Plastoglobule-localized PGM48 is a Zn-metallopeptidase in clade M48D of the M48 family and acts as a positive regulator of leaf senescence
Abstract
Plastoglobuli (PG) are thylakoid-associated monolayer lipid particles with a specific proteome of ∼30 PG core proteins and isoprenoid and neutral lipids. During senescence, PGs increase in size, reflecting their role in dismantling thylakoid membranes. Here, we show that the only PG-localized peptidase PGM48 positively regulates leaf senescence. We discovered that PGM48 is a member of the M48 peptidase family with PGM48 homologs, forming a clade (M48D) only found in photosynthetic organisms. Unlike the M48A, B, and C clades, members of M48D have no transmembrane domains, consistent with their unique subcellular location in the PG. In vitro assays showed Zn-dependent proteolytic activity and substrate cleavage upstream of hydrophobic residues. Overexpression of PGM48 accelerated natural leaf senescence, whereas suppression delayed senescence. Quantitative proteomics of PG from senescing rosettes of PGM48 overexpression lines showed a dramatically reduced level of CAROTENOID CLEAVAGE ENZYME4 (CCD4) and significantly increased levels of the senescence-induced ABC1 KINASE7 (ABC1K7) and PHYTYL ESTER SYNTHASE1 (PES1). Yeast two-hybrid experiments identified PG core proteins ABC1K3, PES1, and CCD4 as PGM48 interactors, whereas several other PG-localized proteins and chlorophyll degradation enzymes did not interact. We discuss mechanisms through which PGM48 could possibly accelerate the senescence process.
INTRODUCTION
Plastoglobuli (PG) are lipoprotein particles found in the plastids of most plant tissues. In chloroplasts, PGs are contiguous with the outer lipid leaflet of the thylakoid membrane, which is compatible with channeling of hydrophobic metabolites between the thylakoid membrane and the PG (Austin et al., 2006). Chloroplast-localized PGs typically range from 45 to 60 nm in size but increase dramatically in size in senescing or stressed chloroplasts (e.g., light stress and nitrogen deprivation) of wild-type plants and in various chloroplast mutants (Austin et al., 2006; Lundquist et al., 2013). The dynamic response of PG suggests that they play a functional role in chloroplast biogenesis, senescence, and stress response (Rottet et al., 2015).
PGs contain various prenyl-lipids, in particular tocopherol, various quinones (plastochromonal 8, phylloquinone, and plastoquinone), carotenoids, and fatty acid phytylesters, as well as triacylglycerols (Gaude et al., 2007; Zbierzak et al., 2009; Eugeni Piller et al., 2011; Lippold et al., 2012). Mass spectrometry analysis of isolated PG proteins and quantitative comparison to the thylakoid, stromal, and total leaf proteomes showed that PGs contain a small but specific proteome of ∼30 proteins, termed the PG core proteome (Vidi et al., 2006; Ytterberg et al., 2006; Lundquist et al., 2012a) and several additional proteins that are recruited to PG under stress conditions and in mutant backgrounds (Lundquist et al., 2013). The most abundant PG proteins are several members of the plastid-specific FIBRILLIN family (FBN) (Singh and McNellis, 2011) and members of the ACTIVITY OF BC1 COMPLEX KINASE (ABC1K) family (Lundquist et al., 2012b). Other PG core proteins include a well-studied tocopherol (vitamin E) cyclase (VTE1) (Porfirova et al., 2002), a key enzyme in tocopherol and plastochromanol biosynthesis, and PHYTYL ESTER SYNTHASE1 (PES1) and PES2 involved in the formation of phytylesters following cleavage of chlorophyll and thylakoid lipids (Lippold et al., 2012). PG-localized NADP(H) dehydrogenase C1 is involved in vitamin K biosynthesis (phyllo-quinone) (Fatihi et al., 2015) and in reduction of oxidized plastochromanols within PG (Eugeni Piller et al., 2011). PG core protein CCD4 and its homologs in various plant species (Martínez et al., 2008) are involved in carotenoid cleavage, in particular 9,10 and 9′,10′ positions, resulting in one or more apocarotenoids, especially β-ionone (Rubio et al., 2008; Huang et al., 2009). An Arabidopsis thaliana genome-wide association study identified CCD4 as a major negative regulator of seed carotenoid content (Gonzalez-Jorge et al., 2013); ccd4 loss-of-function mutants exhibited increased β-carotene content upon seed desiccation and much higher carotenoid levels than the wild type after dark-induced leaf senescence. Arabidopsis CCD4 is also implicated in an apocarotenoid signaling cascade leading to inhibition of chloroplast and leaf development (Avendaño-Vázquez et al., 2014; Hou et al., 2016). White petals of chrysanthemum (Chrysanthemum indicum) were converted into yellow-colored petals by RNAi-mediated suppression of the CCD4 homolog CCD4a, indicating its cleavage of carotenoids into colorless compounds (Zhu et al., 2010). In mandarin fruit (Citrus reticulata), a CCD4 homolog was reported to be involved in cleavage of β-cryptoxanthin and zeaxanthin to yield a red pigment, β-citraurin (Rubio et al., 2008). Collectively, this shows that the CCD4 clade cleaves a variety of carotenoids into various apocarotenoids and that CCD4 in leaves plays a role in leaf development and retrograde signaling; however, the signaling molecules and pathway are not known (Hou et al., 2016). Other PG core proteins have various predicted functional domains, such as two UBI-E methyl-transferases (UBIE-MT1,2; related to Escherichia coli UbiE involved in methylation of reactions in both ubiquinone and menaquinone biosynthesis [Lee et al., 1997]), but their functions have not yet been studied. We also identified a low abundant putative peptidase of the M48 family, here termed PGM48. The functions and targets of PGM48 are unknown, and PGM48 is the focus of this article.
Based on publicly available Arabidopsis genome-wide mRNA data, we previously generated a coexpression network using the PG core proteins as nodes (Lundquist et al., 2012a). This suggested four major coexpression modules, each showing functional enrichment, including senescence, chloroplast proteolysis, carotenoid metabolism, chloroplast redox regulation, the Calvin cycle, and chloroplast biogenesis. The senescence module was formed around genes encoding PG-localized ABC1K7, PES1, PGM48, and PG-ASSOCIATED SENESCENCE GENE (PGSAG; AT1G73750) and included the coexpressors encoding PHEOPHYTENASE (PPH) and PHEOPHORBIDE A OXYGENASE both involved in chlorophyll degradation. Leaf senescence is a highly regulated process, involving expression of thousands of genes and resulting in chlorophyll degradation, loss of photosynthetic activity, and remobilization of the chloroplast constituents for seed development or other tissues (Breeze et al., 2011; Schippers, 2015; Schippers et al., 2015). During senescence, internal structures within chloroplasts are dismantled and the PG increase in size, suggesting their direct involvement in senescence. Comparative, quantitative metabolite analysis of leaf PG isolated from four stages of natural senescence in beech leaves (Fagus sylvatica) showed that in particular, prenylquinones and free fatty acids, but not glycolipids or proteins, accumulated in PG during the senescence process (Tevini and Steinmüller, 1985). Whereas only very small amounts of chlorophylls and carotenoids or lipids were observed in PG prior to senescence, during natural senescence, the triacylglycerol content dramatically declined in PG, whereas carotenoids (mostly in esterified form) and free fatty acid levels strongly increased (Tevini and Steinmüller, 1985). However, the removal of protein complexes must also involve protein degradation, but despite major efforts by labs around the world, no intraplastid peptidase has been shown to be involved in leaf senescence (Martínez et al., 2008; van Wijk, 2015).
Here, we studied the function of PGM48 both in vitro and in vivo in Arabidopsis. This shows that PGM48 is a M48-type metallopeptidase with a conserved HExxH metal binding site. Phylogeny indicates that PGM48 forms a specific clade of peptidases only found in photosynthetic organisms; we named this subfamily M48D. We discuss the likely evolutionary origin of this subfamily and compare it to the M48A, M48C, and M48B subfamilies. Importantly, we discovered that overexpression of PGM48 accelerated natural leaf senescence, whereas suppression of PGM48 by RNA interference delayed senescence. Quantitative proteomics of PG from senescing rosettes of overexpression lines showed a dramatically reduced level of CAROTENOID CLEAVAGE ENZYME4 (CCD4) and significantly increased levels of ABC1K7 and PHYTYL ESTER SYNTHASE1 (PES1). Furthermore, we show that PGM48 physically interacts with PES1, CCD4, and ABC1K3 but not with chlorophyll catabolic enzymes. We discuss how PG-localized PG48 can possibly contribute to leaf senescence. Our research thus directly links this intraplastid protease to the process of leaf senescence.
RESULTS
PGM48 Is a Low-Abundance PG-Localized Protein in Arabidopsis
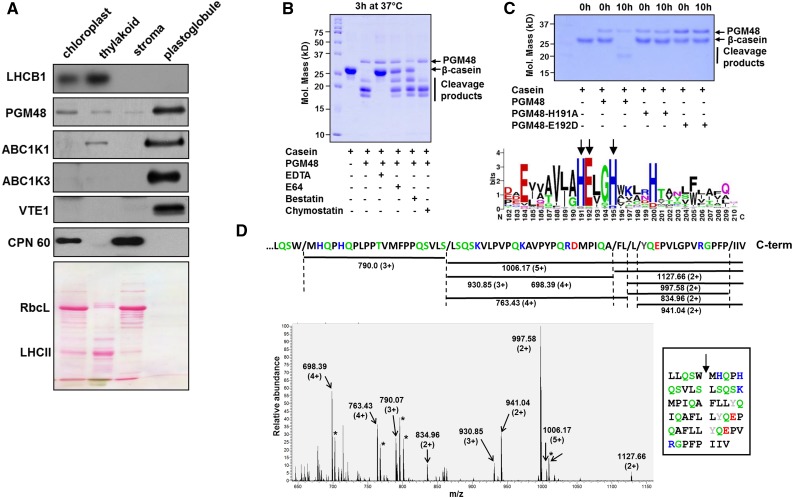
PGM48 was identified by tandem mass spectrometry (MS/MS) in purified PG from leaves using a LTQ-Orbitrap mass spectrometer in Arabidopsis (Lundquist et al., 2012a, 2013), as well as maize (Zea mays; Huang et al., 2013). PGM48 was never observed in any samples other than purified PG, despite hundreds of MS/MS analyses of Arabidopsis or maize leaf, chloroplast, or thylakoid fractions performed in our lab over the last decade (see PPDB) or by others for isolated chloroplast envelope membranes of Arabidopsis (Ferro et al., 2010) or other plant species such as pea (Pisum sativum), alfalfa (Medicago sativa), or maize (Manandhar-Shrestha et al., 2013; Simm et al., 2013; Gutierrez-Carbonell et al., 2014). Furthermore, MS/MS analysis of many PG preparations (Lundquist et al., 2012a, 2013; this article) resulted in high sequence coverage of the mature portion of PGM48. No peptides were detected matching the predicted chloroplast transit peptide (cTP) (Supplemental Figure 1), which is consistent with a plastid location of PGM48; the most N-terminal residue was Arg-61, immediately downstream of a cysteine, as frequently observed for chloroplast proteins (Rowland et al., 2015). To verify localization across chloroplast compartments, we purified thylakoid, stroma, and PG proteomes from isolated chloroplasts and probed distribution with specific antisera for stroma (CPN60), thylakoids (LHCB1), and PG core proteins ABC1K1 and ABC1K3. Consistent with our proteomics data, PGM48 signal was highly enriched in PG similar as ABC1K1, ABC1K3, and VTE1 and unlike CPN60 or LHCB1 (Figure 1A). We conclude that PGM48 is highly enriched in PG and likely carries out its peptidase function in this specialized compartment.
Figure 1.
PG-M48 Is Localized in PGs and Has Zn-Metallo-Dependent Endopeptidase Activity Cleaving Upstream of Hydrophobic Residues.
(A) Proteomes from isolated chloroplasts, thylakoid membranes, stroma, and PGs were separated by SDS-PAGE, transferred to membranes, and blotted with specific antisera against PG core proteins PGM48, ABC1K1, ABC1K3, and VTE1, as well as LHCB1 (marker for thylakoid membrane) and CPN60 (marker for stroma). Protein (15 µg) was loaded for chloroplast, stroma, and thylakoid fractions and ∼5 µg protein was loaded for PGs; due to the extremely high lipid/protein content, protein concentrations are difficult to measure accurately. The Ponceau-stained membrane (lower panel) serves as loaded control. RBCL, Rubisco large subunit (also marker for stromal proteome); LHCII, family of major LHCII proteins.
(B) Recombinant PGM48 (1 µg) was incubated for 3 h at 37°C with β-casein (4 µg) to determine the peptidase activity and the effects of peptidase inhibitors E64 (1 mM), bestatin (1 mM), chymostatin (1 mM), and the metal chelator EDTA (5 mM). EDTA inhibited PGM48 peptidase activity completely.
(C) Metal dependency of PGM48. Upper panel shows in vitro proteolytic activity of recombinant PGM48, PGM48-H191A, and PGM48-E192D mutated in their predicted metal binding site (HExxH). PGM48, but not mutated PGM48, shows peptidase activity. The lower panel shows a sequence logo for the conserved metal binding motif of 11 PGM48 homologs in angiosperms (for a complete list of homologs, see Supplemental Table 1). Color coding: green: Thr, Gly, Ser, Tyr, and Cys, uncharged, polar; pink: Asn and Gln, uncharged polar amine; red: Asp and Glu, acidic; blue: Lys, Arg, and His, basic; black: Ala, Val, Leu, Ile, Pro, Phe, Trp, and Met, apolar.
(D) PGM48 cleavage sites of β-casein as determined by mass spectrometry. The lower panel shows a high-resolution MS spectrum (from the Orbitrap portion of the LTQ-Orbitrap) of a casein digest by PGM48. Positively charged peptides matching to casein peptide fragments are marked; m/z values and charge states are indicated. The upper portion shows the C-terminal portion of β-casein and the identified peptides as indicated by horizontal lines with the respective m/z value and associated charge states. The inset shows the inferred cleavage sites with the β-casein sequence. Collectively, this shows that PGM48 prefers cleavage upstream of hydrophobic residues: methionine (M), phenylalanine (F), leucine (L), isoleucine (I) (in black), or tyrosine (Y) (in gray). Other colors: green: Q,S – uncharged, polar; D,E – acidic in red; K,R,H – basic in blue; remaining residues in black.
PGM48 Is a Functional Metallo Zn-Endopeptidase
To test if PGM48 is a functional peptidase, we expressed mature (i.e., starting from Arg-61; excluding the cTP) Arabidopsis PGM48 with a histidine tag in E. coli and affinity purified the recombinant protein. PGM48 was incubated with β-casein as substrate and degradation was visualized by Coomassie staining after separation by SDS-PAGE (Figure 1B). β-Casein was degraded by PGM48, and this activity was completely inhibited by addition of the metal chelator EDTA (Figure 1B). Other peptidase inhibitors including the cysteine peptidase inhibitor E64 and metallo-aminopeptidase inhibitor bestatin partially inhibited degradation, whereas chymostatin (an inhibitor of various types of peptidases) did not inhibit PGM48 activity. Through site-directed mutagenesis of the conserved metal binding HEXXH in PGM48 (Figure 1C, lower panel with the sequence logo based on angiosperm PGM48 homologs), we replaced His-191 or Glu-192 into Ala-191 or Asp-192, respectively. Recombinant mutated PGM48 (PGM48-H191A and PGM48-E192D) completely lacked peptidase activity (Figure 1C, upper panel), clearly showing that PGM48 is a functional metallo-endopeptidase.
To determine cleavage site specificity, recombinant active PGM48 and inactive PGM48-H191A were each incubated with β-casein, and following desalting, peptide products were analyzed by high-resolution MS through direct infusion. Figure 1D shows an MS spectrum from extracts of the active PGM48. The dominant peaks matched to eight peptides, as determined by MS/MS ranging from 1.7 to 5.0 kD (Figure 1D); these peptides were absent in PGM48-H191A (Supplemental Figure 2). The methionine-containing peptides were also observed with oxidized methionine residues and/or in different charge states (due to the ionization process in the MS source), allowing us to account for almost all ions in the mass spectrum (Supplemental Table 1). The detected fragments correspond to cleavage sites depicted in the sequence of PGM48, and displayed in the Figure 1D inset, with a clear preference for cleavage immediately upstream of hydrophobic residues (Leu, Ile, Met, Phe, or Tyr) and downstream of hydrophobic (Leu, Trp, or Pro) or polar residues (Ala or Ser).
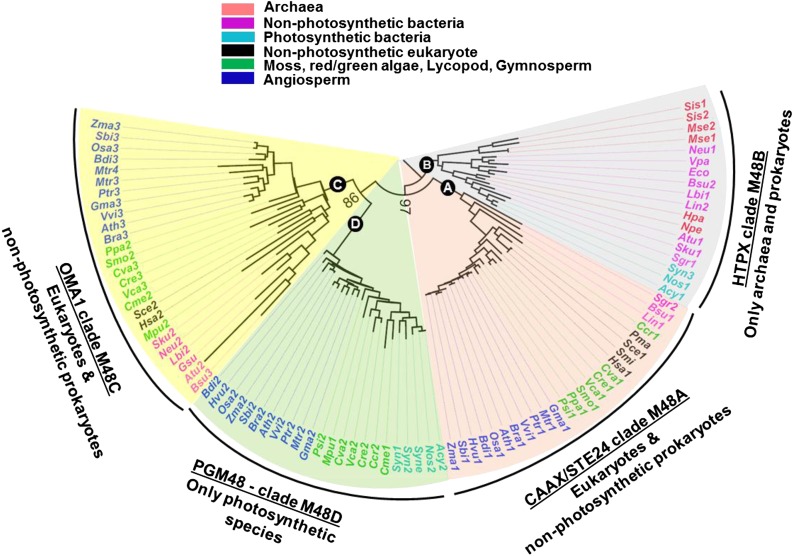
PGM48 Homologs Form a Clade in the M48 Family
PGM48 belongs to the M48 family of ATP-independent metallo (Zn)-endopeptidases as defined in the MEROPS peptidase database (Rawlings et al., 2016). According to MEROPS, the M48 family has three clades, M48A, M48B, and M48C, each represented by a prototypic enzyme named STE24 or CAAX peptidase (Bracha-Drori et al., 2008; Pryor et al., 2013; Quigley et al., 2013; Ast et al., 2016), bacterial HTPX (McBride and Soubannier, 2010; Quiros et al., 2015), and mitochondrial OMA1 (Sakoh et al., 2005; Akiyama, 2009), respectively. However, sequence analysis of M48 proteins in 42 species across the tree of life suggests that there are four clades (Figure 2; Supplemental Table 2; Supplemental File 1 for alignments), with PGM48 and its homologs forming a separate clade that we named M48D. We briefly discuss the distribution of these M48 clades across phylogenetic groups and comment on known functions, followed by a hypothesis for the origin of PGM48 proteins in plants.
Figure 2.
M48 Proteins Are Conserved in Nonphotosynthetic and Photosynthetic Organisms.
Phylogenetic tree of the M48 family based on alignment of amino acid sequences of 92 M48 homologs from diverse archaea, prokaryotes, and eukaryotes. Four clades are assigned: M48A, M48B, M48C, and the new clade M48D. Bootstrap values are indicated at the nodes of the tree. The sequence alignment is shown in Supplemental File 1, and Supplemental Table 1 provides a complete listing of species and number of M48 members. Vitis vinifera (Vvi); Populus trichocarpa (Ptr); Medicago truncatula (Mtr); Brassica rapa (Bra); Arabidopsis thaliana (Ath); Glycine max (Gma); Zea mays (Zma); Brachypodium distachyon (Bdi); Hordeum vulgare (Hvu); Oryza sativa (Osa); Sorghum bicolor (Sbi); Picea sitchensis (Psi); Selaginella moellendorffii (Smo); Physcomitrella patens (Ppa); Cyanidioschyzon merolae (Cme); Chondrus crispus (Ccr); Volvox carteri (Vca); Chlamydomonas reinhardtii (Cre); Chlorella variabilis (Cva); Micromonas pusilla (Mpu); Synechocystis sp PCC6803 (Syne); Anabaena cylindrica (Acy); Nostoc sp PCC7120 (Nos); Synechococcus sp PCC 7942 (Syn); Homo sapiens (Hsa); Penicillium marneffei (Pma); Saccharomyces cerevisiae (Sce); Agrobacterium tumefaciens (Atu); Nitrosomonas europaea (Neu); Escherichia coli (Eco); Sulfuricurvum kujiens (Sku); Streptomyces griseus (Sgr); Bacillus subtilis (Bsu); Leptospira interrogans (Lin); Leptospira biflexa (Lbi); Sulfolobus islandicus (Sis); Metallosphaera sedula (Mse); Haladaptatus paucihalophilus (Hpa); Natrinema pellirubrum (Npe).
Clade M48A consists of well-studied peptidases localized to the endoplasmic reticulum (ER) or inner nuclear membrane in photosynthetic and nonphotosynthetic eukaryotes, and STE24 homologs in nonphotosynthetic bacteria, but absent in Archaea and photosynthetic bacteria. Eukaryotic STE24 are membrane proteins (multiple transmembrane domains) with a large hollow barrel-shaped chamber enclosing the catalytic site. They function to cleave C termini of proteins with a CAAX motif carrying a cysteine linked prenyl group. Recently, STE24 has also been shown to clear clogged SEC protein translocons, in addition to its function in CAAX processing (Ast et al., 2016). Angiosperms, gymnosperms, lycopodia, mosses, and most red/green algae each have a single STE24 homolog (Figure 2; Supplemental Table 2). Arabidopsis STE24 is ER-localized and cleaves prenylated CAAX proteins (Bracha et al., 2002; Bracha-Drori et al., 2008).
Clade M48B is represented by bacterial HTPX Zn-endopeptidase. Members of this clade are present in Archaea and in nonphotosynthetic and photosynthetic bacteria, but not in eukaryotes (Figure 2). Most prokaryotes have one HTPX homolog (Supplemental Table 2). HTPX homologs have been studied in, e.g., Bacillus subtilis and E. coli, and have shown to be integral plasma membrane metallopeptidases with their active side facing the cytosol. HTPX in E. coli complements the ATP-dependent FTSH peptidase in quality control of the plasma membrane proteome (Akiyama, 2009).
Clade M48C is represented by inner membrane mitochondrial OMA1. This clade is found in both photosynthetic and nonphotosynthetic eukaryotes and nonphotosynthetic prokaryotes, but not in photosynthetic bacteria (Figure 2). OMA1 homologs in eukaryotes are (most likely) localized in mitochondria; they have been studied in mammals, zebra fish, and yeast, in particular in proteolysis of OPTIC ATROPHY1 (OPA1/MGM1), a key conserved inner membrane dynamin-like GTPase involved on mitochondrial fusion (Bohovych et al., 2015; Korwitz et al., 2016). OMA1 is present in angiosperms, most green and red algae, lycopods, moss, and angiosperms, but not in the gymnosperm Picea sitchensis, either due to independent gene loss or perhaps incomplete genome sequence annotation (Figure 2; Supplemental Table 2). The OMA1 homolog in Arabidopsis has been observed in several mitochondrial proteomics studies (Finkemeier et al., 2011; Klodmann et al., 2011) and homologs in maize, rice (Oryza sativa), tobacco (Nicotiana tabacum), and other plant species have predicted mitochondrial transit peptides. However, it appears that OMA1 function has not been studied in any photosynthetic eukaryotes nor in prokaryotes.
Based on the cladogram (Figure 2), it is clear that PG-localized M48 homologs form a separate clade; we named this M48D. Proteins in this clade are only found in photosynthetic organisms, including cyanobacteria, green algae, red algae, gymnosperms (conifers), and angiosperms (Figure 2; Supplemental Figure 3 for the M48D clade alone), but absent in the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii. None of the members of the M48 clade have been studied so far, but in addition to Arabidopsis PGM48, we did identify the maize PGM48 protein (GRMZM2G111200) in isolated PG from leaves (Huang et al., 2013). We also identified rice PGM48 (Os01g73910) in isolated chloroplasts with good sequence coverage (in three independent replicates) but not in total leaf extracts (see PPDB). PGM48 in maize, rice, and most other angiosperms have a TargetP predicted cTP (Supplemental Figure 1).
It should be noted that Arabidopsis and other photosynthetic organisms have another peptidase family of CAAX proteases, named M79, that typically cleave a C-terminal tripeptide from an isoprenylated protein; there are five Arabidopsis proteins in this M79 family. One M79 family member in Arabidopsis is the plastid-localized SNOWY COTYLEDON4 (AT5G60750), but there is no evidence that it has CAAX activity (Albrecht-Borth et al., 2013).
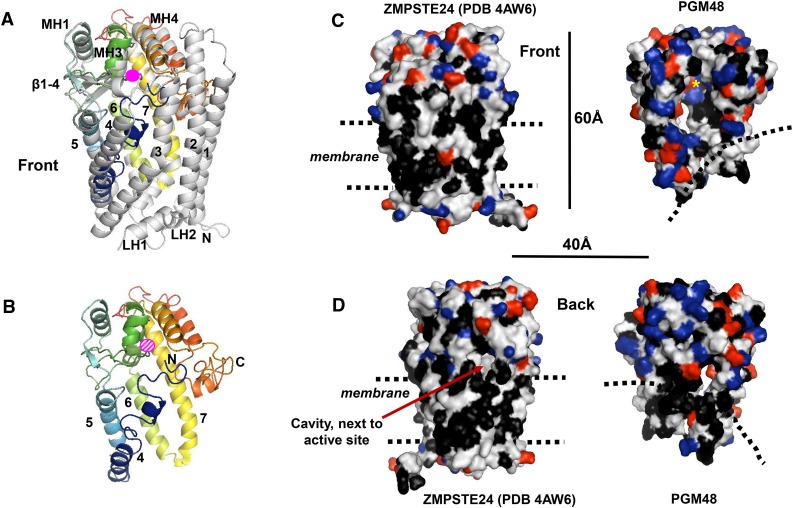
Structural Model of PGM48 and Interaction with the Monolayer PG
It is unknown how PGM48 interacts with PG, how substrates are recognized and interact with PGM48, or how substrates obtain access to the catalytic site. Fortunately, high-resolution x-ray crystallography-based structures for M48 homologs in clade M48A from human (PDB 4AW6) (Quigley et al., 2013) and Saccharomyces mikatae (PDB 4IL3) (Pryor et al., 2013) (37% identity between these two STE24 homologs), clade M48B for Vibrio parahaemolyticus (PDB 3CQB), and from clade M48C for Geobacter sulfurreducens (PDB 3C37) are available in the Protein Data Bank (http://www.rcsb.org/pdb/). Based on these available structures, we built a homology model for mature Arabidopsis PGM48 (Figure 3) to begin answering the key questions stated above. The various scoring parameters of the top scoring i-TASSER model for PGM48 (see legend of Figure 3) suggested a meaningful model that is suitable and sufficient to address general folding, formation of the active site, internal cavity, and interaction with the PG. Lower scoring models were not further considered. The PGM48 model was most similar to the 3.8-Å structure of human STE24. Structural analysis showed that STE24 has a seven transmembrane (TM) α-helical barrel structure surrounding a large, water-filled chamber, capped by the Zn-metallopeptidase domain with the catalytic site facing into the chamber (Quigley et al., 2013). Figure 3A shows this structure overlaid with the PGM48 model, Figure 3B shows PGM48 model alone, and Figures 3C and 3D show it as a space-filling model side-by-side with PGM48. The model quality is highest in the central portion of PGM48, which contains the catalytic site (residues 180 through 265). PGM48 completely lacks the sequence corresponding to TM1, 2, and 3 present in STE24 (Figures 3A and 3B). The PGM48 model shows no transmembrane domains, but it has shorter hydrophobic helices that partially overlay to TM3-7 (Figures 3A and 3B); this is consistent with PGM48 being associated with a monolayer particle. The PGM48 active site residues (His-191, Glu-192, His-195, and Glu-240) are located in the short α-helices 7 and MH3, in close proximity to a 6 × 10-Å diameter cavity, similar to that observed for STE24. Substrates are proposed to enter the STE24 cavity through a pore located close to the lipid surface. These substrates are prenylated at their C termini (through the CAAX motif), and this (hydrophobic) prenyl group helps guide the substrate along the surface of the lipid bilayer into the central pore (Quigley et al., 2013). After two cleavages, the C-terminally truncated substrate is released. Interestingly, the STE24 homolog in yeast was recently shown to clear the ER Sec protein translocon from proteins clogging the translocation pore (Ast et al., 2016); these substrates are probably not prenylated. Similarly, despite older reports of protein prenylation (e.g., palmityolation) in chloroplasts (Mattoo and Edelman, 1987; Parmryd et al., 1997; but see Parmryd et al., 1999), no genetic or further biochemical support for intraplastid prenylation has been found. Hence, PGM48 is unlikely to have CAAX peptidase activity, and it is most likely that PGM48 can cleave proteins without (C-terminal) lipid modifications. The model for PGM48 shows a pore situated at the postulated lipid interface (dashed lines) (Figures 3C and 3D) that could provide substrate access to the central cavity with the active site (Figure 3C, yellow asterisks).
Figure 3.
3D Protein Structural Model for PGM48 and Predicted Interface with the PG Monolayer.
The 3D protein structural model of PGM48 was generated with i-TASSER (Yang et al., 2015) using the predicted mature PGM48 sequence (residues 48 to 344). The top scoring i-TASSER model for PGM48 had a C-score of −0.98 (C-scores range from −5 [poorest] to 2 [best]) and estimated TM score of 0.59 ± 0.14. Analysis of PGM48 protein model using ProSA (Wiederstein and Sippl, 2007) gave a Z-score of −3.73, which is well within the range of scores found for protein structures of this molecular weight generated by crystallography and NMR. The protein model matched most closely to the 3.8-Å structure PDB 4AW6 of the M48 protein STE24 from humans (Quigley et al., 2013). Images were generated with PyMol Version 1.7.4 software (Schrödinger).
(A) Front view of STE24 structure (4AW6) in gray overlaid with the colored modeled PGM48. PGM48 is colored in rainbow from blue N terminus to red C terminus. The Zn metal ion is marked in pink (not to scale). The STE24 helices 1 to 7, lumenal helices (L1 to 3), and cytoplasmic helices (MH1, 3, and 4) are marked using the naming from Quigley et al. (2013). Helices 1, 2, 3, 7A, LH1, and LH2 are absent in the PGM48 model.
(B) Front view of the PGM48 model. Helices are numbered according to 4AW6, and the N and C termini are indicated; the position of the Zn ion is indicated in dashed pink.
(C) and (D) Side-by-side comparison of the structure 4AW6 of STE24 and the PGM48 model using a surface representation. Amino acid side chains are colored in black for hydrophobic residues (Leu, Ile, Val, Phe, Trp, and Ala), red for acidic residues (Glu and Asp), and blue for basic residues (Lys, Arg, and His). Dimensions (in Å) and approximate location of the ER/nuclear membrane lipid bilayer (STE24) and postulated PG lipid surface (PGM48) (dashed lines) are indicated.
Generation of PGM48 Under- and Overexpression Lines
To determine the physiological function and (candidate) substrates of PGM48, we obtained, confirmed, and characterized Arabidopsis T-DNA insertion lines for PGM48 that we named pgm48-1 (insertion in exon) and pgm48-2 (insertion in intron) (Supplemental Figure 4A). As expected, RT-PCR was not able to amplify full-length mRNA for either line due to the relatively large size of the T-DNA, and RT-PCR analysis showed >2-fold reduced mRNA accumulation of the region upstream of the T-DNA insertion for both lines (Supplemental Figure 4B). MS/MS proteome analysis was performed on isolated PG of pgm48-1, which identified PGM48 at 2- to 5-fold reduced levels compared with the wild type (Supplemental Figure 4C). We concluded that neither line is a true loss-of-function (null) mutant. No visible phenotype was observed in developing rosettes under a variety of abiotic conditions, but natural senescence was delayed in both alleles by ∼1 week (Supplemental Figure 4D).
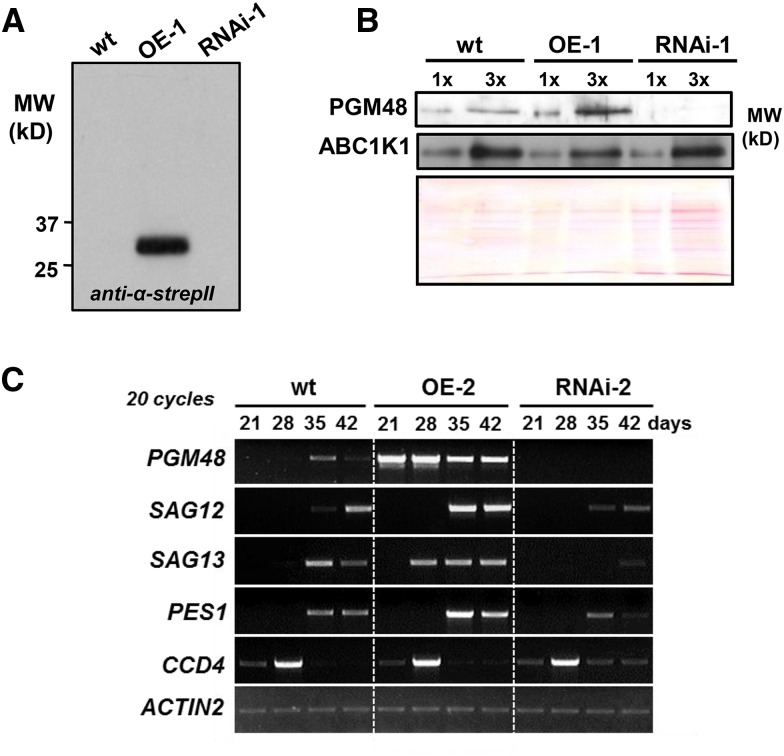
Because both T-DNA alleles were leaky, likely weakening the phenotypes, we generated more complete loss-of-function mutants by silencing PGM48 using RNAi (driven by a 2x35S promoter). Additionally, we generated transgenic plants overexpressing full-length PGM48 with a C-terminal StrepII tag driven by the constitutive 35S promoter. RT-PCR for several independent transformants (18-d-old plants a few days prior to bolting) showed that PGM48 mRNA increased by severalfold in overexpression (OE) lines and decreased to nearly undetectable levels in RNAi plants (even when using a higher cycle number) compared with wild-type plants (Supplemental Figure 5A). Immunoblotting with anti-strepII antiserum confirmed accumulation of intact PGM48-StrepII in the OE lines (Figure 4A). Immunoblotting of purified PG from wild-type, OE, and RNAi lines showed that PGM48 was 2- to 3-fold higher in PG from the OE-1 line but virtually undetectable in the RNAi-1 line (Figure 4B). RT-PCR mRNA analysis (low cycle number) for rosettes of 21-, 28-, 35-, and 42-d-old plants (grown under long daylength, 18 h light) showed that PGM48 mRNA levels were many fold higher in the OE-1 and -2 lines than wild-type plants and that PGM48 mRNA was undetectable in the RNAi-1 and -2 lines throughout these developmental stages (Figure 4C; Supplemental Figure 5A). Extensive MS/MS protein analysis of isolated PG of wild-type and transgenic lines further confirmed the lack of PGM48 accumulation in the RNAi lines (RNAi-1 and -2) and increased levels in the OE line (OE-1 and -2; see below). Thus, we successfully generated Arabidopsis lines with high constitutive levels of PGM48 or very low (undetectable) levels of PGM48.
Figure 4.
Overexpression and Suppression of PGM48 in Arabidopsis and Effect on mRNA Levels during Plant Development and Senescence.
(A) Accumulation of transgenic StrepII-tagged PGM48 proteins in PG isolated from wild-type, OE, and RNAi rosettes as determined by immunoblotting using specific anti-StrepII serum.
(B) Accumulation level of endogenous and transgenic PGM48 and ABC1K1 proteins in PG isolated from wild-type, OE-1, and RNAi-1 rosettes as determined by immunoblotting. The Ponceau-stained blot is shown for loading control. 1x and 3x correspond to ∼4 and 12 µg of loaded isolated PG proteins.
(C) mRNA levels of PGM48, SAG12, SAG13, PES1, and CCD4, in rosettes of wild-type, OE-2, and RNAi-2 lines after 21, 28, 35, and 42 d growth under 18 h light/6 h dark, 130 µmol photons m−2 s−1 light intensity (for additional replicate, see Supplemental Figure 5F).
PGM48 Activates Leaf Senescence
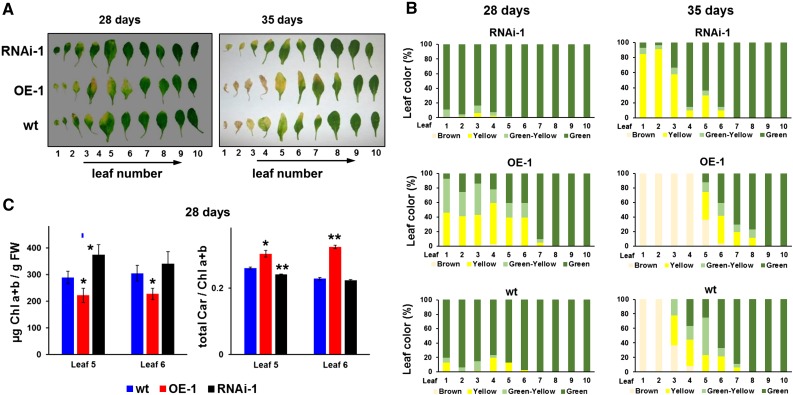
During the vegetative stages of seedling development (i.e., prior to bolting), the under- and overexpression lines did not show a visible phenotype (data not shown). Bolting (the switch to reproductive phase and first appearance of the inflorescence) started between 18 and 21 d in all three genotypes and was completed by day 21 (Supplemental Figure 5B) with the first flowers to appear at 22 to 25 d (under long day, 18-h light period). There was little change in bolting time between the three genotypes (Supplemental Figure 5B). Arabidopsis is a monocarpic plant and in absence of external (stress) factors, the onset of leaf senescence is driven by the developmental switch from vegetative to reproductive phase (Schippers et al., 2015). During these early flowering stages, the first visible signs of leaf senescence can be observed in the oldest leaves of the OE line, as well as wild-type plants (at 21-22 days), whereas the onset of senescence in RNAi lines was delayed by 2 days (Figure 5A; Supplemental Figure 5C). These differences became more pronounced in older rosettes (Figure 5A; Supplemental Figure 5D for additional transgenic lines). Quantification of senescence (categorized as green, green-yellow, yellow, and brown/necrotic) for the individual leaves shows the progression of senescence from 28 to 35 days in all three genotypes, clearly demonstrating the accelerated senescence in the OE line and the delay in the RNAi line (Figure 5B). Consistent with this, leaf chlorophyll concentrations were significantly lower in the OE lines and higher in the RNAi lines as compared with wild-type at 28 days (Figure 5C) and 35 days (Supplemental Figure 5E). Interestingly, the total carotenoid to chlorophyll ratio was significantly higher in the OE line compared with wild-type and the RNAi line (Figure 5C), but there were no significant differences in leaf 5 and 6 at 35 days (Supplemental Figure 5E), likely because these leaves were now in a very advanced stage of senescence (Figure 5A,B; Supplemental Figure 5D).
Figure 5.
Overexpression and Suppression of PGM48 in Arabidopsis Shows That PGM48 Protein Accelerates Natural Leaf Senescence.
(A) Examples of progression of natural leaf senescence of wild-type, OE, and RNAi lines under 18 h light/6 h dark, 130 µmol photons m−2 s−1 light intensity. Ten individual leaves, numbered from oldest to youngest, were separated from a plant rosette for each of the three genotypes at days 28 and 35.
(B) Percentage of leaf color measured from leaf 1 to 10 for each genotype; the average values of for three rosette plants per genotype are presented. Dark green, healthy looking green tissue; light green, light-green tissue undergoing degreening; yellow, yellow leaf section mostly devoid of visible chlorophyll; brown, dead leaf.
(C) Chlorophyll a + b content on fresh weight bases and total carotenoid/total chlorophyll ratios for leaves 5 and 6 in the three genotypes at 28 d for plants as shown in (A) and (B). Significance levels are indicated: *P < 0.05 and **P < 0.01.
The acceleration and delay in senescence in respectively the OE and RNAi lines was also reflected at the mRNA level of the well-known senescence markers SAG12 (encoding for the papain-like cysteine peptidase located in senescence-associated vacuoles (Otegui et al., 2005; Carrión et al., 2013) and SAG13, a dehydrogenase with unknown function (Miller et al., 1999) (Figure 4C; see Supplemental Figure 5F for additional replicate). SAG12 expression was visible in 35 d in the OE lines, but only detected at 42 d in the wild type and at a much lower level in the RNAi-1 line. Similarly, SAG13 was visible at 28 d in the OE lines but at 35 d in the wild type and at very low level in RNAi lines (Figure 4C; Supplemental Figure 5F). Thus, mRNA accumulation patterns confirm accelerated senescence in the PGM48 OE lines and delayed senescence in the RNAi lines. Thus PGM48 is an activator of leaf senescence.
Effects of Natural Leaf Senescence on the PG Transcriptome and Proteome
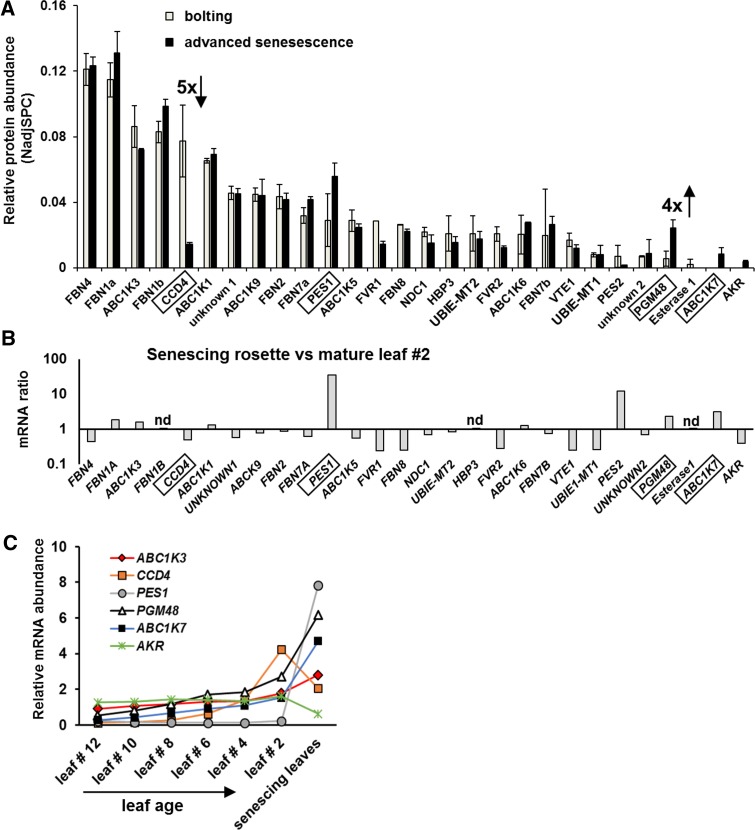
It is well established that the amount and size of PG increases during leaf senescence in many plant species (Besagni and Kessler, 2013). However, it is unknown if the PG proteome changes. To determine if the PG proteome in wild-type plants is affected by natural leaf senescence, we isolated PG from whole rosettes of plants in the bolting and advanced natural senescence stages. The PG proteomes were identified and quantified by label-free spectral counting using MS/MS after SDS-PAGE and tryptic digestion (Figure 6A; Supplemental Data Set 1). Whereas most PG core proteins did not change in relative abundance within the isolated PG, the relative abundance of PGM48 increased 4-fold (Figure 6A). Furthermore, CCD4 stood out for its strong, 5-fold decrease during senescence. ABC1K7 (part of the senescence mRNA expression module, together with PGM48 and PES1) was not detected in the bolting stage but was clearly detected in the senescence stage (with 18 MS/MS spectra) (Figure 6A). To understand if the proteome patterns are also observed at the mRNA level, we extracted the most relevant leaf mRNA expression data from the public domain for all genes encoding for PG proteins. Figure 6B compares mRNA levels between senescing leaves and mature leaf 2 (best representing the bolting stage) for all available PG genes. This shows that in particular PES1, PES2, PGM48, and ABC1K7 are induced during senescence, whereas several others, in particular CCD4, FLAVIN REDUCTASE-RELATED1,2, FBN8, VTE1, and UBIE-MT1, decreased. Figure 6C shows mRNA abundance levels for PGM48, CCD4, ABC1K7, and PES1 in individual leaves from 17-d-old rosettes (continuous light), with leaf 2 being the oldest and leaf 12 the youngest. mRNA for ALDO/KETO REDUCTASE (AKR) is shown for comparison as an example of a gene not induced by senescence. CCD4 mRNA very prominently increases with leaf age (up to approximately the bolting stage) but then clearly declines in senescent leaves. This contrasts to PGM48, which also increases with increasing leaf age but continues to increase in senescing leaves. Indeed, plotting the leaf mRNA expression level for all PG genes shows that CCD4 and PGM48 really stand out in their upregulation during leaf aging (Supplemental Figure 6A).
Figure 6.
PG Proteome Composition in Wild-Type Plants during Natural Leaf Senescence and mRNA Levels for PG Genes.
(A) PGM48 relative protein accumulation levels determined by label-free spectral counting quantitative proteomics of isolated PG at bolting stage and advanced senescence stage in the wild type. Results from two replicates of independent PG preparations were analyzed; relative abundance was normalized to the total amount of PG core protein. Plants were grown under short-day conditions (10 h light/14 h dark). For the complete abundance data of all PG core proteins, see Supplemental Data Set 1. CCD4 and PGM48 showed >4- to 5-fold difference between the bolting and advanced senescence stage.
(B) and (C) mRNA expression data from publicly available microarray data for leaves from the wild type (Col-0); data were downloaded from http://bar.utoronto.ca/. (B) shows the relative abundance ratio between mRNA from mature leaf 2 and senescent leaves for all genes encoding for the PG core proteome, except for FBN1B, HBP3, and ESTERASE1 for which no data are available. (C) shows mRNA abundance levels of individual leaves from 17-d-old rosettes (continuous light), with leaf 2 being the oldest and leaf 12 the youngest. This genes selected are PGM48 and CCD4, showing ABC1K7 and PES1 as senescence-induced genes for comparison and AKR as a gene not induced by senescence. CCD4 mRNA very prominently increases with leaf age but is clearly declining in senescent leaves; this contrasts with PGM48, which also increases with increasing leaf age but continues to increase in senescing leaves.
The microarray data are consistent with the mRNA expression patterns observed experimentally by RT-PCR (Figure 4C), in that CCD4 mRNA level of the rosette peaks around 28 d (when only a few of the oldest leaves on the rosette show some senescence; Figures 5A and 5B) but dramatically decreased in advanced senescing plants (35 d) (Figure 4C; see also Supplemental Figure 5F for independent series). In contrast, mRNA levels of PGM48, PES1, and ABC1K7 peak in later stages of senescence (Figure 4D; Supplemental Figure 5F).
PG Proteome from OE, RNAi, and Wild-Type Lines during Senescence
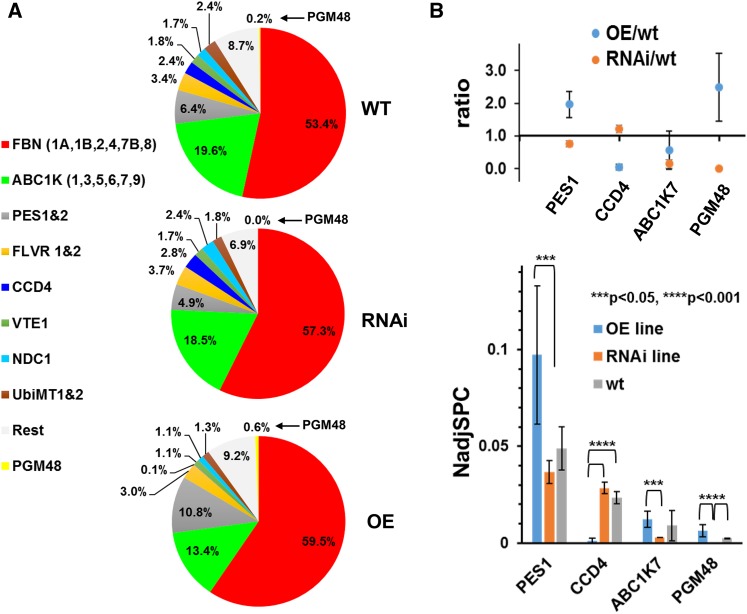
We then determined and compared the proteomes of PG isolated from wild-type, OE, and RNAi lines from 35-d-old senescing rosettes (as in Figure 5 and Supplemental Figure 5) with three biological replicates (using OE-1 and -2 and RNAi-1 and -2) to provide insight in possible targets of PGM48 peptidase activity. This identified 29 proteins out of the 30 known PG core proteins (low-abundance At1G73750 with unknown function was missing) (Supplemental Data Set 2). Generally, the PG proteomes of the three genotypes were similar with 53 to 59% of protein mass invested in the FBN family and 13 to 20% to the ABC1K family (Figure 7A). However, four PG core proteins (PGM48, CCD4, ABC1K7, and PES1) significantly (P < 0.05 or < 0.01) under- or overaccumulated in the OE or RNAi lines (Figure 7B; Supplemental Data Set 2). PGM48 was 2.5-fold higher compared with the wild type and undetected in the RNAi lines, consistent with mRNA levels (Figure 7B). CCD4 decreased ∼20-fold in the OE line (undetected in two replicates and just a few MS/MS spectra in the 3rd replicate) but was unchanged in the RNAi line compared with the wild type. ABC1K7 was 3-fold lower in the RNAi line compared with the wild type, whereas PES1 increased twofold in the OE line and decreased 25% in the RNAi line.
Figure 7.
PG Proteome Composition in Wild-Type, OE, and RNAi Plants during Natural Leaf Senescence Determined by MS/MS-Based Label-Free Spectral Counting of Isolated PG.
(A) Relative mass abundance of PG core proteins and protein families normalized to the abundance of the sum of all observed PG proteins. Twenty-eight PG core proteins were observed. Proteomics data are listed in Supplemental Data Set 2, and spectral data are available at ProteomeXchange (see Methods). PGs were isolated from 35-d-old rosettes of wild-type, OE-1, OE-2, RNAi-1, and RNAi-2 (plants were grown under long-day conditions at 18 h light/6 h dark) and analyzed by MS. PG proteins were determined by label-free quantitative comparative proteomics. PGM48 was not identified in the RNAi lines. The analysis was performed with three sets of independent biological replicates with two replicates for OE-1 and RNAi-1 and one replicate for each OE-2 and RNAi-2 (Supplemental Data Set 2).
(B) Statistically significant pairwise differences of normalized abundance levels (using NadjSPC) for PG core proteins across the three genotypes were determined by Student’s t test. PES1, CCD4, ABC1K7, and PGM48 showed significant pairwise differences (P < 0.05 or P < 0.01) as indicated in the bar diagram. Protein abundances were quantified based on the number of matched adjusted MS/MS spectra (NadjSPC). Data shown here are average of three independent replicate, and bars indicate standard deviations (n = 3). Abundance ratios between OE/wild-type and RNAi/wild-type of PES1, CCD4, ABC1K7, and PGM48 are shown in the upper panel.
PGM48 Interacts with PES1, CCD4, and ABC1K3 in Vitro
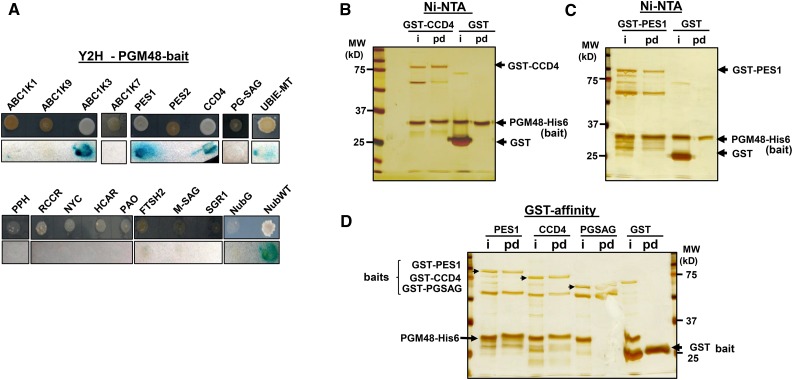
Since PGM48 shows a role in senescence, we tested if PGM48 can interact with the chloroplast chlorophyll catabolic enzymes NYC1, RCCR, HCAR, PAO, and PPH (Sakuraba et al., 2015) in mating-based split ubiquitin yeast two-hybrid assays. This did not identify interactions between these proteins and PGM48 (Figure 8A). However, testing interaction of PGM48 with nine PG core proteins showed that PGM48 interacted with PES1, CCD4, and ABC1K3, and a weak interaction was also observed with UBIE-MET, but no interaction was found for ABC1K1, PES2, ABC1K7, ABC1K9, or PGSAG (Figure 8A). Finally, thylakoid peptidase FTSH2, chloroplast membrane SAG protein (AT2G20920), and senescence-induced stay-green protein 1 (NYE1/SGR1) did not interact with PGM48 either (Figure 8A).
Figure 8.
Interaction of PGM48 with Other PG Proteins.
(A) Yeast two-hybrid assays for interactions between PGM48 and selected proteins. PGM48 was used as bait, and selected candidate proteins were used as prey. Bait plasmid contains Cub-PLV and prey plasmid contains NubG. NubG moiety was fused to the N terminus of prey proteins. The resulting plasmids were transferred into the different yeast strains for bait and prey. The transformed yeasts harboring bait and prey constructs were mated, and resulting transformants were analyzed on selective medium lacking Ade, His, Try, Leu, Ura, and Met (upper lane) and for β-galactosidase (β-Gal) activity (lower lane). Soluble NubG and Nub-WT were used as negative and positive controls, respectively.
(B) Ni-NTA resin-mediated pull-down of PGM48-His with GST-CCD4. Purified GST alone (10 µg) or GST-fused CCD4 (2 µg) was incubated with purified PGM48 fused with His6 (2.0 µg), together with Ni-NTA resin. Input (50% of the reaction) and eluted samples (80%) were loaded into a SDS-PAGE gel and analyzed by silver staining. i, input; pd, pull-down eluate.
(C) Ni-NTA resin-mediated pull-down of PGM48-His with GST-PES1. Purified GST alone (10 µg) or GST-fused PES1 (3 µg) was incubated with purified PGM48 fused with His6 (2.0 µg), together with Ni-NTA resin. Input (50%) and eluted samples (80%) were loaded into a SDS-PAGE gel and analyzed by silver staining.
(D) Glutathione resin-mediated pull-down of GST-PES1 and GST-CCD4 with PGM48-His. Purified GST-PES1 (2 µg), GST-CCD4 (2 µg), GST-PGSAG (2 µg), and GST alone (10 µg) were incubated with purified PGM48-His (3 µg) recombinant protein, together with glutathione resin. Input (50% of the reaction) and eluted samples (80%) were loaded into a SDS-PAGE gel and analyzed by silver staining.
We then tested PGM48 interactions with PES1 and CCD4 using in vitro pull-down assays with purified recombinant proteins expressed in E. coli. Recombinant PGM48 was generated with a His6 tag at the C terminus, whereas recombinant CCD4 and PES1 were N-terminally fused with GST. Recombinant PGM48-His6 was incubated with recombinant GST-CCD4, GST-PES1, or GST (as negative control), followed by affinity purification using Ni-NTA resin. After extensive washing, bait protein (PGM48-His) eluted with GST-CCD4 and GST-PES1, but not with GST (Figure 8B), confirming the observation by the yeast two-hybrid system. Subsequently, we probed for interactions for the same set of proteins, but this time using the GST-fusions as bait, employing glutathione resin to capture GST-CCD4, GST-PES1, and GST (as negative control) and testing if PGM48 is pulled down with these baits (Figure 8C). We included GST-PGSAG as an additional bait. As shown in Figure 8C, PG-M48 eluted with GST-CCD4 and GST-PES1, indicating a positive interaction with PGM48, whereas PGM48 did not elute with GST-PGSAG nor GST alone (Figure 8C). Thus, these reciprocal pull-down experiments further supported interactions of PGM48 with CCD4 and PES1. Coimmunoprecipitation of endogenous PGM48 in PG isolated from naturally senescing leaves using our anti-PGM48 antiserum followed by MS/MS did successfully enrich for PGM48 but did not detect specific protein interactors. Whereas we did apply cross-linkers prior to the co-IP, it is likely that PGM48 interactions to target substrates are highly transient and therefore difficult to capture.
DISCUSSION
PGM48 Represents a Specific Adaptation of Photosynthetic Organisms
PGs are plastid microcompartments with multiple functions integrated in plastid metabolism, developmental transitions, and environmental adaptation (van Wijk and Kessler, 2016). The challenge is to unravel how PG and their molecular components contribute to plastid homeostasis. This study concerns the role of the PG-localized metallopeptidase PGM48 in leaf chloroplasts. Through in vitro assays, we showed that PGM48 is a relatively low-abundance Zn-dependent metallopeptidase and belongs to the peptidase clan MA containing a variety of metallopeptidases. The families in clan MA are united by the presence of an HEXXH motif in which the two His residues are Zn ligands and the Glu has a catalytic function (Rawlings et al., 2016). Importantly, phylogenetic analysis showed that PGM48 is part of a clade within the M48 family, here named M48D, with proteins only present in photosynthetic organisms, including cyanobacteria, algae, and higher plants. Moreover, unlike M48 homologs in the other three clades, members of the M48D clade have no predicted transmembrane domain, further indicating that this subfamily represents a specific adaptation (invention) of photosynthetic organisms. The lack of TMDs is consistent with the complete lack of TMDs in other PG-localized proteins (Lundquist et al., 2012a) and is logical given that PG are monolayer particles. In addition to PGM48 in Arabidopsis PG, we also identified the maize PGM48 homolog in isolated maize PG (Huang et al., 2013); we predict that PGM48 homologs in other plant species, as well as algae, are also located in PG. The phylogeny suggests that plastid PGM48 homologs originate from the cyanobacterial endosymbiont. Most cyanobacteria have both PGM48 (clade M48D) and HTPX homologs (clade M48B), suggesting that the PGM48 members originated from gene duplication of HTPX and obtained a specialized function in photosynthetic organisms. The primitive lycopod S. moellendorffii and moss P. patens lack PGM48, but each possesses a member of the M48A and C clades, indicating independent gene loss in these species, either because these species don’t have PGs or because they developed alternative strategies to carry out PGM48 functions. P. patens chloroplasts can accumulate lipid droplets under stress conditions, but it is not clear if these are indeed PGs (Wang et al., 2009). There are several reports of PGs in cyanobacteria (often named lipid particles or lipid droplets in the cyanobacterial literature) (van de Meene et al., 2006; Peramuna and Summers, 2014), but their proteome composition is not known. High-resolution three-dimensional reconstruction of the cyanobacterium Synechocystis sp PCC 6803 showed that lipid particles were abundant and that their distribution was restricted to thylakoids (van de Meene et al., 2006), similar as in Synechococcus sp PCC 7002 (Nierzwicki-Bauer et al., 1983). Their intracellular location suggests a role in thylakoid maintenance or thylakoid biogenesis, similar as for plastid PGs. The PGM48 homolog in Synechocystis sp PCC 6803 (sll1280) was identified by mass spectrometry (Wegener et al., 2010; Liberton et al., 2016). Experimental studies on PGM48 in cyanobacteria may help further determine why the M48D clade evolved in photosynthetic organisms.
PGM48 Substrate Selection, Interaction, and Cleavage
The PGM48 homology model suggests that the protein in its native state has a hydrophobic surface suitable for interaction with the PG monolayer. The pore providing access to the active site in the cavity is facing the aqueous phase but is not far from the presumed lipid surface. This would make proteins that are part of the PG the most likely substrate candidates. It is so far unclear how a substrate(s) is recognized by PGM48, similar as it is unknown how substrates for members of the other M48 clades are selected (see below). In case of the in vitro substrate β-casein, cleavage by PGM48 occurred at (or from) the C-terminal end. β-Casein has very little native structure (it is classified as an intrinsically disorganized protein) and is highly sensitive to peptidase digestion (Oldfield and Dunker, 2014). The observation that in vitro cleavage of β-casein by recombinant PGM48 occurred at/from the C-terminal end suggests that this is the preferred confirmation of substrate presentation and cleavage. PGM48 and its homologs in the M48D clade likely have unique properties and substrates compared with the other clades, and its location in PG seems to exclude a general role in chloroplast proteostasis.
PGM48 Is Associated with Senescence and Control of Senescence and Stay-Green Mutants
Senescence induction depends on the developmental window. Following cell proliferation and expansion (phase I), the leaf matures and becomes competent for external signals that induce senescence (phase II). That competence increases with leaf age, and in phase III, this will result in the initiation of developmental senescence, independent of external factors (Schippers, 2015; Schippers et al., 2015). A transcriptional network drives senescence in stage III and also involves several plant hormones as positive regulators (ethylene, abscisic acid [ABA], jasmonic acid [JA], and salicylic acid) or negative regulators (brassinosteroids and cytokinin). The impact of these hormones on senescence has been demonstrated though manipulation of transcription factors, signaling/receptor components, or hormone biosynthetic pathways. For example, overexpression of the transcription factor EIN3 causes early senescence (Chen and Bleecker, 1995), whereas ethylene insensitive mutants ein1 and ein2 show delayed senescence (Li et al., 2013). Similarly, manipulation of cytokinin biosynthesis can delay the onset of senescence (Gan and Amasino, 1995) because cytokinin increases sink strength of the tissue (Schippers et al., 2015). During leaf senescence, thylakoid membranes and their protein complexes together with associated cofactors are dismantled in a controlled fashion (Hörtensteiner, 2009; Hörtensteiner and Kräutler, 2011; Besagni and Kessler, 2013; Kusaba et al., 2013; Avila-Ospina et al., 2014; Ishida et al., 2014). Transmission and scanning electron microscopy, documenting dramatic increase of PG volume during this natural senescence process (Lichtenthaler, 1968; Tuquet and Newman, 1980; Tevini and Steinmüller, 1985; Ghosh et al., 1994; Guiamet et al., 1999; van Wijk and Kessler, 2016). The experimental data presented in this study clearly show that over- or underexpression of PGM48 result in respectively an acceleration or delay of natural leaf senescence. One can therefore postulate that PGM48 (1) removes a negative regulator (e.g., associated with brassinosteroid or cytokinin pathways), or (2) results in activation (or increased levels) of positive senescence regulators or signals (e.g., associated with ethylene, ABA, JA, or salicylic acid). However, PGM48 exclusively accumulates in PG, which narrows the immediate substrate pool to the PG proteome or proteins that transiently interact with the PG and eliminates direct effects on nuclear transcription factors or extraplastidic enzymes.
Hypothetical Functional Models for PGM48 Action and Role in Senescence
The early PGM48 overexpression and late underexpression senescence phenotypes indicate that M48 degrades or inactivates a protein that is either an (indirect) repressor of senescence or activates a protein (indirectly) required for senescence by partial cleavage. The metabolite composition of leaf PG isolated from four stages of natural senescence in beech leaves was quantitatively (on weight basis) compared (Tevini and Steinmüller, 1985). In particular, prenylquinones and free fatty acids, but not glycolipids or proteins, accumulated in PG during the senescence process (Tevini and Steinmüller, 1985, and references therein). Interestingly, the PG content of carotenoids and carotenoid esters was very low in green leaves, but increased in early stages of senescence (while total leaf carotenoid content decreases), and then decreased to low levels in more advanced stages of senescence (Tevini and Steinmüller, 1985). Thus, the PG participates in the controlled removal of carotenoids from the thylakoid membrane during the senescence process. The mechanism by which the carotenoid content decreases in PGs is unknown.
Based on our current observations and information from published literature, we propose a functional model that explains how PGM48 affects leaf senescence (Figure 9A). The model proposes that PGM48 degrades or partially cleaves one or more PG-localized or transiently associated PG protein, resulting in modified metabolism and/or metabolite content, thereby accelerating leaf senescence through an unknown retrograde signaling pathway. Possible signals for induction or acceleration of senescence are ABA or JA. Currently, there is no direct evidence for PGM48 substrates, but based on the data presented in this study and insight from the literature, we suggest that CCD4 is a good candidate.
Figure 9.
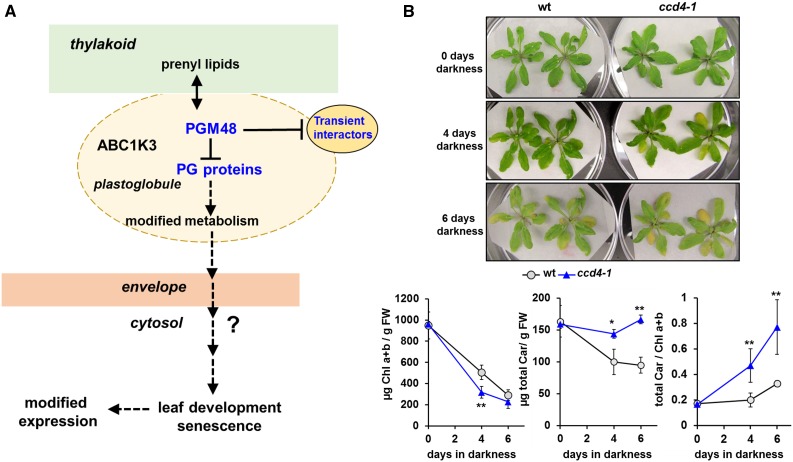
Model for PGM48 Function in Senescing Chloroplasts and Phenotype of the CCD4 Null Mutant during Dark-Induced Senescence.
(A) This model proposes that PGM48 degrades or partially cleaves one of more PG-localized proteins, resulting in modified metabolism and/or metabolite content, thereby accelerating leaf senescence through an unknown retrograde signaling pathway. Possible signals for induction or acceleration of senescence are ABA or JA. So far, no direct substrates for PGM48 have been identified, but we suggest CCD4 as a strong candidate (see Discussion).
(B) The Arabidopsis ccd4 T-DNA insertion null mutant (SALK_097984) shows accelerated dark-induced leaf senescence after 4 d darkness compared with the wild type as evidenced by accelerated chlorophyll degradation, whereas degradation of carotenoids is delayed (lower panels). Throughout dark-induced senescence, but not prior to darkness, carotenoid-to-chlorophyll ratios are higher in the ccd4 mutant, consistent with its role in carotenoid degradation (Gonzalez-Jorge et al., 2013). Student’s t test (*P < 0.1 and **P < 0.05).
Degradation of CCD4 by PGM48 is supported by the fact that (1) both PGM48 and CCD4 locate exclusively to PG and (2) yeast two-hybrid and in vitro protein interaction data suggest physical interaction between PGM48 and CCD4. The comparison of mRNA accumulation patterns for all genes encoding for PG proteins shows a strikingly similar high induction of CCD4 and PGM48, in particular compared with all other PG genes (Figure 6; Supplemental Figure 6); whereas the latter observation constitutes no direct proof that PGM48 degrades CCD4, it does fit the pattern. The very clear observations of the loss of CCD4 protein within the PG during senescence will require proteolytic activity, and PGM48 is an excellent candidate, also because it is the only protease in PG. Recombinant PGM48 did not degrade recombinant GST-CCD4 in vitro (data not shown), but this does not exclude in vivo degradation, perhaps because degradation would require a monolayer lipid interface and/or ABC1K3 dependent activation of PGM48 or phosphorylation of CCD4.
CCD4 as a target for PGM48 is consistent with the observed phenotype for the PGM48 OE and RNAi lines. The Arabidopsis ccd4 T-DNA insertion null mutant (SALK_097984) shows accelerated dark-induced leaf senescence after 4 d of darkness compared with the wild type, as evidenced by accelerated chlorophyll degradation and delayed degradation of carotenoids (Figure 9B). Throughout dark-induced senescence, but not prior to darkness, carotenoid-to-chlorophyll ratios are higher in the ccd4 mutant, consistent with its role in carotenoid degradation (Gonzalez-Jorge et al., 2013). Consistent with this, carotenoid-to-chlorophyll ratios also increased in the PGM48 overexpression line during senescence compatible with reduced CCD4 activity. CCD4 is known to function in degradation of carotenoids resulting in production of various volatile apocarotenoids and not only reducing carotenoid content (Gonzalez-Jorge et al., 2013) but also affecting leaf development though as yet uncharacterized signal transduction pathways (Avendaño-Vázquez et al., 2014; Hou et al., 2016). PGM48-driven degradation of CCD4 would reduce production of these apocarotenoid signals and may also result in increased flux into the downstream ABA biosynthetic pathway; an increase in ABA levels would stimulate leaf senescence. It remains to be determined if PGM48-driven degradation of CCD4 is indeed the primary cause of the senescence phenotypes observed. Future research is needed to obtain direct proof for PGM48 substrates, but identifying such direct substrate-protease relationships is notoriously challenging for many proteases.
METHODS
Plant Materials and Growth Conditions
T-DNA insertions lines pgm48-1 (SALK_082409) and pgm48-2 (GABI_324A06) were obtained from the ABRC and European Arabidopsis stock center (NASC), respectively. Plants were grown in a growth chamber with long-day conditions (18 h light/6 h dark) and temperature at 22°C with 130 µmol photons m−2 s−1 light intensity. Light sources are described by Bhuiyan et al. (2015).
Phylogenetic Analysis and mRNA Expression Analysis
To generate a phylogenetic cladogram, 88 M48 proteins from 39 species across the tree of life were aligned using MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). The alignment is available as Supplemental File 1. The aligned sequences were exported in Clustal format and viewed in Jalview (www.jalview.org/). Sequences were then converted in PHYLIP format, and phylogenetic trees were generated (1000 iterations) using the CIPRES web portal (http://www.phylo.org/) selecting the tool “RAxMLHPC blackbox.” The resulting phylogenetic tree was annotated in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). RAxML bootstrap support values are shown at the nodes of the tree. The in silico mRNA expression Arabidopsis thaliana tissue-specific expression profile was derived from the e-FP browser (http://bar.utoronto.ca/ExpressionAngler/).
Generation of Anti-PGM48 Antiserum
The nucleotide sequence encoding amino acids 72 to 325 of PGM48 were amplified by PCR. The resulting DNA fragment was ligated into restriction sites (BamHI and XhoI) of the C-terminal His affinity tag of the pET21a expression vector. BL21 Escherichia coli cells were transformed by pET21a vector harboring this truncated PGM48 gene, and cells were harvested from liquid culture after addition of 1 mM IPTG for 3 h incubation at 22°C. The overexpressed proteins were solubilized in 200 mM NaCl, 50 mM Tris, and 8 M urea at pH 8.0 and purified on a Ni-NTA resin matrix. A polyclonal antibody against this truncated PGM48 protein was raised in rabbits by injecting purified antigen (Alfa Diagnostic International). Antisera were affinity purified against the same antigen coupled to a HiTrap N-hydroxysuccinimide (NHS) ester-activated column (GE Healthcare Life Science).
T-DNA Insertion Mutants, Genotyping, and RT-PCR
T-DNA insertion lines pgm48-1 and pgm48-2 were identified by genotyping, and insertion was confirmed by DNA sequencing. Details on genotyping primers are listed in Supplemental Table 3. RNA was collected from homozygous plants and RT-PCR was performed using two different primers set. For transcript analysis, total RNA was extracted from Arabidopsis leaves using the RNeasy plant mini kit (Qiagen). RNA was reverse transcribed with random hexamer primers using Superscript III reverse transcriptase from Invitrogen. mRNA was normalized by ACTIN2, and the PCR condition was 28 cycles at 94°C for 2 min, 55°C for 30 s, and 72°C for 1 min. A complete list of primers can be found in Supplemental Table 3.
Generation of Constitutive Overexpression and RNAi Lines
To generate transgenic plants overexpressing PGM48 (OE lines), a full-length PGM48 cDNA was cloned using forward and reverse primers. For a complete list of primers, see Supplemental Table 3. A nucleotide sequence encoding the StrepII tag was added in the reverse primer before the stop codon. PCR products were cloned into pCR8 Topo vector (Invitrogen) and verified by DNA sequencing. This clone was ligated into gateway pEARLYGATE100 vector (ABRC stock center) using LR enzymes (Invitrogen). To suppress PGM48 gene expression through RNAi (RNAi lines), a partial cDNA (nucleotides 1 to 304) was cloned into pCR8 Topo vector, verified by DNA sequencing, and cloned into the pRNAi-GG vector containing the 2x35S promoter (from the ABRC; Yan et al., 2012). These vectors were transformed into Agrobacterium tumefaciens and subsequently transformed into wild-type Arabidopsis by the floral dip method.
Antibiotic Selection, Genotyping, and RT-PCR
Transgenic plants were selected using plates containing BASTA (for pEARLYGATE100 vector) and kanamycin (pRNAi-GG vector). Plants surviving on selective medium were genotyped and confirmed the presence of the transgene. They were then transferred to soil for seed production. For transcript analysis, total RNA was extracted from Arabidopsis leaves using the RNeasy plant mini kit. RNA was reverse transcribed with random hexamer primers using Superscript III reverse transcriptase from Invitrogen. mRNA was normalized to ACTIN2, and the PCR conditions were 20 or 22 cycles at 94°C for 2 min, 55°C for 30 s, and 72°C for 1 min. A complete list of primers can be found in Supplemental Table 3.
Pigment Concentrations
Chlorophyll and carotenoid concentrations were determined by absorbance spectrometry after extraction in 80% acetone (Porra et al., 1989). For chlorophyll and carotenoid determination, we collected specific leaves (leaves 5 and 6) from OE, RNAi, and wild-type plants. In the case of the ccd4-1 mutant, we collected the leaves that turn yellow after 4 and 6 d of dark treatment, and the same leaf number was also collected from the wild type.
PG Isolation
The PG isolation method was from Ytterberg et al. (2006) and Lundquist et al. (2012a). For PG preparations from senescing leaves, plants were grown on soil under 120 µmol photons m−2 s−1 with an 18-h photoperiod for 35 d. Rosettes were harvested and homogenized in grinding buffer (50 mM HEPES-KOH, pH 8.0, 5 mM MgCl2, 100 mM sorbitol, 5 mM ascorbic acid, 5 mM reduced cysteine, and 0.05% [w/v] BSA). Homogenate was filtered through four layers of 20-μm Miracloth, and thylakoid membranes were pelleted by centrifugation for 6 min at 1800g. Thylakoid pellets were washed once in 4 volumes of grinding buffer and resuspended in Medium R (50 mM HEPES-KOH, pH 8.0, 5 mM MgCl2, and cocktail of peptidase inhibitors) containing 0.2 M sucrose. Resuspended thylakoids were sonicated 4 × 5 s at output power 23 W (model 100 sonic dismembrator; Fischer Scientific), returning the samples to ice between each sonication event. Sonicated samples were centrifuged for 30 min at 150,000g, and PGs were released from the thylakoid floated to the surface of the solution. PGs were removed and combined with Medium R with 0.7 M sucrose to achieve a sucrose concentration of 0.5 M, which was then overlaid with Medium R with 0.2 M sucrose and Medium R with no sucrose. The gradient was centrifuged 90 min, 150,000g. The resulting floating pad of PGs was removed, flash frozen in liquid N2, and stored at −80°C.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was performed as previously described (Bhuiyan et al., 2015). The coding sequence of mature PGM48 (amino acid 48 to 344) was cloned in pMetYC-DEST vector (Grefen et al., 2007) and used as bait for interaction studies. Different PG-localized proteins, chlorophyll catabolite enzymes, and other candidate proteins were cloned into pXN22-DEST or pNX32-DEST vector (Grefen et al., 2007) and used as prey for the interaction with bait proteins. Haploid yeast strains THY.AP4 and THY.AP5 were transformed by bait constructs and prey constructs, respectively. Bait plasmid contains Cub-PLV, and prey plasmid contains NubG. NubG moiety was fused to the N terminus of prey proteins. Yeast strains were purchased from the ABRC stock center. After mating THY.AP4 and THY.AP5, diploid cells were selected growing on synthetic medium lacking Try and Leu. Positive colonies were subjected for testing interaction. Interactions between bait and prey proteins were performed according to the protocol described (Grefen et al., 2007). Interactions were verified by growing yeast colonies on synthetic minimal medium lacking Ade, His, Try, Leu, Ura, and Met and by β-galactosidase (β-Gal) activity. Soluble NubG and Nub-WT were used as negative and positive controls, respectively.
PGM48 Site-Directed Mutagenesis and in Vitro Proteolytic Activity Assays
Mature PGM48 (without cTP) was cloned using forward (PGM48-M-FW) and reverse (PGM48-M-RV) primers. The forward primer contains BamHI, and the reverse primer contains SalI sites. The resulting PCR fragment was ligated into pCR8 Topo vector and confirmed by DNA sequencing. pCR8 vector harboring PGM48 gene was digested by BamHI and SalI restriction enzymes. The resulting DNA fragment was ligated into restriction sites (BamHI and SalI) of the C-terminal His affinity tag of the pET21a expression vector.
Two mutants, H191A and E192D of PGM48, were constructed using a PCR method. For the mutant H191A, the C-terminal part of mature protein was amplified from pCR8 plasmid harboring PGM48 gene using forward primer (PGM48H191A-FW) and reverse primers (PGM48-M-RV). Forward primer M48H191AFW contains the mutation site CAT (His) to GCT (Ala). The N-terminal part of mature protein was amplified using forward (PGM48-M-FW-) and reverse (PGM48H191A-RV) primers. Reverse primer PGM48H191A-RV contains the introduced site CAT (His) to GCT (Ala). The amplified two fragments were gel purified and mixed and used as template (1:1) for second-round PCR to amplify mature protein using the forward and reverse primer sets, PGM48-M-FW and PGM48-M-RV, respectively. Second mutant E192D was amplified the same way as H191A except for the different primer sets to introduce the mutation from GAA (Glu) to GAT (Asp). The PCR fragments were ligated into pCR8 Topo vector, and the mutations were confirmed by DNA sequencing. BL21 E. coli cells were transformed by pET21a vector harboring the various PGM48 constructs, and cells were harvested from liquid culture after addition of 1 mM IPTG for 3 h incubation at 22°C.
Overexpressed wild-type and mutants PGM48 were solubilized in 300 mM NaCl, 50 mM Tris, and 10% glycerol at pH 8.0 and purified on a Ni-NTA resin matrix. The purified protein was dialyzed using a dialysis cassette (slide-A-Lyzer; Thermo Scientific) against buffer 100 mM NaCl and 50 mM Tris and 10% glycerol. After dialysis, the protein was concentrated using microcon centrifugal filter units (Millipore). In vitro proteolytic activity was performed by incubating recombinant proteins with β-casein at 37°C for 3 h with or without various peptidase inhibitors. The reaction was stopped by adding 3% SDS and then followed by separation of the protein products by SDS-PAGE followed by staining with Coomassie blue or silver nitrate.
To determine cleavage site specificity, recombinant active PGM48 and inactive PGM48-H191A were incubated with β-casein for 3 h at 37°C, followed by desalting using C4 Ziptip (Millipore) using the manufacturer’s guidelines. The peptide mixture was directly infused into a LTQ-Orbitrap MS instrument at a flow rate of 0.3 μL per minute through a 15-µm fused silica emitter tip (New Objective). Samples were first analyzed using the Orbitrap detector (FT-MS scan) at 7.5k or 15k resolution over a 600 to 2000 m/z range. The AGC target was set to 1.e6, tube lens, and capillary voltages were 200 and 60 V, respectively. The needle voltage was 2000 V. MS/MS spectra were acquired either manually in Tune mode or by running a data-dependent acquisition method identical to that used for proteomics (see below) except without LC separation. As such, MS/MS spectra were acquired for peptides visible in the FT-MS spectra. These MS/MS spectra were searched against the Swiss-Prot proteome (January 2014 version, 542,258 sequences) using MASCOT v 2.4.1 (Matrix Science). For the database search, the MS and MS/MS tolerance was 3 ppm and 0.5 D, respectively; no enzyme was specified and oxidized Met and phosphorylated Ser/Thr were chosen as variable modifications.
In Vitro Protein-Protein Interactions
Mature PES1 and CCD4 were cloned into pCR8 Topo vector and ligated into the pGEX5-1 expression vector using EcoRI and XhoI sites. Recombinant proteins were expressed in E. coli BL21 cells by incubation at 22°C with IPTG for 5 h; solubilized proteins were purified using glutathione resin at 4°C. The purified proteins were dialyzed using a dialysis cassette (slide-A-Lyzer) against 100 mM NaCl and 50 mM Tris-HCl (pH 8) and 10% glycerol. After dialysis, proteins were concentrated using microcon centrifugal filter units. To analyze interactions between PGM48 and PES1 or CCD4, bait and prey proteins were incubated at room temperature for 2 to 3 h and then resin was added to the mixture and incubated for another 1 h. Resin was washed by washing buffer five times, and resin-bound protein was eluted by elution buffer (1.5% SDS, 50 mM Tris-HCl, pH 8, and glycerol 15%) by heating at 75°C for 5 min. Interaction was visualized by SDS-PAGE gel and staining with silver nitrate.
Proteomics, MS, and Display in PPDB and Submission to Public Data Repository
For protein identification and quantification, each gel lane was cut in consecutive gel slices, followed by in-gel digestion using trypsin and subsequent peptide extraction as described previously (Friso et al., 2011). Peptide extracts for each gel band were then analyzed by online nanoLC-MS/MS using an LTQ-Orbitrap (Thermo). Resulting spectral data were searched against the predicted Arabidopsis proteome (TAIR10), including a small set of typical contaminants and the decoy, as described by Nishimura et al. (2013). Only proteins with two or more matched spectra were considered. Protein abundances were quantified according to the number of matched adjusted MS/MS spectra (AdjSPC) as explained by Friso et al. (2011). MS-derived information, as well as annotation of protein name, location, and function for the identified proteins, can be found in the Plant Proteome Database (http://ppdb.tc.cornell.edu). MS-derived information, as well as annotation of protein name, location, and function for the identified proteins, can be found in the PPDB (http://ppdb.tc.cornell.edu/). The MS proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaíno et al., 2014) via the PRIDE partner repository (http://www.ebi.ac.uk/pride) with the data set identifiers PXD003684 and 10.6019/PXD003684.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CCD4, AT4G19170; PGM48, AT3G27110; ABC1K3, AT1G79600; PES1, AT1G54570; and ABC1K7, AT3G07700. Accession numbers for all genes/proteins mentioned in the text or figures in this article are listed in Supplemental Tables 1 and 2.
Supplemental Data
Supplemental Figure 1. The N terminus of Arabidopsis PGM48 and cTP predictions for Arabidopsis and homologs in other dicots and monocots.
Supplemental Figure 2. MS spectra of digests by active PGM48 and mutant inactive PGM48 (PGM48-H191A).
Supplemental Figure 3. Phylogenetic tree of clade M48D with PGM48 homologs.
Supplemental Figure 4. Characterization of T-DNA insertion mutants in PGM48 and PGM48 is a senescence-induced protein in Arabidopsis
Supplemental Figure 5. Additional phenotypic analysis of RNAi and OE lines.
Supplemental Figure 6. mRNA expression data from publicly available microarray data for leaves from the wild type (Col-0) for all genes encoding for the PG core proteome and for ZEP.
Supplemental Table 1. Mass spectrometry analysis of β-casein digests with recombinant wild-type PGM48.
Supplemental Table 2. M48 homologs in the four clades in species across the tree of life.
Supplemental Table 3. Primers used in this study.
Supplemental File 1. Sequence alignment used for the phylogenetic analysis displayed in Figure 2.
Supplemental Data Set 1. The relative accumulation levels for all observed PG core proteins of the wild type after 6 (bolting stage) and 12 (advanced senescence) weeks growth under short-day conditions (10 h light/14 h dark).
Supplemental Data Set 2. PG proteomes of wild-type, RNAi, and OE lines.
Supplementary Material
Acknowledgments
This research was supported by Grant MCB1021963 from the National Science Foundation to K.J.v.W. We thank Vinod Vijayakumar for help in the early stages of this project.
AUTHOR CONTRIBUTIONS
N.H.B., G.F., and E.R. designed and performed the experimental analysis. K.M. carried out coexpression analyses. G.F. and E.R. carried out the mass spectrometry analyses. K.J.v.W. oversaw the project and wrote the article with N.H.B.
Glossary
- PG
plastoglobuli
- MS/MS
tandem mass spectrometry
- ER
endoplasmic reticulum
- OE
overexpression
- ABA
abscisic acid
- JA
jasmonic acid
References
- Akiyama Y. (2009). Quality control of cytoplasmic membrane proteins in Escherichia coli. J. Biochem. 146: 449–454. [DOI] [PubMed] [Google Scholar]
- Albrecht-Borth V., Kauss D., Fan D., Hu Y., Collinge D., Marri S., Liebers M., Apel K., Pfannschmidt T., Chow W.S., Pogson B.J. (2013). A novel proteinase, SNOWY COTYLEDON4, is required for photosynthetic acclimation to higher light intensities in Arabidopsis. Plant Physiol. 163: 732–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T., Michaelis S., Schuldiner M. (2016). The protease Ste24 clears clogged translocons. Cell 164: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J.R. II, Frost E., Vidi P.A., Kessler F., Staehelin L.A. (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño-Vázquez A.O., Cordoba E., Llamas E., San Román C., Nisar N., De la Torre S., Ramos-Vega M., Gutiérrez-Nava M.D., Cazzonelli C.I., Pogson B.J., León P. (2014). An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26: 2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 65: 3799–3811. [DOI] [PubMed] [Google Scholar]
- Besagni C., Kessler F. (2013). A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 237: 463–470. [DOI] [PubMed] [Google Scholar]
- Bhuiyan N.H., Friso G., Poliakov A., Ponnala L., van Wijk K.J. (2015). MET1 is a thylakoid-associated TPR protein involved in photosystem II supercomplex formation and repair in Arabidopsis. Plant Cell 27: 262–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I., Fernandez M.R., Rahn J.J., Stackley K.D., Bestman J.E., Anandhan A., Franco R., Claypool S.M., Lewis R.E., Chan S.S., Khalimonchuk O. (2015). Metalloprotease OMA1 fine-tunes mitochondrial bioenergetic function and respiratory supercomplex stability. Sci. Rep. 5: 13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha K., Lavy M., Yalovsky S. (2002). The Arabidopsis AtSTE24 is a CAAX protease with broad substrate specificity. J. Biol. Chem. 277: 29856–29864. [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Lubetzky T.C., Yalovsky S. (2008). Functional analysis of Arabidopsis postprenylation CaaX processing enzymes and their function in subcellular protein targeting. Plant Physiol. 148: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión C.A., Costa M.L., Martínez D.E., Mohr C., Humbeck K., Guiamet J.J. (2013). In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J. Exp. Bot. 64: 4967–4980. [DOI] [PubMed] [Google Scholar]
- Chen Q.G., Bleecker A.B. (1995). Analysis of ethylene signal-transduction kinetics associated with seedling-growth response and chitinase induction in wild-type and mutant Arabidopsis. Plant Physiol. 108: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugeni Piller L., Besagni C., Ksas B., Rumeau D., Bréhélin C., Glauser G., Kessler F., Havaux M. (2011). Chloroplast lipid droplet type II NAD(P)H quinone oxidoreductase is essential for prenylquinone metabolism and vitamin K1 accumulation. Proc. Natl. Acad. Sci. USA 108: 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatihi A., Latimer S., Schmollinger S., Block A., Dussault P.H., Vermaas W.F., Merchant S.S., Basset G.J. (2015). A dedicated type II NADPH dehydrogenase performs the penultimate step in the biosynthesis of vitamin K1 in Synechocystis and Arabidopsis. Plant Cell 27: 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., et al. (2010). AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics 9: 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkemeier I., Laxa M., Miguet L., Howden A.J., Sweetlove L.J. (2011). Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 155: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G., Olinares P.D.B., van Wijk K.J. (2011). The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. In Chloroplast Research in Arabidopsis, Jarvis R.P., ed (New York: Humana Press; ), pp. 265–282. [DOI] [PubMed] [Google Scholar]
- Gan S., Amasino R.M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988. [DOI] [PubMed] [Google Scholar]
- Gaude N., Brehelin C., Tischendorf G., Kessler F., Dormann P. (2007). Nitrogen deficiency in Arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. The Plant J. 49: 729–739. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Hudak K.A., Dumbroff E.B., Thompson J.E. (1994). Release of photosynthetic protein catabolites by blebbing from thylakoids. Plant Physiol. 106: 1547–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S., et al. (2013). Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Lalonde S., Obrdlik P. (2007). Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr. Protoc. Neurosci. 5: 27. [DOI] [PubMed] [Google Scholar]
- Guiamet J.J., Pichersky E., Nooden L.D. (1999). Mass exodus from senesceing soybean chloroplasts. Plant Cell Physiol. 40: 986–992. [Google Scholar]
- Gutierrez-Carbonell E., Takahashi D., Lattanzio G., Rodríguez-Celma J., Kehr J., Soll J., Philippar K., Uemura M., Abadía J., López-Millán A.F. (2014). The distinct functional roles of the inner and outer chloroplast envelope of pea (Pisum sativum) as revealed by proteomic approaches. J. Proteome Res. 13: 2941–2953. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14: 155–162. [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Kräutler B. (2011). Chlorophyll breakdown in higher plants. Biochim. Biophys. Acta 1807: 977–988. [DOI] [PubMed] [Google Scholar]
- Hou X., Rivers J., León P., McQuinn R.P., Pogson B.J. (2016). Synthesis and function of apocarotenoid signals in plants. Trends Plant Sci. 21: 792–803. [DOI] [PubMed] [Google Scholar]
- Huang F.C., Molnár P., Schwab W. (2009). Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60: 3011–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Friso G., Nishimura K., Qu X., Olinares P.D., Majeran W., Sun Q., van Wijk K.J. (2013). Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J. Proteome Res. 12: 491–504. [DOI] [PubMed] [Google Scholar]
- Ishida H., Izumi M., Wada S., Makino A. (2014). Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 1837: 512–521. [DOI] [PubMed] [Google Scholar]
- Klodmann J., Senkler M., Rode C., Braun H.P. (2011). Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157: 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korwitz A., Merkwirth C., Richter-Dennerlein R., Tröder S.E., Sprenger H.G., Quirós P.M., López-Otín C., Rugarli E.I., Langer T. (2016). Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J. Cell Biol. 212: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Tanaka A., Tanaka R. (2013). Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117: 221–234. [DOI] [PubMed] [Google Scholar]
- Lee P.T., Hsu A.Y., Ha H.T., Clarke C.F. (1997). A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179: 1748–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberton M., Saha R., Jacobs J.M., Nguyen A.Y., Gritsenko M.A., Smith R.D., Koppenaal D.W., Pakrasi H.B. (2016). Global proteomic analysis reveals an exclusive role of thylakoid membranes in bioenergetics of a model cyanobacterium. Mol. Cell. Proteomics 15: 2021–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1968). Plastoglobuli and the fine structure of plastids. Endeavor 27: 144–149. [Google Scholar]
- Lippold F., vom Dorp K., Abraham M., Hölzl G., Wewer V., Yilmaz J.L., Lager I., Montandon C., Besagni C., Kessler F., Stymne S., Dörmann P. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Poliakov A., Bhuiyan N.H., Zybailov B., Sun Q., van Wijk K.J. (2012a). The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 158: 1172–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P., Poliakov A., Giacomelli L., Friso G., Appel M., McQuinn R.P., Krasnoff S.B., Rowland O., Ponnala L., Sun Q., van Wijk K.J. (2013). Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 25: 1818–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Davis J.I., van Wijk K.J. (2012b). ABC1K atypical kinases in plants: filling the organellar kinase void. Trends Plant Sci. 17: 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar-Shrestha K., Tamot B., Pratt E.P., Saitie S., Bräutigam A., Weber A.P., Hoffmann-Benning S. (2013). Comparative proteomics of chloroplasts envelopes from bundle sheath and mesophyll chloroplasts reveals novel membrane proteins with a possible role in c4-related metabolite fluxes and development. Front. Plant Sci. 4: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D.E., Costa M.L., Guiamet J.J. (2008). Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. (Stuttg.) 10 (suppl. 1): 15–22. [DOI] [PubMed] [Google Scholar]
- Mattoo A.K., Edelman M. (1987). Intramembrane translocation and posttranslational palmitoylation of the chloroplast 32-kDa herbicide-binding protein. Proc. Natl. Acad. Sci. USA 84: 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H., Soubannier V. (2010). Mitochondrial function: OMA1 and OPA1, the grandmasters of mitochondrial health. Curr. Biol. 20: R274–R276. [DOI] [PubMed] [Google Scholar]
- Miller J.D., Arteca R.N., Pell E.J. (1999). Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 120: 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierzwicki-Bauer S.A., Balkwill D.L., Stevens S.E. Jr (1983). Three-dimensional ultrastructure of a unicellular cyanobacterium. J. Cell Biol. 97: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Asakura Y., Friso G., Kim J., Oh S.H., Rutschow H., Ponnala L., van Wijk K.J. (2013). ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C.J., Dunker A.K. (2014). Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83: 553–584. [DOI] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.S., Martinez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41: 831–844. [DOI] [PubMed] [Google Scholar]
- Parmryd I., Andersson B., Dallner G. (1999). Protein prenylation in spinach chloroplasts. Proc. Natl. Acad. Sci. USA 96: 10074–10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmryd I., Shipton C.A., Swiezewska E., Dallner G., Andersson B. (1997). Chloroplastic prenylated proteins. FEBS Lett. 414: 527–531. [DOI] [PubMed] [Google Scholar]
- Peramuna A., Summers M.L. (2014). Composition and occurrence of lipid droplets in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 196: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S., Bergmuller E., Tropf S., Lemke R., Dormann P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99: 12495–12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Pryor E.E. Jr., Horanyi P.S., Clark K.M., Fedoriw N., Connelly S.M., Koszelak-Rosenblum M., Zhu G., Malkowski M.G., Wiener M.C., Dumont M.E. (2013). Structure of the integral membrane protein CAAX protease Ste24p. Science 339: 1600–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley A., et al. (2013). The structural basis of ZMPSTE24-dependent laminopathies. Science 339: 1604–1607. [DOI] [PubMed] [Google Scholar]
- Quiros P.M., Langer T., Lopez-Otin C. (2015). New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16: 345–359. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Finn R. (2016). Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44: D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottet S., Besagni C., Kessler F. (2015). The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim. Biophys. Acta 1847: 889–899. [DOI] [PubMed] [Google Scholar]
- Rowland E., Kim J., Bhuiyan N.H., van Wijk K.J. (2015). The Arabidopsis chloroplast stromal N-terminome: Complexities of amino-terminal protein maturation and stability. Plant Physiol. 169: 1881–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A., Rambla J.L., Santaella M., Gómez M.D., Orzaez D., Granell A., Gómez-Gómez L. (2008). Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J. Biol. Chem. 283: 24816–24825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoh M., Ito K., Akiyama Y. (2005). Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli. J. Biol. Chem. 280: 33305–33310. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Park S.Y., Paek N.C. (2015). The divergent roles of STAYGREEN (SGR) homologs in chlorophyll degradation. Mol. Cells 38: 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers J.H. (2015). Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 27: 77–83. [DOI] [PubMed] [Google Scholar]
- Schippers J.H., Schmidt R., Wagstaff C., Jing H.C. (2015). Living to die and dying to live: The survival strategy behind leaf senescence. Plant Physiol. 169: 914–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm S., Papasotiriou D.G., Ibrahim M., Leisegang M.S., Müller B., Schorge T., Karas M., Mirus O., Sommer M.S., Schleiff E. (2013). Defining the core proteome of the chloroplast envelope membranes. Front. Plant Sci. 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., McNellis T.W. (2011). Fibrillin protein function: the tip of the iceberg? Trends Plant Sci. 16: 432–441. [DOI] [PubMed] [Google Scholar]
- Tevini M., Steinmüller D. (1985). Composition and function of plastoglobuli: II. Lipid composition of leaves and plastoglobuli during beech leaf senescence. Planta 163: 91–96. [DOI] [PubMed] [Google Scholar]
- Tuquet C., Newman D.W. (1980). Aging and regreening in soybean cotyledons. 1 Ultrastructural changes in plastids and plastoglobuli. Cytobios 29: 43–59. [PubMed] [Google Scholar]
- van de Meene A.M., Hohmann-Marriott M.F., Vermaas W.F., Roberson R.W. (2006). The three-dimensional structure of the cyanobacterium Synechocystis sp. PCC 6803. Arch. Microbiol. 184: 259–270. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J. (2015). Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu. Rev. Plant Biol. 66: 75–111. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Kessler F. (2016). Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol., in press. [DOI] [PubMed] [Google Scholar]
- Vidi P.A., Kanwischer M., Baginsky S., Austin J.R., Csucs G., Dörmann P., Kessler F., Bréhélin C. (2006). Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281: 11225–11234. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Q., Yang P.F., Liu Z., Liu W.Z., Hu Y., Chen H., Kuang T.Y., Pei Z.M., Shen S.H., He Y.K. (2009). Exploring the mechanism of Physcomitrella patens desiccation tolerance through a proteomic strategy. Plant Physiol. 149: 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener K.M., Singh A.K., Jacobs J.M., Elvitigala T., Welsh E.A., Keren N., Gritsenko M.A., Ghosh B.K., Camp D.G. II, Smith R.D., Pakrasi H.B. (2010). Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteomics 9: 2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35: W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P., Shen W., Gao X., Li X., Zhou P., Duan J. (2012). High-throughput construction of intron-containing hairpin RNA vectors for RNAi in plants. PLoS One 7: e38186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg A.J., Peltier J.B., van Wijk K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbierzak A.M., Kanwischer M., Wille C., Vidi P.A., Giavalisco P., Lohmann A., Briesen I., Porfirova S., Bréhélin C., Kessler F., Dörmann P. (2009). Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 425: 389–399. [DOI] [PubMed] [Google Scholar]
- Zhu C., Bai C., Sanahuja G., Yuan D., Farré G., Naqvi S., Shi L., Capell T., Christou P. (2010). The regulation of carotenoid pigmentation in flowers. Arch. Biochem. Biophys. 504: 132–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.