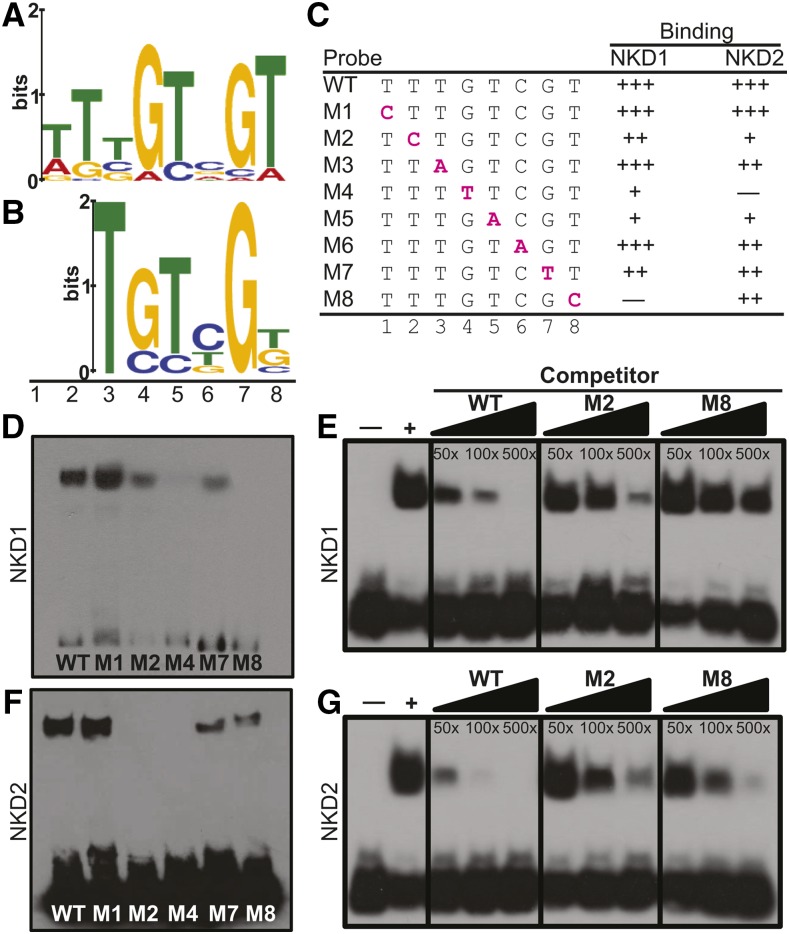
To identify DNA binding sequences for NKD1 and NKD2 proteins, selection and amplification binding (SAAB) was undertaken. Columns were generated with the conserved DNA binding ID domains of NKD1 (NKD1ID) and NKD2 (NKD2ID). Each protein was bacterially expressed as a GST fusion and purified proteins were covalently bound to ester agarose columns. The identity of purified recombinant protein was confirmed by SDS-PAGE and tandem mass spectrometry (Supplemental Figure 7). A column of purified GST from the empty vector served as a negative control for nonspecific binding. Libraries enriched for DNA sequences that interact with the NKDID proteins were generated by passing random double-stranded oligonucleotides over each protein column for six cycles of selection and amplification. After cloning and sequencing selected oligos, a total of 77 unique sequences were recovered for NKD1 and 54 for NKD2. Motif analysis of the recovered sequences identified an 8-bp binding consensus sequence (BCS) of [TA]-T-[TCG]-G-T-[CGA]-G-T for NKD1 produced from 27 of the recovered sequences (Figure 5A; Supplemental Table 3). For NKD2, a 6-bp BCS of T-G-T-[CT]-G-[TG] was identified from 19 recovered sequences, which was similar to the NKD1 BCS (Figure 5B; Supplemental Table 4). No sequences were recovered from the GST negative controls.
Figure 5.
NKD1 and NKD2 DNA Binding.
(A) and (B) SAAB-MEME derived BCSs for NKD1ID (A) and NKD2ID (B).
(C) Oligonucleotide sequences used as probes for EMSAs. The wild type contains the consensus binding sequence, while M1 to M8 contain base substitutions indicated by magenta lettering. Binding, relative to the wild type for a given protein is indicated by +++ (>66%), ++ (33 to 66%), + (<33%), and – (no detected shift).
(D) to (G) EMSAs.
(D) and (F) EMSA of consensus (wild type) and mutant probes for NKD1-ID (D) and NKD2-ID (F) proteins.
(E) and (G) Competition assays using labeled wild-type probe, and 50-, 100-, or 500-fold excess of unlabeled wild type, M2, or M8 oligonucleotide, incubated with NKD1ID (E) or NKD2ID (G) protein. The + indicates reaction with no competitor, and – indicates negative control with GST protein instead of NKD. Overexposed images, additional EMSAs, and negative controls using purified GST are shown in Supplemental Figures 8 and 9.
EMSAs were performed to further examine the DNA binding properties of the NKD1 and NKD2 ID domains. Oligonucleotide probes were designed based on frequency of base occupancy in the SAAB selected BCSs (Supplemental Figures 8A and 8B). To test specific DNA base requirements, each base was substituted individually and assayed with both the NKD1ID-GST and NKD2ID-GST proteins. The GST protein tag purified from the empty vector showed no interaction with any of the oligonucleotide probes (Supplemental Figures 8C and 8E). Relative binding affinity was determined for each mutant by comparing the amount of shifted probe relative to the non-mutant BCS probe. For NKD1ID, binding was abolished by a substitution of cytosine in position 8 and strongly decreased by a substitution of thymine at position 4 or adenine at position 5 (Figures 5C and 5D; Supplemental Figures 8E and 9). Base substitutions at positions 2 and 7 produced moderate decreases in binding and substitutions at positions 3 and 6 produced slight decreases in binding affinity. Although position 1 appeared selected by SAAB, a cytosine substitution did not decrease binding affinity. Taken together, NKD1ID recognizes a 7-bp BCS of T-T-G-T-C-G-T (NKDcore). NKD2ID recognizes the same 7-bp NKDcore BCS but with different binding tolerances for off consensus base substitutions in positions 2, 3, 4, 6, and 8 (Figures 5C and 5F; Supplemental Figure 8E). Although position 2 was not selected by NKD2ID SAAB, binding was abolished in EMSA by a cytosine substitution. Binding was also abolished by a substitution in position 4 and strongly decreased by a substitution in position 5. Moderate decreases in DNA binding were observed for substitutions in positions 7 and 8. Competition EMSA with mutant probes showed binding to the BCS was specific and confirmed that NKD1ID and NKD2ID have different tolerances for sequence variants at positions 2 and 8 (Figures 5E and 5G).
NKD1 and NKD2 Direct Target Gene Prediction and Motif Enrichment
To determine likely direct targets of NKD1 and NKD2, proximal promoter regions (PPRs) of all genes with transcripts detected in SE and AL were searched for the presence of NKD1, NKD2, and NKDcore BCSs using the MEME suite motif analysis tool Find Individual Motif Occurrences (FIMO) (Bailey et al., 2015). Sequence elements shown by EMSA not to bind NKD1 or NKD2 were removed from the analysis. NKDID binding to six of the variant BCSs identified by FIMO were confirmed by EMSA, verifying the efficacy of the FIMO analysis for predicting in vitro binding ability (Figures 5C, 5D, and 5F; Supplemental Figure 8E). A total of 32,621 PPRs were identified from the 15-DAP AL transcriptome, including 2135 DE genes, and 31,389 PPRs were identified from the SE transcriptome, including 2192 DE genes (Supplemental Data Set 3). The occurrences of NKD BCSs are summarized in Table 4, Supplemental Figure 10, and Supplemental Data Set 3. Due to the relatedness among the identified BCSs, there was considerable overlap among the FIMO-identified promoter elements (Supplemental Figure 10). Furthermore, because only six base pairs were selected for NKD2 while eight base pairs were selected for NKD1, there are considerably more potential NKD2 sites throughout the genome. Nearly half the predicted NKD1 and NKDcore sites are included among the NKD2 predicted sites and all the predicted NKD1 sites are included among the NKDcore sites.
Table 4. NKD BCS Enrichment in Differentially Expressed Gene Promoters.
| Motif | DE Genes | Transcriptome | P Valuea | |
|---|---|---|---|---|
| Aleurone | ||||
| NKD1 | 194 | 3,064 | P = 0.70 | |
| NKD2 | 1,050 | 11,200 | P ≤ 0.01 | |
| NKDcore | 816 | 10,684 | P ≤ 0.01 | |
| Starchy endosperm | ||||
| NKD1 | 217 | 2,930 | P ≤ 0.01 | |
| NKD2 | 1,059 | 15,222 | P = 0.95 | |
| NKDcore | 696 | 9,970 | P = 0.982 | |
The number of each motif’s occurrence in aleurone and starchy endosperm differentially expressed and RNA-seq detected (transcriptome) proximal promoter regions was determined. Motif enrichment in DE genes was assessed relative to the transcriptome.
Fisher’s exact test.
To test whether NKD BCSs were enriched in the promoters of genes DE in the nkd1 nkd2 mutant endosperm, the proportion of PPRs with one or more BCS among DE genes was compared with that of the 15 DAP transcriptome. In AL, NKD2 and NKDcore motifs were significantly enriched in DE genes, while the NKD1 motif was not (Table 4; Supplemental Data Set 3). Conversely, in SE, the NKD1 motif was enriched in DE genes, while NKD2 and NKDcore motifs were not significant (Table 4; Supplemental Data Set 3). BCS enrichment controls consisted of three “shuffle motifs” each for NKD1 and NKD2 generated by randomly rearranging bases in the BCSs. Enrichment was not detected for any of the shuffle motifs, indicating that the enrichment of the NKD motifs was specific (Supplemental Figure 11).
Predicted Direct Target Genes are Enriched for Storage Proteins
To determine pathways directly transcriptionally regulated by NKD1 and NKD2, a GO enrichment analysis was performed by comparing occurrences of GO terms among predicted direct target genes to the endosperm-expressed genes. For NKD2 AL, GO enrichment was observed for nutrient reservoir activity (storage proteins), starch biosynthesis, nucleosome and chromatin regulation, and processes related to plastids, vacuoles, and microtubules (Table 5; Supplemental Data Set 4). For NKDCore AL, GO enrichment was detected for nutrient reservoir activity (Table 5; Supplemental Data Set 4). GO enrichment was not detected for NKD1 AL target genes. In the SE, GO enrichment was detected for NKD1 target genes functioning in external encapsulating structure, cell wall, and apoplast (Table 6; Supplemental Data Set 4). For NKD2 SE, GO enrichment was detected for genes in nutrient reservoir activity, cell wall, and plastid-related ontologies (Table 6; Supplemental Data Set 4). GO enrichment was detected for NKDcore SE genes functioning in cell wall and external encapsulating structure (Table 6; Supplemental Data Set 4).
Table 5. Enriched GO Terms in Aleurone Direct Target Genes.
| GO Term | Ontology | Description | DE AL Genesa | AL Transcriptomeb | P Valuec | FDRd |
|---|---|---|---|---|---|---|
| NKD2 | ||||||
| GO:0045735 | F | Nutrient reservoir activity | 22 | 56 | 2.80E-18 | 1.70E-15 |
| GO:0032993 | C | Protein-DNA complex | 14 | 106 | 1.30E-05 | 0.0059 |
| GO:0000786 | C | Nucleosome | 13 | 101 | 3.40E-05 | 0.0077 |
| GO:0034728 | P | Nucleosome organization | 14 | 107 | 1.40E-05 | 0.0085 |
| GO:0006334 | P | Nucleosome assembly | 14 | 107 | 1.40E-05 | 0.0085 |
| GO:0031497 | P | Chromatin assembly | 14 | 109 | 1.80E-05 | 0.0085 |
| GO:0065004 | P | Protein-DNA complex assembly | 14 | 110 | 2.00E-05 | 0.0085 |
| GO:0006333 | P | Chromatin assembly or disassembly | 14 | 111 | 2.20E-05 | 0.0085 |
| GO:0006323 | P | DNA packaging | 14 | 115 | 3.20E-05 | 0.011 |
| GO:0071103 | P | DNA conformation change | 16 | 152 | 5.70E-05 | 0.016 |
| GO:0009536 | C | Plastid | 104 | 2144 | 0.00026 | 0.029 |
| GO:0000785 | C | Chromatin | 13 | 122 | 0.00024 | 0.029 |
| GO:0046982 | F | Protein heterodimerization activity | 15 | 148 | 0.00014 | 0.044 |
| GO:0044427 | C | Chromosomal part | 15 | 173 | 0.00077 | 0.07 |
| GO:0009507 | C | Chloroplast | 96 | 2036 | 0.00095 | 0.072 |
| GO:0005773 | C | Vacuole | 45 | 818 | 0.0011 | 0.072 |
| GO:0005694 | C | Chromosome | 17 | 230 | 0.0021 | 0.12 |
| GO:0019252 | P | Starch biosynthetic process | 5 | 21 | 0.00053 | 0.13 |
| GO:0050660 | F | FAD binding | 13 | 135 | 0.00063 | 0.13 |
| GO:0044435 | C | Plastid part | 57 | 1142 | 0.0025 | 0.13 |
| GO:0003774 | F | Motor activity | 10 | 95 | 0.0013 | 0.16 |
| GO:0003777 | F | Microtubule motor activity | 9 | 79 | 0.0013 | 0.16 |
| GO:0005875 | C | Microtubule associated complex | 9 | 92 | 0.0037 | 0.17 |
| GO:0009579 | C | Thylakoid | 29 | 513 | 0.0049 | 0.17 |
| GO:0044434 | C | Chloroplast part | 55 | 1125 | 0.0044 | 0.17 |
| GO:0015630 | C | Microtubule cytoskeleton | 14 | 189 | 0.0048 | 0.17 |
| GO:0044422 | C | Crganelle part | 135 | 3190 | 0.0058 | 0.18 |
| GO:0044446 | C | Intracellular organelle part | 135 | 3187 | 0.0056 | 0.18 |
| GO:0004553 | F | Hydrolase activity, hydrolyzing O-glycosyl compounds | 24 | 371 | 0.0019 | 0.19 |
| NKD | ||||||
| GO:0045735 | F | Nutrient reservoir activity | 21 | 56 | 6.50E-19 | 3.50E-16 |
The number of each GO term occurrence in differentially expressed AL genes with a motif in proximal promoter region and in RNA-seq AL detected (transcriptome) genes was determined. GO enrichment in DE genes with a motif was assessed relative to the transcriptome. GO enrichment was not detected for NKD1 AL direct target genes. See Supplemental Data Sets 3 and 4 for further details.
Number of putative direct target genes expressed in the aleurone associated with each GO term.
Total number of genes in each GO category expressed in the aleurone transcriptome.
Fisher’s exact test for GO term enrichment.
False discovery rate.
Table 6. Enriched GO Terms in Starchy Endosperm Direct Target Genes.
| GO Term | Ontology | Description | DE SE Genesa | SE Transcriptomeb | P Valuec | FDRd |
|---|---|---|---|---|---|---|
| NKD1 | ||||||
| GO:0030312 | C | External encapsulating structure | 12 | 596 | 0.0011 | 0.12 |
| GO:0005618 | C | Cell wall | 12 | 579 | 0.00084 | 0.12 |
| GO:0048046 | C | Apoplast | 8 | 324 | 0.002 | 0.14 |
| NKD2 | ||||||
| GO:0009536 | C | Plastid | 118 | 2128 | 2.40E-06 | 0.0012 |
| GO:0045735 | F | Nutrient reservoir activity | 12 | 67 | 3.10E-06 | 0.0021 |
| GO:0009507 | C | Chloroplast | 107 | 2023 | 3.80E-05 | 0.0097 |
| GO:0044435 | C | Plastid part | 63 | 1128 | 0.00027 | 0.046 |
| GO:0005618 | C | Cell wall | 36 | 579 | 0.00075 | 0.082 |
| GO:0044434 | C | Chloroplast part | 60 | 1111 | 0.00081 | 0.082 |
| GO:0030312 | C | External encapsulating structure | 36 | 596 | 0.0012 | 0.1 |
| NKD | ||||||
| GO:0005618 | C | Cell wall | 27 | 579 | 0.00053 | 0.15 |
| GO:0030312 | C | External encapsulating structure | 27 | 596 | 0.00081 | 0.15 |
The number of each GO term occurrence in differentially expressed SE genes with a motif in proximal promoter region and in RNA-seq SE detected (transcriptome) genes was determined. GO enrichment in DE genes with a motif was assessed relative to the transcriptome. See Supplemental Data Sets 3 and 4 for further details.
Number of putative direct target genes expressed in the starchy endosperm associated with each GO term.
Total number of genes in each GO category expressed in the starchy endosperm transcriptome.
Fisher’s exact test for GO term enrichment.
False discovery rate.
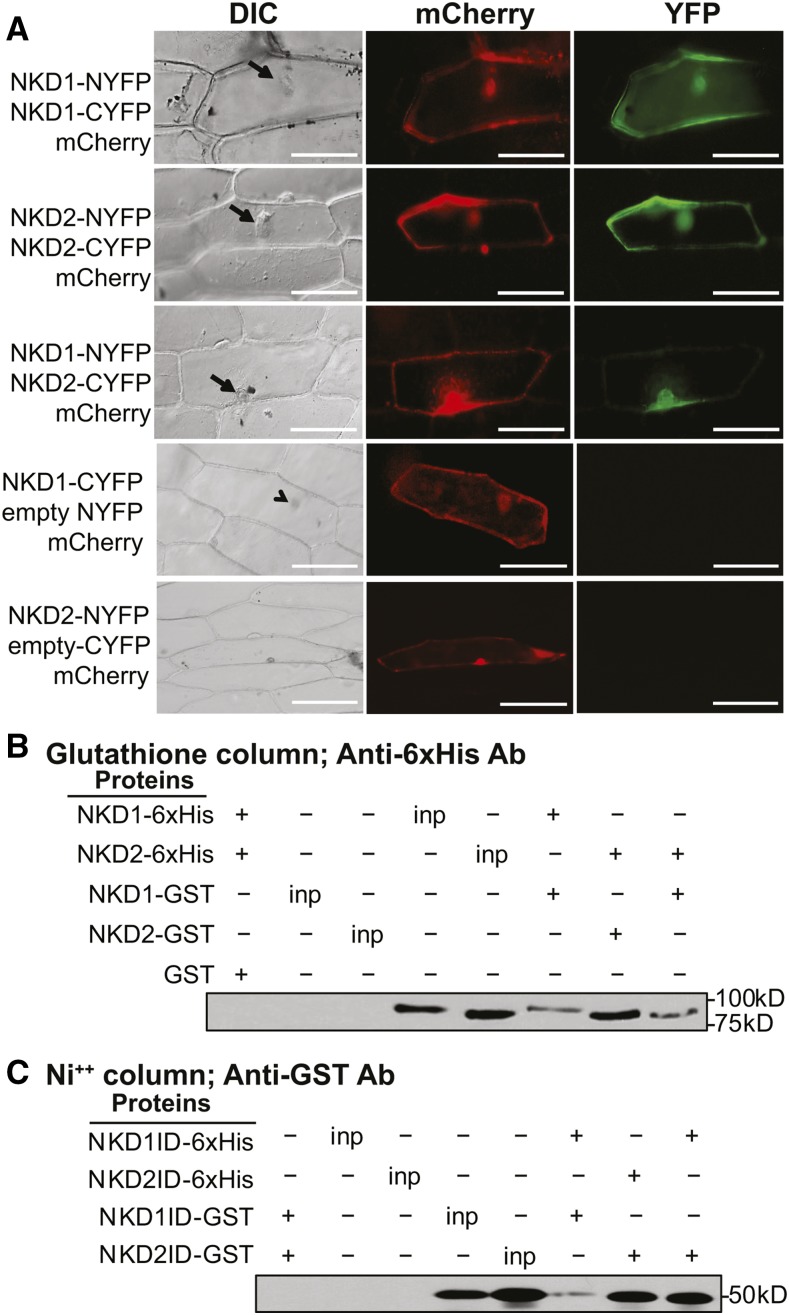
NKD1 and NKD2 Proteins Dimerize via the ID Domain
To determine if NKD1 and NKD2 can dimerize as shown for other IDD protein family members (Seo et al., 2011b; Yoshida et al., 2014), bimolecular florescence complementation (BiFC) and pull-down assays were performed. For BiFC, the N- or C-terminal portion of the YFP protein was fused to the C termini of the NKD1 and NKD2 proteins. Both full-length NKD1 and NKD2 proteins as well as truncated proteins containing only the ID domains were tested. Constructs were biolistically introduced into onion epidermal cells with a 35S:mCherry construct cobombarded to identify transiently transformed cells. For the full-length constructs, reconstituted YFP florescence was detected, indicating homodimerization of NKD1 and NKD2, as well as heterodimerization between NKD1 and NKD2 (Figure 6A). Likewise for the ID domain truncations, NKD1ID and NKD2ID could homo- and heterodimerize (Supplemental Figure 12). Control bombardments consisting of combinations of NKD full-length-nYFP+ empty vector cYFP or NKD full-length-cYFP+ empty vector nYFP did not produce YFP fluorescence, indicating that YFP reconstitution was dependent on interactions among the NKD1 and NKD2 proteins (Figure 6; Supplemental Figure 12).
Figure 6.
NKD1 and NKD2 Protein BiFC and Co-Pull-Down.
(A) Transient BiFC assays in onion epidermal cells for full-length NKD1 and NKD2 proteins. Vectors used for each BiFC assay are listed to the left of each row. Arrows designate nuclei viewed under differential interference contrast (DIC). mCherry florescence marks transient cells transformation and YFP florescence indicates a positive protein-protein interaction. Control bombardments containing NYFP + NKD1-CYFP or NKD2-NYFP + CYFP did not produce YFP fluorescence. Bars = 100 μm. Additional controls are shown in Supplemental Figure 12.
(B) and (C) Co-pull-down immunoblots for full-length NKD1 and NKD2 proteins (B) and NKD1-ID and NKD2-ID proteins (C). Affinity column and antibody (Ab) used for each assay are indicated at the top of each panel. Total soluble bacterial lysates (inp) were immunoblotted as a positive control for protein expression. The first set of control lanes show no cross-reaction between NKD-GST protein with anti-6xHis antibody or NKD-6xHis protein with anti-GST antibody.
Pull-down assays were performed on NKD1 and NKD2 full-length and ID domain proteins expressed in Escherichia coli cells with a C-terminal 6x His-tag or a N-terminal GST-tag. Co-pull-downs were performed by mixing lysates of NKD-6x His and GST-NKD proteins, precipitating protein complexes using Ni resin for 6x His and immunoblotting with GST antibodies or precipitating with glutathione Sepharose for GST tags and immunoblotting with 6x His antibodies. Coprecipitation was observed in all combinations of NKD1 and NKD2 full-length proteins, but not with the empty vector controls (Figure 6B; Supplemental Figure 13). Likewise, for the ID domain truncated protein constructs, coprecipitation was observed for all NKD1ID and NKD2ID combinations (Figure 6C; Supplemental Figure 13). Thus, NKD1 and NKD2 homo- and heterodimerize, and these interactions are mediated by residues in the ID domain.
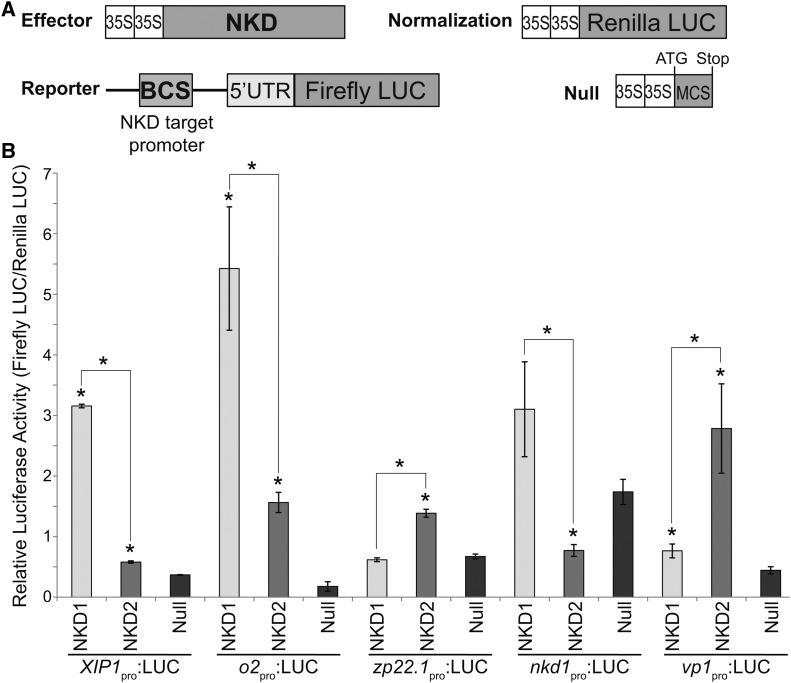
NKD1 and NKD2 Function as Transcription Factors
To test the transcriptional regulatory activity of NKD1 and NKD2, transcription assays were designed using the promoter regions of select predicted direct targets of NKD1 and NKD2. Reporter constructs contained the promoters of interest cloned upstream of the firefly luciferase (LUC) coding sequence. Promoter regions tested included xylanase inhibitor protein1 (zmX1P-1pro:LUC), opaque2 (o2pro:LUC), 22-kD zein protein 22.1 (zp22.1pro:LUC), nkd1 (nkd1pro:LUC), viviparous1 (vp1pro:LUC), jasmonate induced protein (JIPpro:LUC), mother of FT-like (MTFpro:LUC), and WRKY transcription factor 29 (wrky29pro:LUC). Effector constructs contained the NKD1 or NKD2 cDNAs under the regulation of the cauliflower mosaic virus 35S promoter (35Spro:NKD1 and 35Spro:NKD2). The empty effector vector (35Spro:null) was used as a control. A 35S promoter:Renilla luciferase construct (35Spro:RLUC) served as the internal normalization standard. The reporter, effector, and normalization constructs were cointroduced into protoplasts of nkd1-Ds nkd2-Ds0297 mutant endosperms using polyethylene glycol (PEG) calcium transfection. Transcriptional activity for each effector and test promoter construct pair was determined by comparing the firefly/Renilla luminescence ratio to the 35Spro:null effector. Transcriptional activation was detected for the NKD1 effector from the zmX1P-1pro (P < 0.0001), o2pro (P = 0.0409), and vp1pro reporter constructs (P = 0.038; Figure 7B). No significant transcriptional regulation was observed for NKD1 effector from the zp22.1pro or nkd1pro reporters (P = 0.3368 and P = 0.1008, respectively; Figure 7B). Transcriptional activation was detected for the NKD2 effector from the zmX1P-1pro (P = 0.0009), o2pro (P = 0.0025), zp22.1pro (P = 0.0014), and vp1pro reporters (P = 0.0317; Figure 7B). Transcriptional repression was detected for NKD2 effector from the nkd1pro reporter (P = 0.0140; Figure 7B). No significant transcriptional activation or repression was detected for NKD1 or NKD2 from the JIPpro, MTFpro or wrky29pro reporters (P > 0.05).
Figure 7.
NKD1 and NKD2 Transcription Assays.
(A) Schematic of constructs transfected into aleurone protoplasts for transient reporter assays of NKD1 and NKD2 transregulatory activity on selected direct target promoters. Effector, reporter, and normalization constructs were cotransfected. The null construct was substituted for the effector construct as a negative control. See Supplemental Table 14 for details on promoters used for each reporter construct.
(B) Relative luciferase activities. Reporter activity (firefly LUC) is shown in proportion to the normalization standard (Renilla LUC). Error bars represent standard errors of the means for three biological replicates. Reporter and effector constructs used for each assay are listed. Asterisk designates statistically significant difference (P = 0.05, Student’s t test) relative to control (35Spro:null) or between NKD1 (35Spro:NKD1) and NKD2 (35Spro:NKD2) effectors.
For each reporter, the relative luciferase ratios were compared between NKD1 and NKD2 effectors. NKD1 showed significantly higher activation of the zmX1P-1pro (P ≤ 0.001) and o2pro (P = 0.048) reporters, while NKD2 transcriptional activities were higher for zp22.1pro (P ≤ 0.001) and vp1pro (P = 0.020) reporters. For the nkd1pro (P = 0.025) reporter, NKD2 effector repressed activity, while activation by NKD1 was not statistically significant (Figure 7B). These results suggest possible functional differences between NKD1 and NKD2 transcriptional activities, consistent with observed differences in DNA binding specificities (Figure 5; Supplemental Figure 8). Overall, these results were consistent with direct target gene predictions and confirmed NKD1 and NKD2 function to regulate gene transcription.
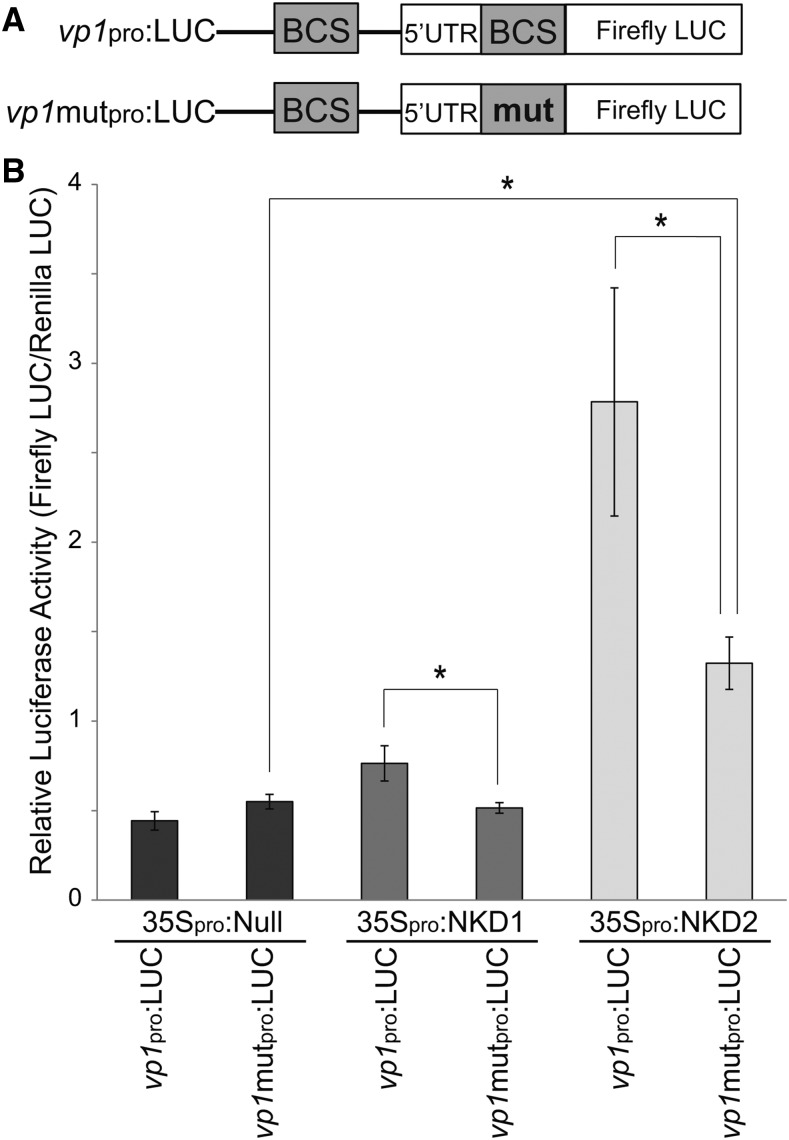
To test whether NKD1 and NKD2 require their BCS to regulate transcription, a mutant vp1 promoter construct (vp1-mutpro:LUC) was cloned with the thymine in the 7th position of the second NKDcore BCS substituted with a cytosine (TTGTCGT to TTGTCGC). This substitution abrogated binding in EMSAs (Figures 5D and 5F). Transcriptional activation of the vp1-mutpro:LUC reporter construct was not detected for NKD1 effector (P = 0.3120; Figure 8B), while NKD2 was still able to activate transcription (P = 0.0377). The level of vp1-mutpro reporter activity was decreased compared with wild-type vp1pro for both NKD1 and NKD2 effectors (P = 0.0139 and P = 0.0437, respectively). Also, transcriptional activity from the vp1-mutpro reporter was significantly different between NKD1 and NKD2 effectors (P = 0.007). These results demonstrate that NKD1 and NKD2 transcriptional activity is mediated by binding to their BCS in the promoter regions of target genes, validates the bioinformatic predictions of NKD direct target genes and further indicate NKD1 and NKD2 have differences in transcriptional activity.
Figure 8.
A NKD Binding Motif Is Required for Transcriptional Activation.
(A) Reporter constructs used to test the requirement of a NKD binding site for transcriptional activation of the vp1 promoter by NKD1 and NKD2. The mutant vp1 promoter construct (vp1-mutpro:LUC) was cloned with the thymine in the 7th position of the second NKDcore BCS in the vp1pro:LUC construct substituted with a cytosine [TTGTCG(T) to TTGTCGC].
(B) Activities of NKD1 and NKD2 on expression of wild-type and mutant vp1 promoter constructs. Reporter and effector constructs are listed. Error bars represent standard errors of the means. Asterisk designates statistically significant (P = 0.05, Student’s t test) difference relative to control (35Spro:null) or between NKD1 and NKD2 effectors.
DISCUSSION
Functions of NKD1 and NKD2 in Developing Endosperm
In this study, we revealed the functions of the NKD1 and NKD2 transcription factors in maize endosperm development by analyzing the AL and SE transcriptomes of the wild type compared with nkd1 nkd2 double mutant. Previous genetic analysis indicated that nkd1 and nkd2 are largely redundant with nkd1 single mutants, producing a mild opaque phenotype and no discernable nkd2 mutant phenotype (Yi et al., 2015). Therefore, we chose to focus on the double mutant for this study. Pathway analyses showed NKD1 and NKD2 are regulators of diverse biological processes, including cell growth, division, and differentiation, hormone metabolism and signaling, resource reserve deposition, and seed maturation. These results were largely consistent with the nkd1 nkd2 mutant’s pleiotropic endosperm phenotype (Becraft and Asuncion-Crabb, 2000; Yi et al., 2015), which provides biological validity to the transcriptomic and bioinformatic analyses.
NKD1 and NKD2 are required to restrict AL to a single cell layer and to promote AL cell identity (Becraft and Asuncion-Crabb, 2000; Yi et al., 2015). Consistent with these functions, NKD1 and NKD2 were found to regulate genes associated with cell cycle, cell growth and division. NKD1 and NKD2 are predicted to be direct transcriptional repressors of retinoblastoma-related1 and mitotic cyclin 3B-like (Figure 2; Supplemental Data Sets 1 to 3). The defective kernel1 gene is required for aleurone differentiation (Becraft et al., 2002; Lid et al., 2002) and regulates cell wall orientation and cell division plane via the regulation of cell cycle (Liang et al., 2015). These results suggest the regulation of cell division is important for aleurone development and that the multiple peripheral cell layers in the nkd1 nkd2 mutant may result in part from the misregulation of cell proliferation genes.
Grain weight of nkd1 nkd2 mutants was decreased compared with wild type, which is likely related to NKD1 and NKD2 functions in multiple aspects of resource reserve deposition including starch and storage protein accumulation. Starch is the major product for grain yield, and starch structure can greatly affect physical properties that influence end use. The nkd1 nkd2 mutant endosperm displayed a reduction in total starch abundance as well as altered glucan chain length distribution. Decreased starch accumulation is consistent with decreased expression of starch biosynthetic genes (Table 3, Figure 2). Notably, transcript levels of shrunken-2 (sh2) and brittle-2 (bt2) were both decreased (Figure 2); these genes encode subunits of ADP-glucose pyrophosphorylase, which catalyzes a rate-limiting step in starch biosynthesis (Tuncel and Okita, 2013). Other key starch biosynthetic genes showing decreased transcript abundance included sucrose synthase2, waxy1, starch synthase1 (ss1), and sugary2 (su2) (Figure 2). Likewise, altered glucan chain length distribution (Figure 3B) could result from decreased expression of amylose-extender1 (ae1), which encodes starch branching enzyme IIb (BEIIb) (Nishi et al., 2001), and ss1 and su2 genes, which catalyze glucan chain elongation. Direct target analysis predicted that NKD1 and NDK2 are direct transcriptional activators of ss1, su1, and wx1 (Supplemental Data Set 3).
The nkd1 nkd2 mutant starch granules showed irregular morphology that could not be obviously attributed to specific genes (Figures 2, 3C, and 3D). While wild-type starch granules are nearly spherical, mutants showed a mild faceting somewhat reminiscent of other starch mutants like opaque5 or various starch branching mutants (Myers et al., 2011; Zhao et al., 2015). The pitted surface of nkd1 nkd2 mutant starch granules resembled starch grains subjected to amylase digestion (Dhital et al., 2014). Given the propensity for vivipary in nkd1 nkd2 mutant kernels and decreased ABA signaling, it is possible that amylase expression is prematurely activated (Yi et al., 2015). Also, the irregular hollow core of nkd1 nkd2 mutant starch granules is fascinating, and it is unclear whether this might be caused by internal amylase digestion or whether this reflects aberrant starch grain initiation.
Pathway analyses indicated that NKD1 and NKD2 promote storage protein accumulation, both directly and indirectly, consistent with decreased storage protein content in the nkd1 nkd2 mutant. NKD2 activated transcription from the 22KD zein protein 22.1 promoter (Figure 7B), while NKD1 and NKD2 both activated transcription of the o2 promoter. Furthermore, prolamin binding factor1 (pbf1) is a predicted direct target of NKD1 and NKD2. O2 and PBF1 are TFs both well known to promote expression of zein storage protein genes (Zhang et al., 2015, 2016). The FL2-RFP and GLB3-RFP transgenes showed decreased expression in nkd1 nkd2 mutant endosperm corroborating the positive regulation of storage protein gene expression by NKD1 and NKD2 (Table 3, 5, and 6, Figures 4A and 4B).
NKD1 and NKD2 promote carotenoid accumulation because mutant endosperm is pale yellow, sometimes almost white. While carotenoid biosynthesis did not meet the pathway criterion of having at least three DE genes, two key genes showed decreased expression in the mutant; yellow endosperm1 (y1) encodes phytoene synthase and viviparous5 (vp5) encodes phytoene desaturase (Buckner et al., 1996; Hable et al., 1998). Both represent rate-limiting steps in carotenoid biosynthesis and mutations in either gene cause carotenoid deficiency. Carotenoids are precursors for ABA biosynthesis, so this is also likely to contribute to the vivipary phenotype. Additionally, the nkd1 nkd2 mutant showed decreased expression of aldehyde oxidase1 and 2 genes encoding enzymes that catalyze the final step in ABA synthesis (Seo et al., 2000), which is also consistent with vivipary and the decreased expression of ABA response pathways (Figure 2; Supplemental Data Sets 1 and 2).
The nkd1 nkd2 mutant was originally identified based on its anthocyanin-deficient phenotype (Becraft and Asuncion-Crabb, 2000), and our analysis suggests the regulation of anthocyanin biosynthesis is indirect. The colored1 (r1) and colored aleurone1 (c1) genes encode transcription factors that together activate expression of anthocyanin biosynthetic genes, and VP1 is a transcriptional activator of c1 (Cone, 2007). NKD1 and NKD2 directly activate vp1 transcription and are predicted to directly activate r1 (Figure 7B; Supplemental Data Sets 1 to 3).
A phenotype we have not systematically evaluated is the proclivity of nkd1 nkd2 mutant kernels to develop fungal infections. It is unclear whether this is a direct mutant defect or an indirect effect caused by soft texture or impaired seed maturation. However, the defense response pathway showed decreased activity in the nkd1 nkd2 mutant endosperm (Supplemental Figures 2 and 3), and genome-wide promoter scan analysis suggests NKD1 and NKD2 are direct positive regulators of defense responses (Supplemental Data Sets 1 to 3). The aleurone is the outermost cell layer of the endosperm and the only cell type alive at maturity; thus, it is fundamentally important for defense to pathogens (Jerkovic et al., 2010). Among predicted direct targets was xylanase inhibitor protein-1 (XIP-1), which is involved in fungal defense (Moscetti et al., 2013) and whose promoter showed transcriptional activation by NKD1 and NKD2 (Figure 7B). These results implicate NKD1 and NKD2 as potentially having a direct regulatory role in mediating defense.
NKD1 and NKD2 Are Key Regulators of Endosperm Gene Expression
NKD1 and NKD2 regulate widespread gene expression in developing endosperm and direct target analysis revealed ∼6% of maize gene promoters contain a NKDcore BCS. Eighty DE genes in AL and 86 in SE were TF-related, with 36 and 33, respectively, predicted as direct NKD targets (Figure 2; Supplemental Data Sets 1 to 4). Additionally, epigenetic regulation was enriched in GO terms of DE genes. Taken together, NKD1 and NKD2 appear to function as central regulators in the gene networks governing multiple aspects of maize endosperm development.
Several key TFs were shown or predicted to be direct targets of NKD regulation, including O2, PBF1, VP1, and R1. O2 is an important promoter of zein storage protein gene expression, significant for its importance in quality protein maize with enhanced lysine content (Gibbon and Larkins, 2005). PBF1 is a Dof TF that also promotes expression of storage protein genes (Marzábal et al., 2008). O2 binds a DNA motif known as GCN4 while PBF1 binds the prolamin box. These motifs are frequently combined into a bifactorial “endosperm box” conserved among storage protein genes in all cereals (Marzábal et al., 1998). Recent analyses showed O2 and PBF1 also function broadly, controlling multiple classes of zeins, as well genes involved in carbon and nitrogen metabolism, pathogen and stress responses, as well as other TFs (Li et al., 2015; Zhang et al., 2016). Comparison of the direct target genes of O2 with NKD1 and NKD2 revealed 23 genes related to resource reserve deposition in common (Supplemental Tables 5 and 6). Furthermore, GO terms associated with protein synthesis and storage were overrepresented in both NKD and O2 regulated genes (Supplemental Tables 7 and 8), suggesting O2, NKD1, and NKD2 may function together to regulate storage protein accumulation. VP1 (ABI3 ortholog) is a B3 TF required for ABA responses to promote seed maturation and inhibit germination (Suzuki et al., 2003). As mentioned, VP1 is also required for anthocyanin accumulation in the aleurone via its transcriptional activation of the c1 gene, which encodes a myb TF (Cone, 2007). C1 heterodimerizes with R1, a bHLH TF, to activate expression of structural genes in anthocyanin biosynthesis (Cone, 2007). The r1 gene is also predicted to be directly regulated by NKD1 and NKD2. The c1 and r1 genes are historically important in early genetic studies by McClintock and others.
It was previously reported that the nkd genes were subject to feedback regulation because nkd1 transcript levels increased in a nkd2 single mutant and vice versa (Yi et al., 2015). Here, we showed that NKD2 repressed expression of the nkd1 gene promoter; thus, the observed feedback appears to be due to direct transregulation between the nkd1 and nkd2 duplicate factors.
The nkd1 and nkd2 genes were recently shown to be direct targets of transcriptional activation by DOF3 (Qi et al., 2016), hereafter referred to as DOF36 (GRMZM2G137502) in accordance with NCBI and MaizeGDB annotations. RNAi knockdown of dof36 generated similar effects as the nkd1 nkd2 double mutant including abnormal starch deposition and multiple layers of compromised aleurone cells. Some of the same genes involved in sugar and starch metabolism were identified as direct (and indirect) targets for DOF36 as for NKD1 and NKD2. Interestingly, dof36 transcript shows decreased expression in nkd1 nkd2 mutant (Supplemental Data Set 1), although it was not identified as a direct target. This indicates dof36 belongs to the same GRN as nkd1 and nkd2 and that dof36 expression is reinforced by feedback, albeit indirect, from nkd genes.
Functions of NKD1, NKD2, and Other IDD Family Members
NKD1 and NKD2 bind DNA and regulate transcription via a consensus motif (TTGTCGT) similar to the ID1 binding site (TTTGTCGTTTT) although shorter (Kozaki et al., 2004). Despite 97% amino acid identity (118 of 122 residues) between their ID domains, NKD1 and NKD2 showed differences in their DNA binding and transcriptional regulation activities. Mutation of position 1 in the NKDcore motif to C decreased binding by NKD1 but abolished NKD2 binding, whereas mutation of position 7 to C abolished NKD1 binding but NKD2 could still bind (Figure 5). In transcription assays, NKD1 showed higher activity than NKD2 on the XIP-1 and o2 promoters, whereas NKD2 showed greater activation of vp1 and zp22.1 promoters (Figure 7). On the nkd1 promoter, NKD2 repressed transcription, whereas NKD1 activated, though not statistically significant. Whether these functional differences relate to differences in DNA binding activity or interactions with unknown cofactors remains to be explored, as do the functional differences between NKD1 and NKD2 on a genome-wide scale. These differences indicate that NKD1 and NKD2 are not completely redundant at the molecular level and that they have undergone subfunctionalization since the most recent maize genome duplication event that generated these loci.
Different IDD protein family members function directly as DNA binding transcriptional regulators, indirectly as cofactors for other transcription factors, or both. DELLA proteins, such as Arabidopsis RGA1, are GRAS family TFs that regulate gene expression in response to GA signaling, among other things. RGA1 activates transcription of SCARECROW-LIKE3 (SCL3) via interaction with any of five IDD proteins, AtIDD3, -4, -5, -9, and -10. The IDD proteins bind DNA and function as scaffolds for the DELLA proteins, which lack intrinsic DNA binding activity (Yoshida et al., 2014; Yoshida and Ueguchi-Tanaka, 2014). SCL3 is another GRAS protein that interacts with the IDD proteins to competitively inhibit the binding and action of RGA1. This provides feedback to modulate gene expression responses to GA signaling. Similarly, the IDD protein JACKDAW (JKD) regulates transcription of SCARECROW (SCR) in Arabidopsis root development. JKD activity is enhanced by the GRAS protein SHORTROOT (SHR), as well as by the target gene product, SCR, another GRAS protein (Ogasawara et al., 2011). Furthermore, the activity of JKD is also modulated by interactions with other IDD proteins, MAGPIE and BALDIBIS (Long et al., 2015; Ogasawara et al., 2011). In addition to modulating transcriptional activity of this complex, these interactions prevented intercellular trafficking of the mobile protein SHR.
We found NKD1 and NKD2 could each homodimerize as well as heterodimerize with one another. The functional significance of this is unknown, and we do not yet know whether they can dimerize with other IDD family members. Arabidopsis IDD14 also homo- and heterodimerizes, and this dimerization modulates function (Seo et al., 2011b). Under standard laboratory temperatures, IDD14α binds DNA and regulates transcription of target genes, but under cold temperatures, the IDD14 gene produces an alternatively spliced transcript that encodes a product, IDD14β, that lacks the DNA-binding domain. IDD14β can heterodimerize with IDD14α and decreases DNA binding activity.
IDD gene family members are implicated in many biological functions, including the regulation of carbohydrate metabolism, gravitropism, seed germination, lateral organ morphogenesis, cellular patterning, flowering time, and hormone signaling (Morita et al., 2006; Welch et al., 2007; Tanimoto et al., 2008; Hassan et al., 2010; Feurtado et al., 2011; Ogasawara et al., 2011; Seo et al., 2011a; Cui et al., 2013; Wu et al., 2013; Yoshida et al., 2014; Yoshida and Ueguchi-Tanaka, 2014; Jöst et al., 2016). It is striking how many similar processes are regulated by the nkd genes during seed development. For example, Arabidopsis IDD8 and IDD14 transcriptionally regulate carbohydrate metabolism and starch accumulation by modulating expression of some of the same target genes as NKD1 and NKD2, such as ss1 (Seo et al., 2011a, 2011b) (Figures 2, 3, and 4A).
An intriguing functional analogy is with IDD proteins (aka BIRD proteins) in Arabidopsis root development. Four IDD proteins, JKD, BALDIBIS (BIB), NUTCRACKER (NUC), and MAGPIE, have overlapping functions in the specification of the cortical cell layer, and jkd bib double mutants contained extra layers of cells with indistinct identity (Ogasawara et al., 2011; Long et al., 2015). The normal pattern of cell division is mediated in part through the transcriptional repression of the CYCLIND6 gene (Sozzani et al., 2010; Long et al., 2015). Similarly, in nkd1 nkd2 mutants of maize, the normally single layer of aleurone cells is replaced by multiple layers of cells with compromised identity. Furthermore, scl1 is predicted to be directly activated by NKD1 and NKD2, while the cell cycle-related genes retinoblastoma-related1 and mitotic cyclin 3B-like are predicted to be negatively regulated by NKD1 and NKD2 (Figure 2; Supplemental Data Set 3). Future work will seek to resolve potential mechanistic conservation among these processes.
METHODS
Plant Materials
Plants were grown in the field at Iowa State University experimental farms near Ames, IA for RNA isolation and quantitative RT-PCR and for analysis of kernel composition, starch structure and morphology, and expression of transgenic fluorescent markers (FL2-RFP, GLB3-RFP, and RAB17-YFP). Protoplast isolation was performed on developing endosperm from plants grown in the greenhouse at 28°C under natural lighting, supplemented with high-intensity sodium halide lamps for 16 h.
The nkd1-R and nkd2-R alleles and all the fluorescent marker transgenes were backcrossed into the B73 inbred background at least four generations and B73 served as the wild-type control. The nkd1-Ds and nkd2-Ds0297 alleles arose and were maintained in a W22 background and W22 was used for the wild-type control.
LCM-RNA-Seq Maize v3 Transcriptome Assembly and DE Analyses
The LCM-RNA-seq was previously reported and involved isolation of RNA from three biological replicates of AL and SE cells from B73 wild-type and nkd1 nkd2 mutant kernels (Yi et al., 2015). The RNA-seq reads were aligned to maize (Zea mays) reference genome B73 V3 assembly (AGPv3) using TopHat 2 (version 2.1.0; http://ccb.jhu.edu/software/tophat/index.shtml) with default settings. The sequence aligned files (SAM/BAM format) were analyzed using coverageBed in the BEDTools package (Quinlan and Hall, 2010) for coverage of transcript models reported in the filtered gene set (Zea_mays.AGPv3.28.gff3.gz) to generate an integer count of transcript data for each sample. Differentially expressed genes were identified using DESeq2 (Love et al., 2014) at an adjusted P value cutoff of 0.01. MA plots were generated using the M (log intensity ratios or fold change) values against the A (mean average or mean normalized read count between the two samples). Multidimensional scaling analysis was performed to assess the relative similarity in the sample data, wherein the information contained in the DE data sets was transformed into distance matrices in two dimensions. The number of genes expressed in any endosperm cell type was computed based on the presence of a detected read in any one of the replicates. The Genesect web analysis tool (Katari et al., 2010) was used for testing the statistical significance of overlap between two DE data sets.
RNA Isolation and Quantitative RT-PCR
Aleurone was hand dissected from 15-DAP endosperms and tissues collected from 10 kernels off the same cob were pooled to constitute one biological replicate. Three replicates were sampled from independent cobs for each genotype, wild type, and homozygous nkd1-R nkd2-R mutant. Total RNA was extracted as described (Wang et al., 2012). Samples of 12-DAP pooled BETL enriched endosperm was also collected from three independent cobs and RNAs extracted from five pooled kernels from the same cob constituted a single biological replicate. For BETL qRT-PCR, samples were enriched for BETL by endosperm dissecting the approximately one-third basal end of each kernel. The RNA samples were subjected to DNAase treatment using RQ1 RNase-free DNase (Promega), and 2 µg of RNA was reverse transcribed to cDNA with SuperScript III reverse transcriptase (Invitrogen) using the manufacturer’s protocol. The PrimerQuest Tool (http://www.idtdna.com/Primerquest/Home/Index) was used to design gene-specific RT-PCR primers corresponding to coding region that spans an intron, overlaps an intron-exon junction, or in some instances including 3′ untranslated regions. Supplemental Table 9 shows the gene-specific primer sequences and the respective amplicon sizes.
Quantitative RT-PCR was performed using iQ SYBR Green Master Mix (Bio-Rad) on an Applied Biosystems StepOnePlus real-time PCR system with 250 nM of gene-specific primers and cDNA template. The thermal cycle applied was 95°C for 10 min, 40 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, followed by dissociation curve analysis. Melting curves of samples were examined for the absence of multiple peaks/nonspecific amplification and those of nontemplate control samples were checked for possible primer dimers. Threshold cycle (CT) was automatically calculated for each reaction using the StepOnePlus qRT-PCR machine default parameters. Data were normalized to the expression level of transcript for UBIQUITIN CONJUGATING ENZYME (GRMZM2G132759 for AL qRT and GRMZM2G027378 for BETL qRT), and fold changes in nkd1 nkd2 mutant were computed relative to the wild-type control using the comparative threshold cycle (2−ΔCT) method (Livak and Schmittgen, 2001).
Construction of GST-Tag and 6x His Expression Vectors
The GST-Tag and 6x His NKD1 and NKD2 fusion proteins were constructed by cloning the full-length coding sequences (CDSs) or ID domains into pGEX-4T (GE Healthcare Life Sciences) and pET-34b (Novagen), respectively, using primers listed in Supplemental Table 10. Protein expression constructs were transformed into ORIGAMI (Novagen) or BL21 (Promega) chemically competent cells. Protein expression was induced with IPTG and NKD-GST or NKD-6xHis were purified using Glutathione Sepharose 4B (GE Healthcare) or HIS-Select HF Nickel Affinity Gel resins (Sigma-Aldrich P6611), respectively, according to the manufacturer’s instructions. Fusion protein identity was confirmed by immunoblotting with GST antibody (Thermofisher MA4-004-HRP) and His antibody (Thermofisher MA1-21315-HRP) and by QSTAR MS/MS (Supplemental Figure 7).
SAAB
SAAB was performed using purified NKD1ID-GST and NKD2ID-GST fusion proteins as described (Kozaki et al., 2004) with the following alterations: The library of randomly synthesized dsDNA was generated via Klenow fill-in reaction of a single-stranded oligo library. Oligos had 5′ flanking sequence CAGGGTCGCTGGTACGAA and 3′ flanking sequence CGTACCAGCGACCCTG with 20 random bases in between (CAGGGTCGCTGGTACGAA[N20]TTCGTACCAGCGACCCTG). Forward (CAGGGTCGCTGGTACGAA) and reverse (CAGGGTCGCTGGTACGAA) primers were used to PCR amplify selectant oligos after each round of selection. NKD1ID and NKD2ID GST proteins were purified as described above and were covalently cross-linked to active ester agarose Affi-Gel 10 (Bio-Rad) following the manufacturer’s instructions and loaded into columns (Bio-Rad). The library of random oligos in binding buffer (described below) was passed over NKDID columns, washed, eluted, and PCR amplified as previously described (Kozaki et al., 2004). After six cycles of selection and amplification, selectant oligo libraries were blunt end cloned into pGEM-T Easy (Promega) and sequenced. Primer sequences were trimmed and SAAB selected sequences were then analyzed for a recurring pattern (motif) with MEME-suit tool Multiple Em for Motif Elicitation (Bailey et al., 2015) using parameters previously described (Kozaki et al., 2004).
EMSA
NKD1ID-GST, NKD2ID-GST, and empty vector GST proteins were purified as described above. The 5′ biotinylated oligos (Integrated DNA Technologies) were made double stranded by annealing with reverse complement oligos listed in Supplemental Table 11. EMSA was performed as previously described (Kozaki et al., 2004) with the following alterations: 100 ng of purified NKD1-GST, NKD2-GST, or empty vector GST fusion proteins were incubated in binding buffer (10 mM Tris-HCl, pH 7.5, 75 mM NaCl, 1 mM DTT, 6% glycerol, 1% BSA, 1% Nonidet P‐40, poly[d(I,C)], and 10 µM ZnCl) with 50 fmol of 5′ biotin-labeled BCS or point-mutated BCS variant oligos. Purified GST from induction of the empty vector served as a negative control for nonspecific interactions. After incubation on ice for 30 min, loading dye (without EDTA) was added and samples were loaded onto a 10% native polyacrylamide gel and run in Tris-borate buffer (without EDTA) on ice. DNA and DNA-protein complexes were transferred onto an ImmobilonNy+ membrane and UV cross-linked. Biotin-labeled oligos were detected with a LightShift chemiluminescent EMSA kit (Thermo Scientific). Relative binding affinity for each oligo was determined by the intensity of the shifted oligo relative to unmutated BCS quantified with ImageJ.
Competition EMSA was performed using the biotinylated wild-type BCS oligo with the addition of 50-, 100-, and 500-fold excess (relative to labeled) nonbiotinylated (unlabeled) wild-type or point mutant oligos. Sequences are listed in Supplemental Table 11. Oligonucleotide probes were added to the binding reaction before the addition of NKD1ID-GST, NKD2ID-GST, or GST proteins.
NKD Motif Enrichment Analysis
The differentially expressed gene PPRs were searched for the presence of NKD1, NKD2, and NKDcore BCSs via the Meme Suite tool FIMO (http://meme-suite.org/tools/fimo) using NKD1, NKD2 SAAB-MEME derived BCS probability matrices, and the NKDcore motif. PPR was defined as −600 bp upstream of the transcriptional start site extending to the transcriptional start site, and PPR sequences were downloaded from GRAMENE BIOMART (http://ensembl.gramene.org/biomart/martview/, last accessed 1/3/2016). Motif enrichment was determined by comparing the number of genes in the AL or SE transcriptome with one or more motif in its PPR to the number of DE genes in nkd1 nkd2 mutant with one or more motif in its PPR by use of Fisher’s exact test. The AL and SE transcriptomes were defined as all genes detected in either B73 or nkd1 nkd2 mutant LCM-RNA-seq data sets with one or more read counts. To control for the specificity of NKD BCS enrichment, three shuffle control SAAB-MEME BCS were generated for NKD1 and NKD2 from SAAB-MEME-selected sequences using the MEME Suit shuffle sequence option (http://meme-suite.org/tools/meme). Enrichment was determined as described above. Direct target GO enrichment was determined by comparing the number of GO terms in DE AL or SE genes with one or more motif in PPR to the number of GO terms in the in the AL or SE transcriptome via agriGO Singular Enrichment tool (http://bioinfo.cau.edu.cn/agriGO/, last accessed 6/14/16).
GO Enrichment and RNA-Seq Pathway Analysis
GO term enrichment was determined by comparing the number of GO terms in DEGs to the number of GO terms in the endosperm transcriptomes via agriGO Singular Enrichment Analysis tool with default parameters and a critical cutoff value of false discovery rate ≤ 0.2 (Genome version Zea mays AGPv3.30, http://bioinfo.cau.edu.cn/agriGO/, last accessed on 6/14/2016). The AL and SE transcriptomes were defined as all genes detected with one or more read counts in RNA-seq. The AL transcriptome was compared with the AL DEGs and the SE transcriptome to the SE DEGs. Fisher exact test P values were calculated by agriGO using default parameters. Pathway analysis was performed on DEGs using MaizeCyc, CornCyc, and GO tools and databases (Monaco et al., 2013). Pathways with a significant cutoff value of 3 nodes were analyzed for up- or downregulation by overlaying differentially expressed gene log2 fold change expression onto each pathway. For nonmetabolic pathways, GO was used with the maize sequence v3 ontologies (agriGO v3.3, http://bioinfo.cau.edu.cn/agriGO/; maizeGDB, http://www.maizegdb.org/). Pathways were visualized using heat maps generated by the R pheatmap program.
Epifluorescence Microscopy
The nkd-R alleles were crossed into GL3B-RFP, FL2-RFP, and RAB17-YFP transgenic lines (Maize Cellgenomics Database, http://maize.jcvi.org). Transgenic individuals were selected by LIBERTY herbicide and self-pollinated to produce nkd-R segregating cobs. Mature kernels were harvested and sectioned using a Leica Vibratome or by hand. Three kernels each, from the same segregating cob, of the wild type and nkd-R within each transgenic line were viewed using an Olympus BX-60 microscope under bright-field or epifluorescence. Tissues were visualized by autofluorescence using a Chroma narrow violet (filter (excitation, 400 to 410 nm; dichroic mirror and barrier filter, 455 nm). YFP was observed with a Chroma EYFP filter set (excitation, 495 nm; dichroic barrier filter, 515 nm; emission, 540 nm), and mCherry was observed using a Chroma mCherry filter set (excitation, 560 nm; dichroic barrier filter, 600 nm; emission, 635 nm). Micrography was performed with a Jenoptik C-5 camera, and constant gain and exposure time settings were used for each filter set to compare expression of each respective transgene reporter protein in wild type versus nkd1 nkd2 mutant kernels. Standard PCR genotyping of transgene and nkd1-R and nkd2-R alleles was performed using primers described (Yi et al., 2015) to confirm kernel genotypes.
Endosperm Starch Extraction, Quantification, and Chain Length Distribution Assay
Wild-type (W22) and nkd1-Ds nkd2-Ds0297 kernels were collected from the same segregating ear, with six individual kernels of each genotype serving as biological replicates. Mature kernels were soaked in 0.45% (w/v) sodium metabisulfite at 50°C overnight. Pericarp and embryo were removed and the total endosperm starch was isolated. The extraction procedure (Dinges et al., 2001) was modified as follows: The endosperm starch was washed with chilled deionized water twice and with chilled 80% ethanol once, and centrifuged 3000g at 4°C for 10 min after each liquid addition. The final pellet was dissolved in 100% DMSO and boiled in water bath for 1 h.
To measure endosperm starch content, DMSO-dissolved starch was diluted 10-fold with amyloglucosidase buffer (0.1 M sodium acetate, pH 5, and 5 mM calcium chloride), and digested by amyloglucosidase (60 units/reaction; Megazyme E-AMGDF100), followed by incubation at 50°C for 100 min, producing glucose. Glucose was measured by a GOPOD assay kit (Megazyme K-GLUC) following the manufacturer’s instructions to determine the weight of starch.
For glucan chain length distribution, <1 mg of starch was precipitated in 5 volumes of 100% ethanol at 4°C overnight, followed by centrifugation at 13,000g for 10 min. The pellet was resolubilized in deionized water and pH was adjusted to 4.5 with 0.5 M sodium acetate. Starch was debranched with 4 units of isoamylase (Megazyme E-ISAMY) at 42°C overnight. The chain length distribution of each sample was analyzed via a Dionex HPAEC-PAD system (Thermo Fisher Scientific) as described (Dinges et al., 2001).
Scanning Electron Microcopy
For scanning electron microscopy, six wild type and six nkd1 nkd2 mutant mature kernels from F2 nkd1-Ds nkd2-Ds0297 segregating ears were used. Kernel genotypes were verified by standard PCR genotyping using primers described by Yi et al. (2015). Mature kernels were cracked and freshly planed with a razor blade, cleaned with ethanol, and placed on a specimen stub with a carbon coated adhesive. Specimen stubs were then painted with silver paint and air-dried for 10 min at room temperature. Kernels were sputter coated with gold and imaged with a digital JEOL 5800LV scanning electron microscope.
Total Pericarp, Endosperm, Embryo, and Seed Dry Weight Analysis
For pericarp, endosperm, embryo, and total seed weight analysis, 31 wild-type and 31 nkd1 nkd2 mutant mature kernels from the same F2 segregating nkd1-Ds nkd2-Ds0297 segregating ear were randomly selected. This was done on three independent ears for a total of 93 wild-type kernels and 93 nkd1 nkd2 mutant kernels. Mature kernels were imbibed in double-distilled water for 12 h and were frozen solid at 4°C and thawed to allow for efficient dissection. Pericarp, endosperm, and embryos were then dissected into pools of tissue from 31 kernels and placed in a 55°C oven for 36 h to redesiccate the materials. Samples were then weighed to obtain pooled kernel pericarp, endosperm, and embryo dry weights. Total seed weights were determined by adding the pericarp, endosperm, and embryo dry weights together for each independent ear. Average pericarp, endosperm, embryo, and total seed weights per kernel for the wild type and nkd1 nkd2 mutant were determined, and statistical analysis was performed using a Student’s t test.
Total Nitrogen and Protein Analysis
For total nitrogen analysis, 10 wild-type and 10 nkd1-Ds nkd2-Ds0297 kernels were randomly selected from an F2 segregating nkd1-Ds nkd2-Ds0297 ear. This process was repeated for a total of three randomly selected 10-kernel pools each of the wild type and nkd1-Ds nkd2-Ds0297 from the same ear. This process was performed on a total of three independent ears representing 90 wild type and 90 nkd1-Ds nkd2-Ds0297 kernels for a total of 9 wild type and 9 nkd1 nkd2 mutant kernel 10-kernel pools. Each kernel pool was imbibed in double dilute water for 12 h, frozen at −20°C, and thawed to allow efficient dissection. Endosperm dissections were then performed and 10-kernel pools and were ground in a mortar and pestle with liquid nitrogen to a particle size of <1 mm in diameter. Ground samples were placed in a 55°C oven for 48 h to redesiccate the materials and then weighed. Total nitrogen content was determined from 0.15 g of each of the nine wild-type and nine nkd1 nkd2 mutant pooled endosperm samples using the Dumas N combustion procedure with a Leco Truspec CN analyzer and Elementar Variomax CNS analyzer. Protein content (%) per sample was determined by multiplying percentage of nitrogen content by 6.25. Total grams protein per sample was determined by multiplying percentage of total protein by dry weight. Sample means were compared using Student’s t test.
PCA
PCA was performed using the MetaboAnalyst 3.0 web tool (Xia et al., 2015). Due to experimental design differences, PCA was performed separately for endosperm weight combined with the total protein to account for the effects of independent ears (cob) and for starch branch chain length distribution combined with the total starch to account for the effect of independent kernels within the same segregating ear.
BiFC
BiFC assays were performed as described (Citovsky et al., 2006). nkd1 and nkd2 ID domain and full-length CDSs were cloned in-frame to the N-terminal half and the C-terminal half of YFP in vectors pSAT1A-nEYFP-N1 and pSAT1A-cEYFP-N1, respectively, using primers listed in Supplemental Table 12. A 35S:mCherry construct was generated by cloning the mCherry CDS into a modified pSAT1A vector from which the EYFP fragment had been removed. The 35S:mCherry, nYFP-NKD, and cYFP-NKD constructs were biolistically introduced into onion epidermal cells and incubated in the dark for 24 to 36 h. 35S:mCherry was cobombarded with each experiment and used as an internal control to identify transiently transformed cells. Expression of the mCherrry marker and reconstitution of YFP florescence was observed by epifluorescence microcopy as described above.
Reciprocal Co-Pull-Down
Pull-down assays were performed using the NKD-GST and NKD-6x HIS fusion proteins. Fusion protein expression was induced by IPTG for 2 h, and total soluble protein extracts were collected as previously described (Kozaki et al., 2004). NKD-GST and NKD-6x His-tagged total soluble protein extracts were mixed and incubated in PBS buffer with 0.15 mM PMSF for 1 h at room temperature with gentle rotation. Samples were then passed though Glutathione Sepharose 4B (GE Healthcare) or HIS-Select HF Nickel Affinity Gel resins (Sigma-Aldrich) and then washed with GST wash buffer (1× PBS, pH 7.4) or 6x His wash buffer (50 mM phosphate buffer, pH 7.0, 300 mM NaCl, and 1 mM imidizole). Proteins were eluted with GST elution buffer (50 mM Tris-HCl and 10 mM reduced glutathione, pH 8.0) or 6x His elution buffer (50 mM phosphate buffer, pH 7.0, 300 mM NaCl, and 150 mM imidizole) and detected by SDS-PAGE immunoblotting using GST antibody (product no. MA4-004-HRP) or His antibody (product no. MA1-21315-HRP).
Protoplast Isolation and Transformation
To avoid confounding effects from endogenous NKD1 and NKD2 protein, 18- to 20-DAP nkd1-Ds nkd2-Ds0297 kernels were used for aleurone protoplast isolation as described (Bethke and Jones, 2001). Aleurone peels from 30 to 40 kernels (∼0.5 g) of the same ear were harvested and placed immediately in 15 mL of TVL solution (0.3 M sorbitol and 50 mM CaCl2). Twenty milliliters of enzyme solution (0.5 M sucrose, 10 mM MES-KOH, pH 5.7, 20 mM CaCl2, 40 mM KCl, 1% Cellulase Onozuka R-10, and 1% Macerozyme R10) was then added and the tissue shaken at 35 rpm at room temperature for 16 to 18 h. Protoplasts were collected by passing through 10-μm nylon mesh (spectrum labs) with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7). Protoplasts were recovered from the nylon mesh by rinsing with 15 mL of W5 solution followed by centrifugation for 5 min at 60g. Protoplast pellets were gently resuspended in 15 mL of fresh W5 solution, centrifuged for 5 min at 60g, and then resuspended 1 mL MMg solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7). Protoplast integrity and quantification were determined by optical visualization on a light microscope.
Protoplast transformation was performed following the protocol outlined by Bethke and Jones (2001). For each transformation, 100 μL of fresh protoplasts (∼2 × 104) was added to a 2-mL microfuge tube. Effector, reporter, and normalization plasmids in 10 μL total volume (10 to 20 μg DNA total) were added and 110 μL of PEG solution (40% [w/v] PEG 4000, 0.2 M mannitol, and 100 mM CaCl2) was added, gently mixed, and incubated at room temperature for 10 min without shaking. W5 solution (400 μL) was added to stop the transfection process, tubes were centrifuged at 100g for 2 min at room temperature, and protoplast pellets gently resuspended in 1 mL WI solution (0.5 M mannitol, 4 mM MES, pH 5.7, and 20 mM KCl) and incubated at room temperature on a rotator for 14 h.
Transcriptional Activity Assays
To test the transcriptional activity of NKD1 and NKD2, a series of reporter, effector, and normalization constructs (Figure 8A) were designed and cloned using primers listed in Supplemental Table 13. For reporter vectors, the promoter regions of select putative direct targets of NKD were cloned upstream of the firefly luciferase coding sequence in the pGL3 vector (Promega). The proximal promoter regions of xylanase inhibitor protein 1 (zmX1P-1; GRMZM2G328171), opaque2 (o2; GRMZM2G015534), zein protein 22.1 (zp22.1; GRMZM2G044625), nkd1 (GRMZM2G129261), viviparous1 (vp1; GRMZM2G133398), jasmonate induced protein (JIP; GRMZM2G112238), Mother of FT-like (MTF; GRMZM2G059358), and WRKY transcription factor 29 (wrky29; GRMZM2G040298) constituted the reporter plasmids (Supplemental Table 14). For the effector constructs, 35Spro:NKD1 and 35Spro:NKD2, nkd1 and nkd2 CDSs were cloned into a modified pSAT1A vector described above using primers listed in Supplemental Table 13. The empty vector was used as 35S:null control. The normalization construct was generated by cloning the Renilla luciferase CDS into a modified pSAT1A vector using primers listed in Supplemental Table 13. The reporter, effector, and normalization constructs were cotransformed into nkd1-Ds nkd2-Ds0297 AL protoplasts as described above. Sets of transactivation assays were performed on protoplasts from the same isolation and protoplasts from independent isolations constituted biological replicates. A control using the 35S-null construct as effector was included for each set of assays. Three biological replicates were performed for each treatment. Firefly and Renilla luciferase activity assays were performed using a dual luciferase assay system kit following the manufacturer’s recommended instructions (Promega E1910). Luminescence was measured using a microplate reader (BioTek), and three technical replicates were used for each assay.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: RNA-seq reads were deposited in NCBI Gene Expression Omnibus under accession number GSE61057. Gene models are provided for differentially expressed genes in Supplemental Data Set 1 for the aleurone and Supplemental Data Set 2 for the starchy endosperm.
Supplemental Data
Supplemental Figure 1. Maize V3 LCM-RNA-seq transcriptome assembly and DE gene confirmation.
Supplemental Figure 2. Disrupted pathways in nkd1 nkd2 mutant aleurone.
Supplemental Figure 3. Disrupted pathways in nkd1 nkd2 mutant starchy endosperm.
Supplemental Figure 4. BETL qRT-PCR.
Supplemental Figure 5. nkd1 nkd2 mutant starch granule scanning electron microscopy.
Supplemental Figure 6. Principal component analysis of nkd1 nkd2 mutant and wild-type resource reserve data sets.
Supplemental Figure 7. Verification of NKD-ID GST fusion proteins.
Supplemental Figure 8. EMSA controls and additional tested mutant probes.
Supplemental Figure 9. Scanning mutagenesis EMSA overexposure.
Supplemental Figure 10. Overlap in NKD motifs and predicted direct target genes.
Supplemental Figure 11. NKD BCS enrichment shuffled controls
Supplemental Figure 12. BiFC controls.
Supplemental Figure 13. Reciprocal tag co-pull-downs.
Supplemental Table 1. Summary of RNA sequencing reads.
Supplemental Table 2. Summary of gene expression analysis in endosperm cell types.
Supplemental Table 3. NKD1 SAAB-selected sequences
Supplemental Table 4. NKD2 SAAB-selected sequences.
Supplemental Table 5. NKD1, NKD2, and O2 aleurone coregulated genes.
Supplemental Table 6. NKD1, NKD2, and O2 starchy endosperm coregulated genes.
Supplemental Table 7. Enriched gene ontologies in NKD1, NKD2, and O2 aleurone coregulated genes.
Supplemental Table 8. Enriched gene ontologies in NKD1, NKD2, and O2 starchy endosperm coregulated genes.
Supplemental Table 9. Primer sequences used for qRT-PCR.
Supplemental Table 10. Primers used to generate NKD GST and 6x His tag expression constructs.
Supplemental Table 11. Oligonucleotides used in electrophoretic mobility shift assays.
Supplemental Table 12. Primers used for cloning of constructs used in BiFC.
Supplemental Table 13. Primers used for cloning of transcription assay constructs.
Supplemental Table 14. Constructs cloned in this study.
Supplemental Data Set 1. nkd1 nkd2 mutant aleurone differentially expressed genes and pathway analyses.
Supplemental Data Set 2. nkd1 nkd2 mutant starchy endosperm differentially expressed genes and pathway analyses.
Supplemental Data Set 3. NKD direct target gene analyses.
Supplemental Data Set 4. NKD direct target gene ontologies and enrichment.
Supplementary Material
Acknowledgments
We thank members of the Becraft lab for helpful discussions of the manuscript, Siva Chudalayandi (ISU) for help with RNA-seq data analysis, and Alan Myers for help with starch branch chain length assays. This research was supported by the National Science Foundation (Grants IOS-1121738 and IOS-1444568). The following Iowa State University facilities provided technical assistance: the DNA Facility and the Microscopy and NanoImaging Facility assisted with microscopic preparations and imaging, and the Soil and Plant Analysis Laboratory assisted with total nitrogen analysis.
AUTHOR CONTRIBUTIONS
P.W.B., B.C.G., A.K.N., and H.W. designed experiments. A.K.N. performed LCM (Yi et al., 2015), RNA-seq transcriptome assembly, and differential expression analyses. A.K.N. and B.C.G. performed qRT-PCR and transgene fluorescent microscopy experiments. B.C.G. performed RNA-seq DE gene enrichment and pathway analyses, seed composition analyses, PCA, SAAB and MEME analyses, EMSAs, NKD direct target and enrichment analyses, BiFC and coimmunoprecipitation assays, transcription assays, and cloned all constructs used in the manuscript. H.W. and B.C.G. performed starch branch chain length distribution and kernel starch content experiments. B.C.G. was the main contributor to the writing of the manuscript. A.K.N., H.W., and P.W.B. also contributed to writing.
Glossary
- SE
starchy endosperm
- BET
basal endosperm transfer
- AL
aleurone
- TF
transcription factor
- LCM
laser capture microdissection
- DAP
days after pollination
- DE
differential expression
- GO
Gene Ontology
- ABA
abscisic acid
- PCA
principal component analysis
- SAAB
selection and amplification binding
- BCS
binding consensus sequence
- FIMO
Find Individual Motif Occurrences
- PPR
proximal promoter region
- BiFC
bimolecular florescence complementation
- PEG
polyethylene glycol
- CDS
coding sequence
References
- Bailey T.L., Johnson J., Grant C.E., Noble W.S. (2015). The MEME Suite. Nucleic Acids Res. 43: W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft P.W., Asuncion-Crabb Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048. [DOI] [PubMed] [Google Scholar]
- Becraft P.W., Gutierrez-Marcos J. (2012). Endosperm development: dynamic processes and cellular innovations underlying sibling altruism. Wiley Interdiscip. Rev. Dev. Biol. 1: 579–593. [DOI] [PubMed] [Google Scholar]
- Becraft P.W., Li K., Dey N., Asuncion-Crabb Y. (2002). The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225. [DOI] [PubMed] [Google Scholar]
- Bethke P.C., Jones R.L. (2001). Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 25: 19–29. [DOI] [PubMed] [Google Scholar]
- Buckner B., Miguel P.S., Janick-Buckner D., Bennetzen J.L. (1996). The y1 gene of maize codes for phytoene synthase. Genetics 143: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Lee L.Y., Vyas S., Glick E., Chen M.H., Vainstein A., Gafni Y., Gelvin S.B., Tzfira T. (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Yuan Z., Sundaresan V. (1998). The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- Colasanti J., Tremblay R., Wong A.Y., Coneva V., Kozaki A., Mable B.K. (2006). The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.E., Clore A.M., Ranch J.P., Higgins R., Lopes M.A., Larkins B.A. (1997). Expression of a mutant α-zein creates the floury2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. USA 94: 7094–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone K.C. (2007). Anthocyanin synthesis in maize aleurone tissue. In Endosperm: Development and Molecular Biology, Olsen O.-A., ed (Berlin, Heidelberg: Springer-Verlag; ), pp. 121–140. [Google Scholar]
- Cui D., Zhao J., Jing Y., Fan M., Liu J., Wang Z., Xin W., Hu Y. (2013). The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 9: e1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhital S., Warren F.J., Zhang B., Gidley M.J. (2014). Amylase binding to starch granules under hydrolysing and non-hydrolysing conditions. Carbohydr. Polym. 113: 97–107. [DOI] [PubMed] [Google Scholar]
- Dinges J.R., Colleoni C., Myers A.M., James M.G. (2001). Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 125: 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath A., Bethke P., Lonsdale J., Meza-Romero R., Jones R. (2000). Programmed cell death in cereal aleurone. Plant Mol. Biol. 44: 255–266. [DOI] [PubMed] [Google Scholar]
- Feurtado J.A., Huang D., Wicki-Stordeur L., Hemstock L.E., Potentier M.S., Tsang E.W., Cutler A.J. (2011). The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler-Lee J., Gallie D.R. (2005). Aleurone cell identity is suppressed following connation in maize kernels. Plant Physiol. 139: 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbon B.C., Larkins B.A. (2005). Molecular genetic approaches to developing quality protein maize. Trends Genet. 21: 227–233. [DOI] [PubMed] [Google Scholar]
- Gruis D.F., Guo H., Selinger D., Tian Q., Olsen O.-A. (2006). Surface position, not signaling from surrounding maternal tissues, specifies aleurone epidermal cell fate in maize. Plant Physiol. 141: 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable W.E., Oishi K.K., Schumaker K.S. (1998). Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol. Gen. Genet. 257: 167–176. [DOI] [PubMed] [Google Scholar]
- Hassan H., Scheres B., Blilou I. (2010). JACKDAW controls epidermal patterning in the Arabidopsis root meristem through a non-cell-autonomous mechanism. Development 137: 1523–1529. [DOI] [PubMed] [Google Scholar]
- Jerkovic A., Kriegel A.M., Bradner J.R., Atwell B.J., Roberts T.H., Willows R.D. (2010). Strategic distribution of protective proteins within bran layers of wheat protects the nutrient-rich endosperm. Plant Physiol. 152: 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jöst M., Hensel G., Kappel C., Druka A., Sicard A., Hohmann U., Beier S., Himmelbach A., Waugh R., Kumlehn J., Stein N., Lenhard M. (2016). The INDETERMINATE DOMAIN protein BROAD LEAF1 limits barley leaf width by restricting lateral proliferation. Curr. Biol. 26: 903–909. [DOI] [PubMed] [Google Scholar]
- Katari M.S., Nowicki S.D., Aceituno F.F., Nero D., Kelfer J., Thompson L.P., Cabello J.M., Davidson R.S., Goldberg A.P., Shasha D.E., Coruzzi G.M., Gutiérrez R.A. (2010). VirtualPlant: a software platform to support systems biology research. Plant Physiol. 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizis D., Pagès M. (2002). Maize DRE-binding proteins DBF1 and DBF2 are involved in rab17 regulation through the drought-responsive element in an ABA-dependent pathway. Plant J. 30: 679–689. [DOI] [PubMed] [Google Scholar]
- Kozaki A., Hake S., Colasanti J. (2004). The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res. 32: 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux B.M., Goodyke A.J., Schumacher K.I., Abbott C.P., Clore A.M., Yadegari R., Larkins B.A., Dannenhoffer J.M. (2014). Maize early endosperm growth and development: from fertilization through cell type differentiation. Am. J. Bot. 101: 1259–1274. [DOI] [PubMed] [Google Scholar]
- Li C., Qiao Z., Qi W., Wang Q., Yuan Y., Yang X., Tang Y., Mei B., Lv Y., Zhao H., Xiao H., Song R. (2015). Genome-wide characterization of cis-acting DNA targets reveals the transcriptional regulatory framework of opaque2 in maize. Plant Cell 27: 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Brown R.C., Fletcher J.C., Opsahl-Sorteberg H.-G. (2015). Calpain-mediated positional information directs cell wall orientation to sustain plant stem cell activity, growth and development. Plant Cell Physiol. 56: 1855–1866. [DOI] [PubMed] [Google Scholar]
- Lid S.E., Gruis D., Jung R., Lorentzen J.A., Ananiev E., Chamberlin M., Niu X., Meeley R., Nichols S., Olsen O.-A. (2002). The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99: 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Long Y., Smet W., Cruz-Ramírez A., Castelijns B., de Jonge W., Mähönen A.P., Bouchet B.P., Perez G.S., Akhmanova A., Scheres B., Blilou I. (2015). Arabidopsis BIRD zinc finger proteins jointly stabilize tissue boundaries by confining the cell fate regulator SHORT-ROOT and contributing to fate specification. Plant Cell 27: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzábal P., Busk P.K., Ludevid M.D., Torrent M. (1998). The bifactorial endosperm box of gamma-zein gene: characterisation and function of the Pb3 and GZM cis-acting elements. Plant J. 16: 41–52. [DOI] [PubMed] [Google Scholar]
- Marzábal P., Gas E., Fontanet P., Vicente-Carbajosa J., Torrent M., Ludevid M.D. (2008). The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol. Biol. 67: 441–454. [DOI] [PubMed] [Google Scholar]
- Mohanty A., Luo A., DeBlasio S., Ling X., Yang Y., Tuthill D.E., Williams K.E., Hill D., Zadrozny T., Chan A., Sylvester A.W., Jackson D. (2009). Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 149: 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco M.K., et al. (2013). Maize metabolic network construction and transcriptome analysis. Plant Genome 6: 10.3835/plantgenome2012.09.0025. [Google Scholar]
- Morita M.T., Sakaguchi K., Kiyose S., Taira K., Kato T., Nakamura M., Tasaka M. (2006). A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. Plant J. 47: 619–628. [DOI] [PubMed] [Google Scholar]
- Moscetti I., Tundo S., Janni M., Sella L., Gazzetti K., Tauzin A., Giardina T., Masci S., Favaron F., D’Ovidio R. (2013). Constitutive expression of the xylanase inhibitor TAXI-III delays Fusarium head blight symptoms in durum wheat transgenic plants. Mol. Plant Microbe Interact. 26: 1464–1472. [DOI] [PubMed] [Google Scholar]
- Myers A.M., James M.G., Lin Q., Yi G., Stinard P.S., Hennen-Bierwagen T.A., Becraft P.W. (2011). Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 23: 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A., Nakamura Y., Tanaka N., Satoh H. (2001). Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127: 459–472. [PMC free article] [PubMed] [Google Scholar]
- Ogasawara H., Kaimi R., Colasanti J., Kozaki A. (2011). Activity of transcription factor JACKDAW is essential for SHR/SCR-dependent activation of SCARECROW and MAGPIE and is modulated by reciprocal interactions with MAGPIE, SCARECROW and SHORT ROOT. Plant Mol. Biol. 77: 489–499. [DOI] [PubMed] [Google Scholar]
- O’Shea M.G., Morell M.K. (1996). High resolution slab gel electrophoresis of 8-amino-1,3, 6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis 17: 681–686. [DOI] [PubMed] [Google Scholar]
- Qi X., Li S., Zhu Y., Zhao Q., Zhu D., Yu J. (2016). ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli P.A., Larkins B.A. (2009). The development of endosperm in grasses. Plant Physiol. 149: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler F.V., Knighton R.E. (1999). Sample preparation for total nitrogen and 15N‐ratio analysis by the automated Dumas combustion method. Commun. Soil Sci. Plant Anal. 30: 1315–1324. [Google Scholar]
- Sekhon R.S., Briskine R., Hirsch C.N., Myers C.L., Springer N.M., Buell C.R., de Leon N., Kaeppler S.M. (2013). Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Peeters A.J., Koiwai H., Oritani T., Marion-Poll A., Zeevaart J.A., Koornneef M., Kamiya Y., Koshiba T. (2000). The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc. Natl. Acad. Sci. USA 97: 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Ryu J., Kang S.K., Park C.-M. (2011a). Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. Plant J. 65: 418–429. [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Ryu J.Y., Jeong E.Y., Park C.M. (2011b). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2: 303. [DOI] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dewitte W., Murray J.A.H., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A., Nield H., Lott J.N.A. (1988). An investigation of the mineral content of barley grains and seedlings. Plant Physiol. 86: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Ketterling M.G., Li Q.-B., McCarty D.R. (2003). Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 132: 1664–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto M., Tremblay R., Colasanti J. (2008). Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Mol. Biol. 67: 57–69. [DOI] [PubMed] [Google Scholar]
- Thompson R.D., Hueros G., Becker H., Maitz M. (2001). Development and functions of seed transfer cells. Plant Sci. 160: 775–783. [DOI] [PubMed] [Google Scholar]
- Tuncel A., Okita T.W. (2013). Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci. 211: 52–60. [DOI] [PubMed] [Google Scholar]
- Wang G., Wang G., Zhang X., Wang F., Song R. (2012). Isolation of high quality RNA from cereal seeds containing high levels of starch. Phytochem. Anal. 23: 159–163. [DOI] [PubMed] [Google Scholar]
- Welch D., Hassan H., Blilou I., Immink R., Heidstra R., Scheres B. (2007). Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev. 21: 2196–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.Y., Colasanti J. (2007). Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J. Exp. Bot. 58: 403–414. [DOI] [PubMed] [Google Scholar]
- Woo Y.M., Hu D.W., Larkins B.A., Jung R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Tang D., Li M., Wang K., Cheng Z. (2013). Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 161: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Sinelnikov I.V., Han B., Wishart D.S. (2015). MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 43: W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi G., Neelakandan A.K., Gontarek B.C., Vollbrecht E., Becraft P.W. (2015). The naked endosperm genes encode duplicate INDETERMINATE domain transcription factors required for maize endosperm cell patterning and differentiation. Plant Physiol. 167: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Ueguchi-Tanaka M. (2014). DELLA and SCL3 balance gibberellin feedback regulation by utilize INDETERMINATE DOMAIN proteins as transcriptional scaffolds. Plant Signal. Behav. 9: e29726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 111: 7861–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J., Thakare D., Ma C., Lloyd A., Nixon N.M., Arakaki A.M., Burnett W.J., Logan K.O., Wang D., Wang X., Drews G.N., Yadegari R. (2015). RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27: 513–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yang J., Wu Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of OPAQUE2, PROLAMINE-BOX BINDING FACTOR, and O2 heterodimerizing proteins. Plant Cell 27: 1162–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zheng X., Yang J., Messing J., Wu Y. (2016). Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc. Natl. Acad. Sci. USA 113: 10842–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Li N., Li B., Li Z., Xie G., Zhang J. (2015). Reduced expression of starch branching enzyme IIa and IIb in maize endosperm by RNAi constructs greatly increases the amylose content in kernel with nearly normal morphology. Planta 241: 449–461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.