Abstract
The N-methyl-D-aspartate (NMDA) receptor is a key target of ethanol action in the central nervous system. Alcohol inhibition of NMDA receptor function involves small clusters of residues in the third and fourth membrane-associated (M) domains. Previous results from this laboratory have shown that two adjacent positions in the M3 domain, F636 and F637, can powerfully regulate alcohol sensitivity and ion channel gating. In this study, we report that these positions interact with one another in the regulation of both NMDA receptor gating and alcohol action. Using dual mutant cycle analysis, we detected interactions among various substitution mutants at these positions with respect to regulation of glutamate EC50, steady-state to peak current ratios (Iss:Ip), mean open time, and ethanol IC50. This interaction apparently involves a balancing of forces on the M3 helix, such that the disruption of function due to a substitution at one position can be reversed by a similar substitution at the other position. For example, tryptophan substitution at F636 or F637 increased or decreased channel mean open time, respectively, but tryptophan substitution at both positions did not alter open time. Interestingly, the effects of a number of mutations on receptor kinetics and ethanol sensitivity appeared to depend upon subtle structural differences, such as those between the isomeric amino acids leucine and isoleucine, as they could not be explained on the basis of sidechain molecular volume or hydrophilicity.
1. Introduction
Ethyl alcohol, or ethanol, causes a range of behavioral effects including ataxia and impaired cognition, and is widely abused, contributing to over 5% of all diseases and injuries worldwide (World Health Organization, 2014). Although ethanol produces its effects in the brain by acting on multiple target proteins (Vengeliene et al., 2008), one of the most important of these targets is the NMDA receptor. The NMDA receptor is a glutamate-gated ion channel most commonly composed of GluN1 and GluN2(A–D) subunits, with allosteric sites for several endogenous modulators located in the extracellular N-terminal domain, and multiple sites for phosphorylation and interaction with intracellular proteins located in the C-terminal domain (Traynelis et al., 2010). Although NMDA receptor ethanol sensitivity can be modulated by sites in both the N-terminal and C-terminal domains (Smothers et al., 2013;Woodward, 2000), the primary site of ethanol action is not located in either of these regions, because removal of the C-terminus does not abolish or decrease ethanol inhibition (Peoples & Stewart, 2000), and the influence of sites in the N-terminus on ethanol sensitivity is subtle (Smothers et al., 2013). Furthermore, although glycine concentration can in some cases modulate ethanol inhibition, ethanol does not act at the agonist binding domains for glutamate or glycine (Woodward, 2000; Peoples, 2003). Ethanol appears to inhibit NMDA receptor function via specific amino acids in the M3 and M4 domains (Ronald et al., 2001; Ren et al., 2003b, 2007, 2012, 2013; Honse et al., 2004; Smothers & Woodward, 2006; Zhao et al., 2015) , as mutations at these positions powerfully regulate, and in some cases can essentially eliminate, ethanol sensitivity (Ren et al., 2012). The M3 domain forms the upper part of the ion channel lumen and is essential for gating of the ion channel (Kohda et al., 2000; Jones et al., 2002; Sobolevsky et al., 2002), and the M4 domain also influences gating (Ren et al., 2003a; Schorge & Colquhoun, 2003), likely by interacting with M3. Ethanol acts by altering NMDA receptor gating, by decreasing ion channel mean open time and opening frequency (Wright et al., 1996). Thus, alcohol inhibits the NMDA receptor by biasing the ion channel toward the closed state. We consider it likely that alcohol does so by binding in or near the clusters of alcohol-sensitive positions in the M3 and M4 domains, because mutations at these positions can profoundly alter alcohol sensitivity. For example, mutation of only two of these positions, one each in the GluN1 and GluN2A subunits, results in an ethanol IC50 value of over 1 M (Ren et al., 2012).
Of the alcohol-sensitive residues in M3 and M4 of GluN1/GluN2A NMDA receptors, mutations at two phenylalanines in the GluN2A M3 domain, F636 and F637, have the greatest effect on alcohol inhibition (Ren et al., 2012; Ren et al., 2013). Recent studies have shown that these positions and positions in the GluN1 M4 subunit interact to regulate both ion channel gating and alcohol sensitivity (Ren et al., 2012; Xu et al., 2015). In the present study we report that the side chains at positions 636 and 637 in M3 interact with one another in the regulation of ion channel kinetics and alcohol inhibition. In addition, for some substituents there is an apparent balancing effect between the two positions, such that the changes in ion channel gating and ethanol sensitivity caused by a substitution at one position can be reversed by an identical substitution at the adjacent position.
2. Materials and Methods
2.1. Materials
Ethanol (95%, prepared from grain) was obtained from Aaper Alcohol & Chemical Co. (Shelbyville, KY, USA), and all other drugs and chemicals were obtained from Sigma Chemical Co. (St. Louis, MO, USA).
2.2. Site-directed mutagenesis, cell culture, and transfection
Site-directed mutagenesis in plasmids containing GluN2A subunit cDNA was performed using the QuikChange II kit (Stratagene, La Jolla, CA, USA), and all mutants were verified by double-stranded DNA sequencing. Human embryonic kidney (HEK) 293 cells obtained from the American Type Culture Collection (Manassas, VA) were cultured as previously described (Ren et al., 2012), with minor modifications. Cells were cultured in an incubator at 37 °C in 5% CO2 in DMEM containing 10% heat-inactivated fetal bovine serum, Earle’s salts, non-essential amino acids, sodium pyruvate, and L-glutamine. Cells were allowed to grow to 50 – 90% confluence, and were suspended with EDTA-trypsin. Suspended cells were plated at low density in 35-mm poly-D-lysine coated dishes and transfected with cDNA for the GluN1 and wild-type or mutant GluN2A subunits and green fluorescent protein (pGreen Lantern; Invitrogen, Carlsbad, CA) at a ratio of 2:2:1 using a calcium phosphate transfection kit (Invitrogen). The NMDA receptor antagonists dl-2-amino-5-phosphonovaleric acid (APV), 200 μM, ketamine, 100 μM, or MgCl2, 1 mM, were added to the culture medium of each dish to protect cells from receptor-mediated excitotoxicity post-transfection. Recordings were made within 48 hours following transfection.
2.3. Electrophysiological recording
Transfected cells were extensively washed to remove NMDA receptor antagonists before use in experiments. Patch-clamp recording was performed at room temperature using an Axopatch 1D or Axopatch 200B (Molecular Devices, Sunnyvale, CA, USA) amplifier. For whole-cell recordings, patch pipettes pulled from thin-walled glass capillaries with open tip resistances of 1 – 6 MΩ were used; GΩ seals were formed and series resistances of 2 – 8 MΩ were compensated by 80%. For single-channel recordings, patch pipettes pulled from thick-walled glass capillaries with tip resistances of 8 – 20 MΩ were fire polished and coated with R6101 elastomer (Dow-Corning). Cells were voltage-clamped at −50 mV and superfused in an external recording solution containing (in mM): 150 NaCl, 5 KCl, 0.2 CaCl2, 10 HEPES, 10 glucose, and 10 sucrose (pH 7.4). Low calcium was used to minimize calcium-dependent inactivation (Zilberter et al., 1991). In glutamate concentration-response experiments, glycine, 50 μM, was added to the external solution to saturate the glycine binding site, EDTA, 10 μM, was added to minimize Zn2+ -dependent desensitization (Erreger and Traynelis, 2005), and cells were lifted off the surface of the dish to increase the speed of the solution exchange. Under these conditions 10 – 90% rise times for solution exchange are ~1.5 ms. The intracellular recording solution in whole-cell experiments contained (in mM): 140 CsCl, 2 Mg4ATP, 10 BAPTA, and 10 HEPES (pH 7.2). Solutions of agonists and ethanol were applied to cells using a stepper motor-driven solution exchange apparatus (Warner Instruments, Hamden, CT, USA) and 600 μm id square glass tubing.
Data from whole-cell recordings were filtered at 2 kHz (8-pole Bessel) and acquired at 5 kHz on a computer using a DigiData interface and pClamp software (Molecular Devices). Single-channel data were filtered at 10 kHz (8-pole Bessel) and acquired at 50 kHz.
2.4. Data analysis
In concentration-response experiments, IC50 or EC50 and n (slope factor) were calculated using the equation: , where y is the measured current amplitude, x is concentration, n is the slope factor, and Emax is the maximal current amplitude. A complete concentration-response curve was obtained from each cell, and log-transformed EC50 or IC50 values calculated from curve fits from individual cells containing mutant subunits were compared to those for wild-type subunits using one-way ANOVA followed by the Dunnett test. Linear relations of mean values of log EC50, log IC50, or maximal steady-state to peak current ratio (Iss:Ip) for the various mutant subunits were tested using linear regression analysis. All values are reported as the mean ± S.E.
Data from single-channel recordings were idealized using the segmentation K-means algorithm in the QUB software suite (Qin, 2004). Dwell time histograms were fitted with two or three exponential components using ChannelLab software and mean open times were obtained from the proportionally weighted averages of the individual components. Data were obtained from 5 – 22 patches for each GluN2A receptor mutant tested.
The apparent interaction free energies (ΔΔGINT) between amino acid side chains in dual mutants were calculated from natural logarithm-transformed values of glutamate EC50, mean open time, and ethanol IC50 values using the equation: , where R is the gas constant and T is the temperature in degrees Kelvin (Venkatachalan and Czajkowski, 2008). Mean values of ΔΔGINT ± S.E. were analyzed for statistically significant differences from zero energy using one-sample t tests with degrees of freedom df = NWT + NMUT1 + NMUT2 + NMUT1,MUT2 − 4, where NX was the number of cells used for wild-type and mutant receptors and S.E. was determined from propagated errors. Values for mutants containing single substitutions of alanine, isoleucine, or tryptophan at F636 or F637 are those reported previously (Ren et al., 2007, 2013).
3. Results
3.1. Effects of dual mutations at F636 and F637 in M3 of the GluN2A subunit on glutamate potency and desensitization
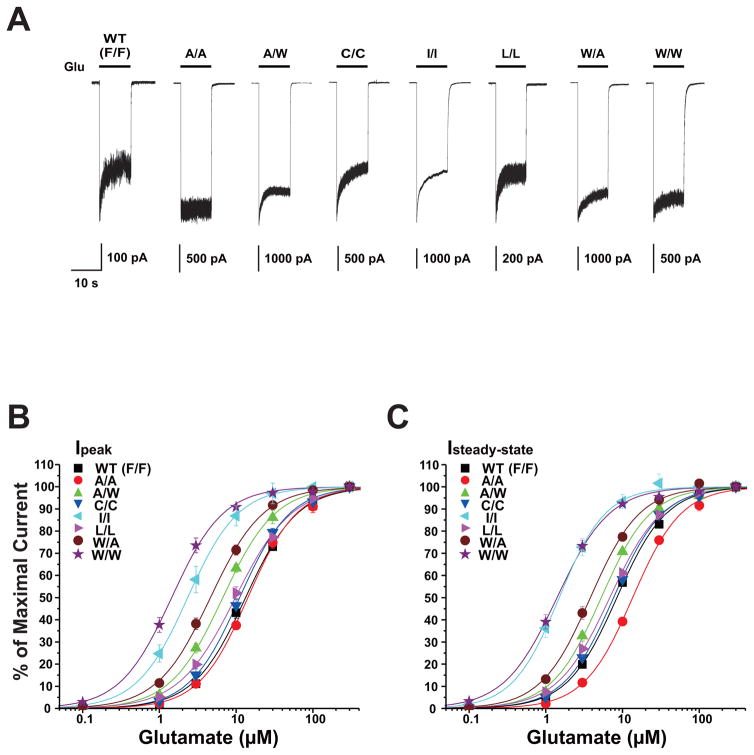
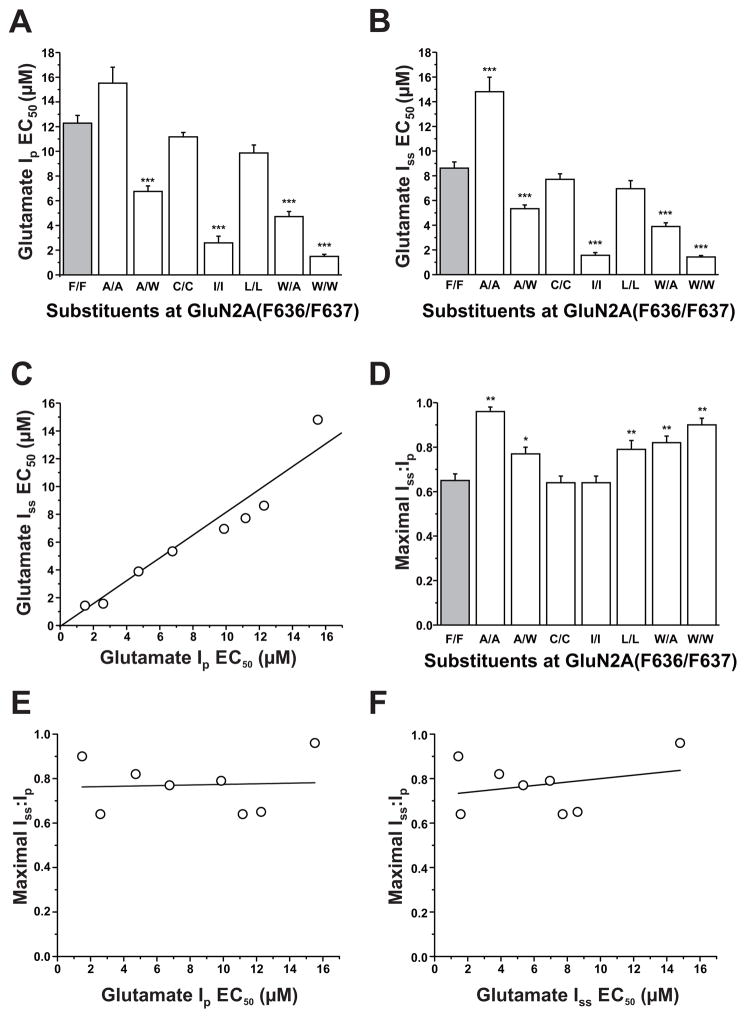
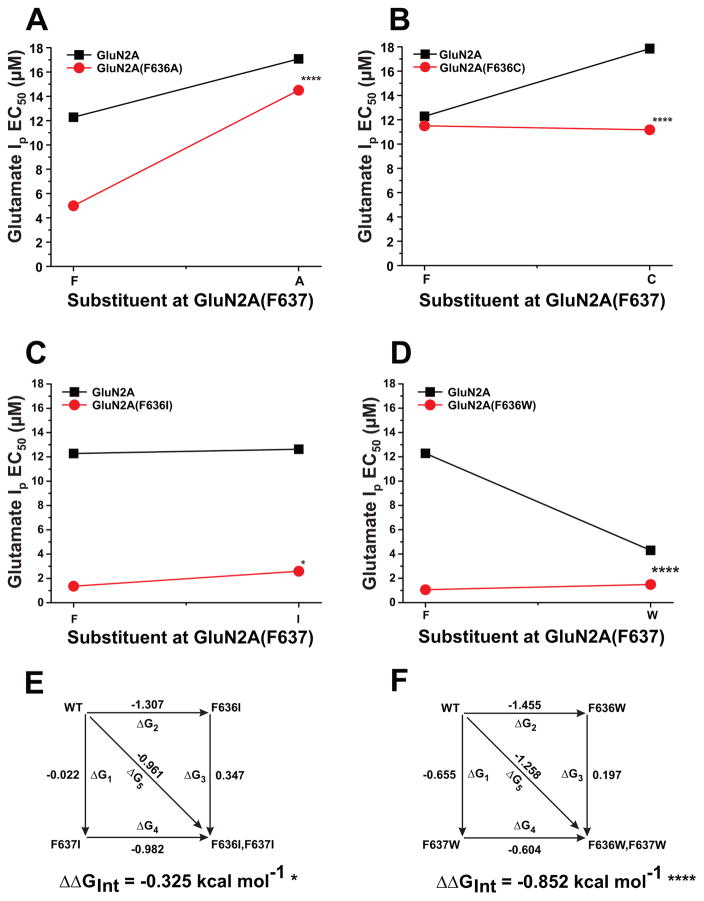
As we have previously reported, both F636 and F637 in the GluN2A subunit M3 domain regulate NMDA receptor function (Ren et al., 2007, 2012, 2013). To determine whether these two positions might interactively influence NMDA receptor kinetics, we constructed and tested a series of dual substitution mutants at these positions. All tested dual mutant combinations were functional, with no obvious differences in receptor kinetics other than changes in current amplitude (Table 1) and macroscopic desensitization in some cases (Fig. 2A). Concentration-response curves among the various mutant subunits were essentially parallel (Fig. 2B), as none of the mutations changed the Hill coefficient (p > 0.05; ANOVA, data not shown). A number of mutant subunit combinations shifted concentration-response curves for glutamate-activated peak and steady-state currents to either the right or left (Fig. 2B,C), altering EC50 values for glutamate peak (ANOVA; p < 0.0001; Fig. 3A) and steady-state currents (ANOVA; p < 0.0001; Fig. 3B). Four of the seven mutant subunits tested decreased glutamate peak and steady-state current EC50 values (ANOVA and Dunnett’s test; p < 0.05, p < 0.01; Fig. 3A,B), whereas the only increase observed was in the glutamate EC50 for steady-state current in the GluN2A(F636A/F637A) mutant. In previous studies, we found that single substitution mutations at either F636 or F637 in the M3 domain of the GluN2A subunit showed a strong correlation between peak and steady-state EC50 values (Ren et al., 2007, 2013). In the present study, we observed a similar correlation for dual mutants at these positions (R2 = 0.930, p < 0.0001; Fig. 3C).
Table 1.
Current amplitudes of GluN2A(F636/F637) mutants
| Substituent at F636/F637 | Ipeak (pA) | Isteady-state (pA) | n |
|---|---|---|---|
| F/F (WT) | 3580 ± 1010 | 2080 ± 538 | 13 |
| A/F | 2170 ± 683 | 2040 ± 630 | 5 |
| C/F | 2280 ± 840 | 1830 ± 617 | 6 |
| I/F | 947 ± 375*F/A, I/I | 865 ± 336*I/I | 6 |
| L/F | 1790 ± 805 | 1280 ± 578 | 5 |
| W/F | 893 ± 261 | 825 ± 234*I/I | 5 |
| F/A | 6090 ± 2120 | 2840 ± 860 | 6 |
| F/C | 1540 ± 425*F/A, I/I | 771 ± 226*C/C,**I/I | 9 |
| F/I | 1950 ± 835 | 711 ± 298*C/C,**I/I | 6 |
| F/W | 928 ± 333*F/A, I/I | 709 ± 262*C/C,**I/I | 6 |
| A/A | 1380 ± 452 | 1250 ± 362 | 6 |
| A/W | 4100 ± 1070 | 3040 ± 754 | 6 |
| C/C | 5610 ± 1150 | 3430 ± 622 | 7 |
| I/I | 6520 ± 1330 | 4100 ± 884 | 5 |
| L/L | 642 ± 182*C/C, F/A, I/I | 415 ± 105*C/C, **I/I | 6 |
| W/A | 3000 ± 1040 | 2100 ± 700 | 6 |
| W/W | 1010 ± 384*I/I | 886 ± 320*I/I | 6 |
The single-letter code F for phenylalanine is shown in italic typeface to denote that it is the native residue at either position. Asterisks indicate significant differences from the current amplitudes of the indicated mutants (ANOVA and Tukey/Kramer test; *p < 0.05, **p < 0.01). None of the current amplitudes differed from that of the wild-type receptor (ANOVA and Dunnett’s test; p > 0.05).
Fig. 2. Dual mutations at F636 and F637 in the M3 domain of the GluN2A subunit influence both peak and steady-state glutamate EC50 values.
A, Traces are currents activated by 300 μM glutamate in the presence of 50 μM glycine in lifted HEK 293 cells expressing wild-type GluN1/GluN2A (WT) or various GluN1/GluN2A(F636/F637) mutant subunits. Labels indicate mutations at 636/637 using single-letter amino acid codes. B–C, Concentration-response curves for glutamate-activated peak (Ip) and steady-state (Iss) currents in the presence of 50 μM glycine in lifted HEK 293 cells expressing wild-type GluN1/GluN2A (WT) or various GluN1/GluN2A(F636/F637) mutant subunits. Data points are the means ± S.E. of five to twelve cells, error bars not visible were smaller than the size of the symbols, and the curves shown are the best fits to the equation given in the “Materials and Methods.”
Fig. 3. Comparison of glutamate potency and desensitization among GluN2A F636/F637 dual mutant subunits.
A–B, Bar graphs show the average EC50 values for glutamate-activated peak (Ip, A) and steady-state (Iss, B) currents in lifted HEK 293 cells expressing GluN1 and wild- type GluN2A subunits (F/F, gray) or various GluN1/GluN2A(F636/F637) mutant subunits. Asterisks indicate EC50 values that differed from the wild-type value (***P < 0.001; one-way ANOVA). Results are the means ± S. E of 5–12 cells. C, Graph plots values of glutamate log EC50 for peak current in the series of mutants versus values of glutamate log EC50 for steady- state current; these values were significantly correlated (R2 = 0.93, P < 0.001). The line shown is the least-squares fit to the data. D, Bar graph shows the average values of maximal steady-state to peak current ratio (Iss:Ip) in lifted cells coexpressing GluN1 and wild-type GluN2A subunits (F/F, gray) or various GluN1/GluN2A(F636/F637) mutant subunits. Currents were activated by 300 μM glutamate in the presence of 50 μM glycine. Asterisks indicate values that differed significantly from the wild-type value (*P < 0.05, **P < 0.01; one-way ANOVA). Results are the means ± S. E of 5–12 cells.
Dual mutations at GluN2A(F636/F637) altered macroscopic desensitization as assessed by steady-state to peak current ratio (Iss:Ip; ANOVA, p < 0.0001; Fig. 3D). Five of seven dual mutants tested increased Iss:Ip values compared to that of the wild-type subunits (ANOVA and Dunnett’s test; p < 0.05 or p < 0.01; Fig. 3D), and the remaining mutants did not alter Iss:Ip values. Although both glutamate potency and apparent desensitization were changed by mutations at GluN2A(F636/F637), maximal Iss:Ip values were not correlated with either glutamate peak or steady-state EC50 values (r2 = 0.0030 and r2 = 0.076 for peak and steady-state current EC50, respectively; correlation analysis; p > 0.05; Fig. 3E–F).
3.2. Effects of dual mutations at GluN2A F636 and F637 on NMDA receptor mean open time
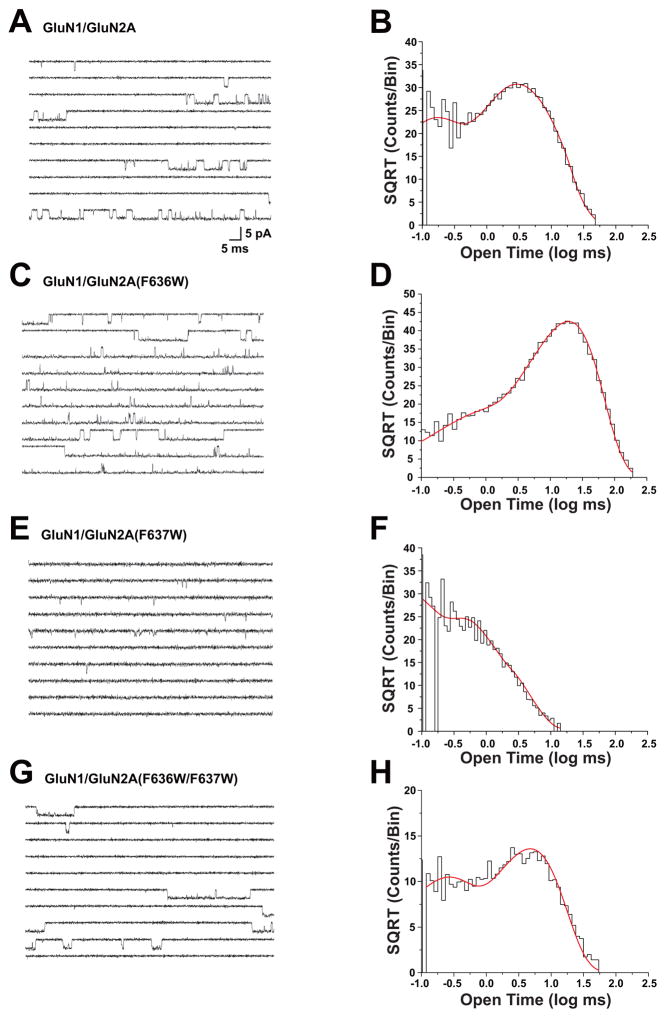
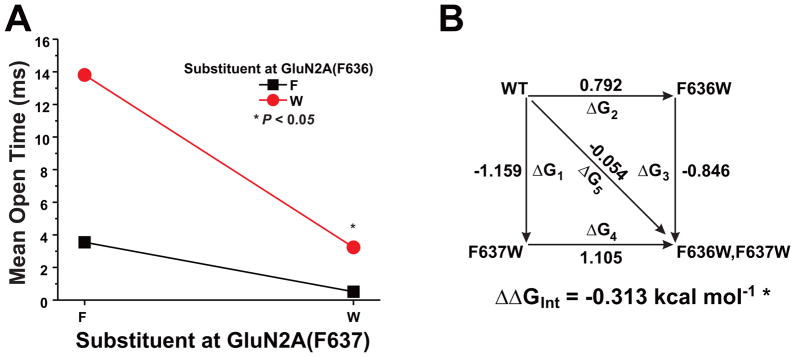
Previous studies from this laboratory have shown that both GluN2A(F636) and GluN2A(F637) regulate ion channel gating (Ren et al., 2007, 2013). To investigate whether dual mutations at these positions can interact to modulate mean open time, we tested the effect of selected mutants on mean open time by using single-channel recording in outside-out patches (Fig. 4 and Table 2). Open time distributions in cells expressing wild-type GluN1 and GluN2A subunits (Fig. 4A, B) were best fitted with two exponential components with average time constants of 0.10 ± 0.0077 ms (37%) and 4.5 ± 0.23 ms (63%; n = 22 patches), yielding a mean open time of 3.4 ± 0.21 ms. The open time distributions in cells expressing F636W mutant subunits (Fig. 4C, D) also were best fitted with two exponential components with average time constants of 0.27 ± 0.032 ms (11%) and 15 ± 1.7 ms (89%; n = 10 patches). The mean open time of 14 ± 1.6 ms in F636W mutant subunits was over four times that in wild-type receptors (ANOVA and Dunnett’s test, p < 0.001). Open time distributions for the GluN2A(F637W) mutation (Fig. 4E, F) were best fitted with two exponential components with average time constants of 0.12 ± 0.0094 ms (65%) and 0.97 ± 0.13 ms (35%; n = 9 patches). In contrast to GluN2A(F636W), the mean open time of 0.51 ± 0.055 ms in the GluN2A(F637W) subunit was less than one-sixth of the value in the wild-type subunit (ANOVA and Dunnett’s test; p < 0.001). The dual mutant GluN2A(F636W/F637W) subunit (Fig. 4 G, H) could be fitted with two time constant values of 0.17 ± 0.010 ms (39%) and 4.6 ± 0.38 ms (61%; n = 6 patches). Interestingly, mean open time in the dual tryptophan mutant subunit was 3.2 ± 0.36 ms (n = 6), which was not different from the value of 3.4 ± 0.21 ms (n = 22) in the wild-type GluN1/GluN2A receptor (ANOVA and Dunnett’s test, p > 0.05). Mutant subunits bearing dual isoleucine substitutions at these positions had decreased mean open times (Table 2; ANOVA; p < 0.01). Mean open time was also decreased in the dual alanine mutant GluN2A(F636A/F637A) as well as in the GluN2A(F636A/F637W) mutant (Table 2; ANOVA; p < 0.01), but was unchanged in GluN2A(F636W/F637A) subunits (Table 2; ANOVA; p > 0.05).
Fig. 4. Tryptophan mutations at F636 and F637 positions interactively regulate mean open time of the NMDA receptor.
Traces (A, C, E, G) and open time distributions (B, D, F, H) of single-channel currents activated by 100 nM -1 μM glutamate and 50 μM glycine in outside-out patches from cells expressing wild-type GluN1 subunits and GluN2A wild-type (A,B), F636W (C,D), F637W (E,F), or F636W/F637W (G,H) mutant subunits. Open time distributions were fitted with two exponential components; average values of τ and percentage for all patches are given in the text. Traces and corresponding open time distributions were from the same outside-out patches. Similar results were obtained in 5 to 21 additional patches for each subunit combination.
Table 2.
Mean open times in GluN2A(F636/F637) mutants
| Substituents at GluN2A(F636/F637) | Mean Open Time (ms) | N |
|---|---|---|
| WT (F/F) | 3.4 ± 0.21 | 22 |
| A/F | 3.1 ± 0.42 | 5 |
| F/A | 0.82 ± 0.084*** | 6 |
| A/A | 0.77 ± 0.034 *** | 5 |
| I/I | 1.4 ± 0.25** | 5 |
| W/F | 14 ± 1.6*** | 10 |
| F/W | 0.51 ± 0.055*** | 9 |
| W/W | 3.2 ± 0.36 | 6 |
| A/W | 0.81 ± 0.080*** | 5 |
| W/A | 3.2 ± 0.47 | 6 |
WT(F/F), wild-type. Values are the means ± S.E. obtained from 5 to 22 patches. Significant differences from values obtained for the wild-type receptor (ANOVA and Dunnett’s test; **p < 0.01, ***p < 0.001). Values for wild-type, A/F, and W/F receptors are from Ren et al. (2013).
3.3. Effects of dual mutations at GluN2A F636 and F637 on NMDA receptor ethanol sensitivity
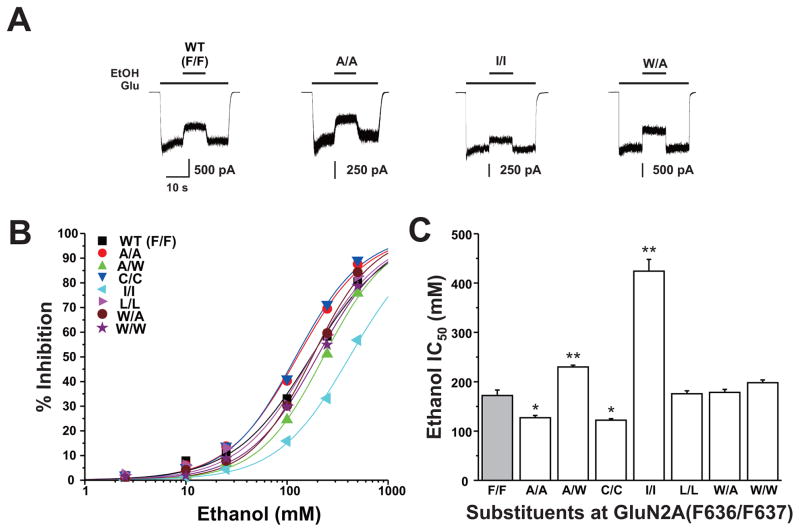
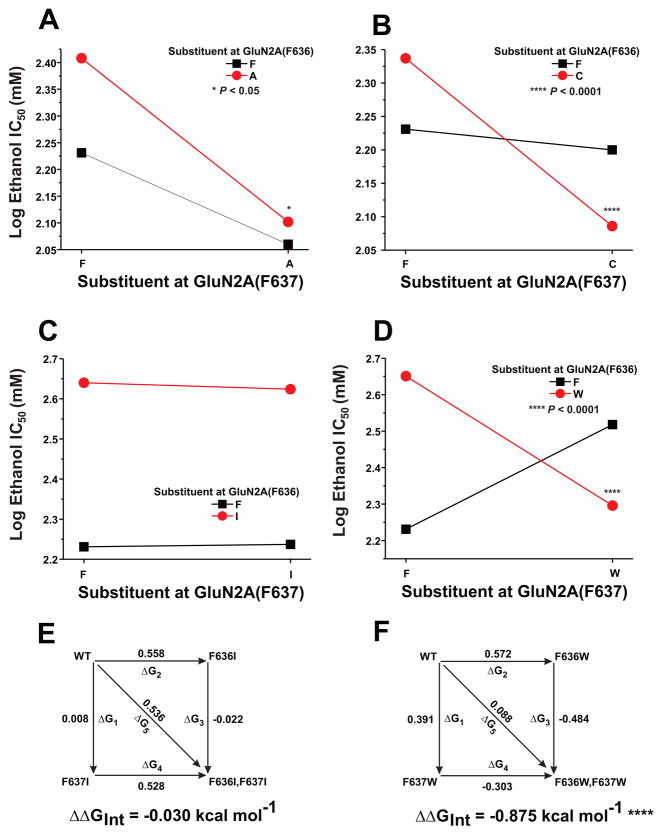
We have previously reported that four pairs of positions at the intersubunit interfaces of the M3 and M4 domains of the GluN1 and GluN2A subunits interactively regulate ethanol sensitivity (Ren et al, 2012). In these studies, we tested the effect of dual mutations at GluN2A F636 and F637 in regulating NMDA receptor ethanol inhibition using concentration-response analysis. All GluN2A mutants were inhibited by ethanol in a concentration-dependent manner (Fig. 5B). Ethanol sensitivity was altered in four of seven mutant subunits (Fig. 5C): IC50 values were decreased in the dual alanine and dual cysteine mutants (ANOVA and Dunnett’s test; p < 0.05), but were increased in the dual isoleucine and F636A/F637W mutants (ANOVA and Dunnett’s test; p < 0.01). Interestingly, ethanol sensitivity was unchanged in the dual leucine and tryptophan mutant subunits (ANOVA and Dunnett’s test; p > 0.05).
Fig. 5. Dual mutations at GluN2A(F636/F637) can alter NMDA receptor ethanol sensitivity.
A, Traces are currents activated by 10 μM glutamate and 50 μM glycine and their inhibition by 100 mM ethanol (EtOH) in cells expressing GluN1 and GluN2A wild-type (F/F) or 636/637 mutant subunits. B, Concentration-response curves for ethanol inhibition of glutamate-activated current in cells expressing various substitution mutations at GluN2A(F636/F637). Data are the means ± S.E. of 5–12 cells; error bars not visible were smaller than the size of the symbols. Curves shown are the best fits to the equation given in the Materials and Methods. The black curve shows the fit for the wild-type GluN2A subunit. C, Bar graph shows the average IC50 values for ethanol inhibition of glutamate-activated current in cells expressing GluN1 and wild-type GluN2A subunits (F/F) or GluN2A subunits containing various mutations at F636 and F637. Asterisks indicate IC50 values that are significantly different from the wild- type value (*P < 0.05; **P < 0.01; one-way ANOVA). Results are the means ± S. E of 5–12 cells. The wild-type value is from Ren et al. (2012).
3.4. Interactions between GluN2A F636 and F637 on NMDA receptor ion channel function and ethanol sensitivity
To determine whether GluN2A F636 and F637 could functionally interact to regulate NMDA receptor ion channel function and ethanol sensitivity, we performed two-way interaction analysis of variance and mutant cycle analysis on concentration-response and single-channel data among the substitution mutants at these two positions. Both two-way analysis of variance and mutant cycle analysis detected strong interactions between the positions in regulating glutamate peak EC50 values (Fig. 6, Table 3) for substitution of alanine, cysteine, or tryptophan (p < 0.0001), and an apparently weaker interaction for substitution of isoleucine (p < 0.05). Similar results were obtained for glutamate steady-state EC50 values (Table 3). GluN2A positions 636 and 637 also interacted with respect to regulation of mean open time in tryptophan substitution mutants (Fig. 7), as indicated by both types of analysis (p < 0.05). Although a majority of the mutant subunits altered values of Iss:Ip, an interaction between GluN2A 636 and 637 was detected only in the leucine mutants (Table 3). We also tested for the presence of functional interactions between the positions with respect to regulation of ethanol inhibition (Fig. 8; Table 3). Despite the marked decrease in ethanol sensitivity in the GluN2A(F636I) mutant, no functional interaction between positions 636 and 637 was detected for regulation of ethanol IC50 by isoleucine substitutions using either type of analysis (p > 0.05). In contrast, both two-way analysis of variance and mutant cycle analysis indicated functional interactions with respect to ethanol sensitivity for alanine mutants (p < 0.05), as well as cysteine and tryptophan mutants (p < 0.0001) at these positions.
Fig. 6. Positions F636 and F637 in the GluN2A subunit interactively regulate glutamate apparent potency.
A-D, Graphs plot Ip EC50 values for glutamate versus the substituent at F637 position for various mutants at position F636. Asterisks indicate that interaction free energy (ΔΔGINT) values are significantly different from zero energy (*, P < 0.05; ****, P < 0.0001). E–F, Mutant cycle analysis of glutamate Ip EC50 values in dual isoleucine and tryptophan mutations at GluN2A F636/F637. Apparent free energy values associated with the various mutations (ΔGx) are given in kcal mol−1. Values of ΔΔGINT are the means ± S. E. (*, P < 0.05; ****, P < 0.0001; ). The equation used to calculate ΔΔGINT is given in the Materials and Methods.
Table 3.
Mutant cycle analysis of glutamate Ip EC50, Iss EC50, Iss:Ip, and ethanol IC50
| Mutant (F636/F637) | Ip EC50 | Iss EC50 | Iss:Ip | Ethanol IC50 | ||||
|---|---|---|---|---|---|---|---|---|
| ΔΔGINT | df | ΔΔGINT | df | ΔΔGINT | df | ΔΔGINT | df | |
| A/A | 0.44 ± 0.094**** | 24 | 0.47 ± 0.10**** | 24 | 0.082 ±0.079 | 25 | −0.19 ± 0.071* | 23 |
| C/C | −0.24 ± 0.054**** | 27 | −0.32 ± 0.067**** | 27 | −0.13 ± 0.064 | 28 | −0.30 ± 0.056**** | 20 |
| I/I | 0.33 ± 0.14* | 24 | 0.42 ± 0.11** | 24 | −0.043 ± 0.062 | 24 | −0.030 ± 0.061 | 20 |
| L/L | 0.043 ± 0.089 | 25 | 0.055 ± 0.13 | 25 | 0.46 ± 0.081**** | 25 | −0.12 ± 0.054* | 23 |
| W/W | 0.85 ± 0.14**** | 25 | 0.89 ± 0.11**** | 25 | −0.091 ± 0.048 | 25 | −0.88 ± 0.065**** | 23 |
Ip: peak current; Iss: steady-state current; Iss/Ip: maximal peak to steady-state current ratio; ΔΔGINT : interaction free energy; df: degrees of freedom.
Values for GINT are means ± S.E in kcal mol−1. Asterisks indicate significant differences from zero energy (one sample t- test as described in the Materials and Methods; * p < 0.05, ** p <0.01, **** p < 0.0001). Values for wild-type and single mutations used in calculations are from Ren et al. (2007, 2013).
Fig. 7. Positions F636 and F637 in the GluN2A subunit interactively regulate ion channel mean open time.
A, Graph plots the mean open time values (in ms) versus the substituent at F637 for the substituent at position F636. Asterisks indicate that interaction free energy (ΔΔGINT) values are significantly different from zero energy (*, P < 0.05). B, Mutant cycle analysis of mean open time for dual tryptophan mutants at GluN2A F636/F637. Apparent free energy values associated with the various mutations (ΔGx) are given in kcal mol−1. Values of ΔΔGINT are the means ± S. E. (*, P < 0.05). The equation used to calculate ΔΔGINT is given in the Materials and Methods.
Fig. 8. Positions F636 and F637 in the GluN2A subunit interactively regulate ethanol sensitivity.
A-D, Graphs plot IC50 values for ethanol versus the substituent at F637 for various mutants at position F636. Asterisks indicate that interaction free energy (ΔΔGINT) values are significantly different from zero energy (*, P < 0.05; ****, P < 0.0001). E–F, Mutant cycle analysis of ethanol IC50 values in dual isoleucine and tryptophan mutations at GluN2A F636/F637. Apparent free energy values associated with the various mutations (ΔGx) are given in kcal mol−1. Values of ΔΔGINT are the means ± S. E. (****, P < 0.0001). The equation used to calculate ΔΔGINT is given in the Materials and Methods.
4. Discussion
Previous studies from this laboratory have found that two adjacent phenylalanines at positions 636 and 637 in M3 domain of the NMDA receptor GluN2A subunit can regulate ion channel gating and ethanol action (Ren et al., 2007, 2012, 2013). The results of the present study provide evidence that these residues can interact with each other to regulate receptor function, such that the side chain at one position alters the influence of the side chain at the adjacent position on ethanol sensitivity and ion channel gating.
We observed marked differences in maximal amplitudes of glutamate-activated current in the present study. The changes in current amplitude observed among mutants could arise from changes in open probability, as we have observed for a position in the M4 domain (Ren et al., 2003a). Of particular interest, the maximal glutamate-activated current in cells expressing the F636I/F637I mutant was ten-fold larger than that in cells expressing the F636L/F637L mutant. This difference, however, does not appear to be attributable to differences in ion channel gating. Although we do not have single-channel data for the F636L/F637L mutant, the current amplitude in cells expressing the wild-type channel was approximately half that of cells expressing the F636I/F637I mutant, whereas the mean open time (Table 2) and open probability (data not shown) of the wild-type channel were both approximately twice that of the F636I/F637I mutant. It thus appears most likely that the observed changes in current amplitude among the mutants in the present study are largely attributable to variations in receptor density due to factors such as transfection efficiency.
The majority of mutant combinations at GluN2A(F636/F637) tested altered glutamate EC50 values for both peak and steady-state current. We and others have previously shown that single mutations at either position can affect glutamate potency in the GluN2A subunit (Ren et al., 2007, 2012, 2013), and mutations at GluN1(F639), one of the corresponding positions in the GluN1 subunit, can affect glycine potency (Ronald et al., 2001; Smothers & Woodward, 2006). These changes in glutamate apparent affinity are attributable to changes in ion channel gating that reciprocally affect agonist binding (Colquhoun, 1998; Ren et al., 2007), as has been shown for mutations at other positions in or near the M3 domain in AMPA and NMDA receptors (Klein et al., 2004; Kohda et al., 2000; Yelshansky et al., 2004; Harms et al., 2014). As was the case for both of the individual positions, peak and steady-state EC50 values were highly correlated among dual mutants at F636/F637, suggesting that peak and steady-state current were similarly affected by the changes in gating produced by the mutations. Desensitization, as assessed using Iss:Ip, was altered in five of the seven dual mutants tested, but was not correlated with steady- state EC50 values. Furthermore, macroscopic desensitization in these five mutants was decreased, whereas glutamate affinity was increased in three of these five mutants. These results, taken together with the strong correlation between peak and steady-state glutamate EC50 values, suggests that increases in glutamate affinity in the dual mutants were not due to trapping of agonist in a desensitized state of the receptor, as was observed for single mutations at GluN2A(M823) in the M4 domain (Ren et al., 2003a). Similarly, changes in glutamate affinity observed in the dual mutants do not appear to be due to changes in mean open time, because glutamate potency was increased in GluN2A mutants in which mean open time was prolonged (F636W), shortened (F637W), or unchanged (F636W/F637W). The precise changes in ion channel gating responsible for the alterations in glutamate affinity are unclear at present, and could well differ for the two positions.
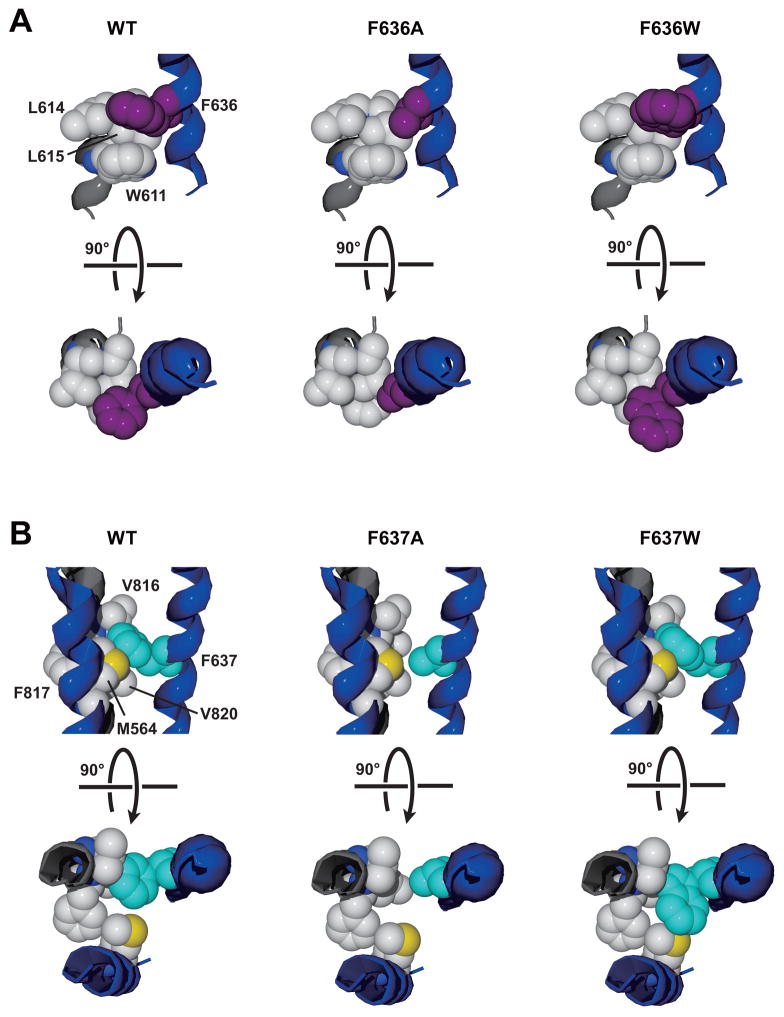
Although the changes in glutamate affinity that we observed among mutants at GluN2A positions F636 and F637 are likely due to altered ion channel gating, we also observed that these positions could interact in the regulation of mean open time, a direct measure of ion channel gating. Previous studies from this laboratory reported that substitution of various amino acids at either position could alter mean open time. For example, at GluN2A(F636), mean open time was unchanged by alanine substitution but increased over four-fold by tryptophan substitution (Ren et al., 2013), whereas at GluN2A(F637), mean open time was decreased significantly by either alanine or tryptophan substitution (Ren et al., 2007). The conformational changes associated with opening of the NMDA receptor ion channel pore are not known at present, but likely involve rotation and tilting of the M3 helices away from the central axis of the pore (Karakas & Furukawa, 2014; Kazi et al., 2013; Sobolevsky et al., 2009). Because mean open time of an ion channel reflects its closing rate (Hille, 2001), tryptophan substitution at GluN2A(F636) stabilizes, and alanine or tryptophan substitution at GluN2A(F637) destabilizes, the open state. Stabilization of the open state by GluN2A(F636W) may involve either strengthening of interactions that delay closing or weakening of interactions that promote closing, or both, while the reverse may be true of GluN2A(F637A) and GluN2A(F637W). A possible molecular basis for these observations is illustrated in figure 9. Based on the structure of the closed GluN1/GluN2B receptor (Karakas and Furukawa, 2014), the native phenylalanine at 636 is surrounded by hydrophobic side chains from the GluN1 subunit M2 domain: two leucine residues at 614–615, with which it may form hydrophobic interactions, and a tryptophan residue at 611, with which it may form an aromatic bond. Introduction of an alanine at this position would be predicted to decrease the strength of hydrophobic interactions and eliminate the aromatic bond, whereas introduction of a tryptophan, provided that its additional molecular volume could be accommodated, could strengthen these interactions. The changes in gating associated with these mutations may indicate that weakening the interactions at GluN2A position 636 does not influence open time, but strengthening of these interactions slows the closing rate. The native phenylalanine at 637 in GluN2A is surrounded by the side chains of a phenylalanine at 817 and two valine residues at 816 and 820 from the GluN1 M4 domain, as well as a methionine at 564 in the GluN2A M1 domain. An alanine substitution at 637 would likely greatly reduce the strength of hydrophobic interactions with the GluN1 M4 side chains, which may account for the decrease in mean open time. The apparently paradoxical observation that tryptophan substitution at 637, which would be predicted to increase the strength of hydrophobic interactions, also decreases mean open time may be explained by a steric clash with the methionine at 564. Although full resolution of the molecular mechanisms underlying these observations will likely require the solution to the structure of the M domains in the open state, further mutagenesis studies should be able to give important insights into this question.
Fig. 9. Altered interactions of phenylalanine side chains at GluN2A positions 636 and 637 with hydrophobic side chains from adjacent helices may explain effects of alanine and tryptophan mutations on mean open time.
A, The environment around the phenylalanine at GluN2A 636 (purple) includes two leucine residues at 614–615 and a tryptophan residue at 611 from the GluN1 M2 domain (left). Introduction of an alanine at 636 (center) may decrease the strength of hydrophobic and aromatic interactions, while introduction of a tryptophan (right) could strengthen these interactions. We propose that weakening the interactions at GluN2A position 636 does not influence open time, but strengthening these interactions slows the closing rate, prolonging open time. B, The environment around the phenylalanine at GluN2A 637 (cyan) includes a phenylalanine at 817 and two valine residues at 816 and 820 from the GluN1 M4 domain, and a methionine at 564 in the GluN2A M1 domain (left). Alanine substitution at 637 may diminish hydrophobic interactions with the GluN1 M4 side chains (center), decreasing mean open time. Tryptophan substitution at 637 (right) may decrease mean open time by introducing a steric clash with the methionine at GluN2A 564.
Experiments using multiple substituents revealed that the M3 domain positions GluN2A F636 and F637 interactively regulate glutamate potency and mean open time. Dual mutant cycle analysis and two-way interaction ANOVA revealed interactions between F636 and F637 regulating glutamate EC50 values following substitution of alanine, cysteine, isoleucine, or tryptophan into one or both positions. Although the uncertainty regarding the underlying mechanism by which these positions individually regulate glutamate affinity prevents a detailed analysis of the molecular mechanism for its interactive regulation, the observation that interactions could be detected by either decreasing the molecular volume at these positions by alanine, cysteine, or isoleucine substitution, or increasing molecular volume by tryptophan substitution, suggests a possible role for molecular volume of the substituent at these positions (Fig. 9). Interestingly, the interaction that appeared to be weakest was that for isoleucine, which represented the smallest change in molecular volume among the substituents tested compared to the native phenylalanine. We also observed interactions with respect to mean open time. Open time histograms were fitted with two components corresponding to short and long openings, and changes in mean open time in both the GluN2A(F636W) and GluN2A(F637W) mutants were attributable to changes in the relative proportions and durations of the two components. Tryptophan substitution at both positions, however, restored mean open time, as well as the durations and relative proportions of the two open time components, to the normal ranges for the wild-type NMDA receptor. Both mutant cycle analysis and two-way interaction ANOVA of mean open time values revealed an interaction between the positions, but the molecular mechanism underlying this interaction is unclear. Because GluN2A F636 and F637 are adjacent positions in the M3 domain alpha helix, the nature of this interaction differs from that of interactions we and others have reported between closely apposed side chains on separate M domains at intersubunit interfaces (Ren et al., 2012; Xu et al., 2015). In the latter instance, the distance between the interacting positions, as well as their orientation to one another, would change along with conformational changes associated with ion channel gating. For GluN2A F636 and F637, however, any functional interaction between the positions would be dependent upon additional bonds of one or both positions with nearby residues on other parts of the protein, as discussed above. The presence of these additional bonds appears likely, because positions in the M3 domain have been shown to interact with multiple positions in the M1 and M2 domains in non-NMDA glutamate receptors and NMDA receptors (Ogden and Traynelis, 2013; Lopez et al., 2013; Siegler Retchless et al., 2012; Xu et al., 2015; Wilding et al., 2014). We envision the interactions involving tryptophan as “balancing” this part of the M3 helix, such that the interactions of 636 and 637 with other side chains to alter channel closing rate effectively offset each other (Fig. 10A). For example, in the dual tryptophan mutant, the increased hydrophobic interactions at 636 may be opposed by the steric hindrance at 637. It should be noted that in this instance, the balancing of the M3 helix would be more complex than the simple two-dimensional lateral shift represented in Fig. 10. For alanine substitution mutants, however, the alteration of mean open times differs from tryptophan mutants. GluN2A(F637A) decreased mean open time (Ren et al., 2007), whereas GluN2A(F636A) did not alter mean open time (Ren et al., 2013) or reverse the effect of F637A on mean open time in the F636A/F637A dual mutant (Table 2). Furthermore, mutant cycle analysis and two-way ANOVA showed that alanine substitutions at these two positions do not interact. Similar results were obtained using tryptophan and alanine dual mutants: F636A/F637W decreased mean open time, but F636W/F637A did not, and these mutants did not interact to regulate mean open time (Table 2). These results likely indicate the inability of the smaller, aliphatic alanine side chains to form hydrophobic or aromatic bonds with other positions to exert the effects seen in phenylalanine and tryptophan mutants.
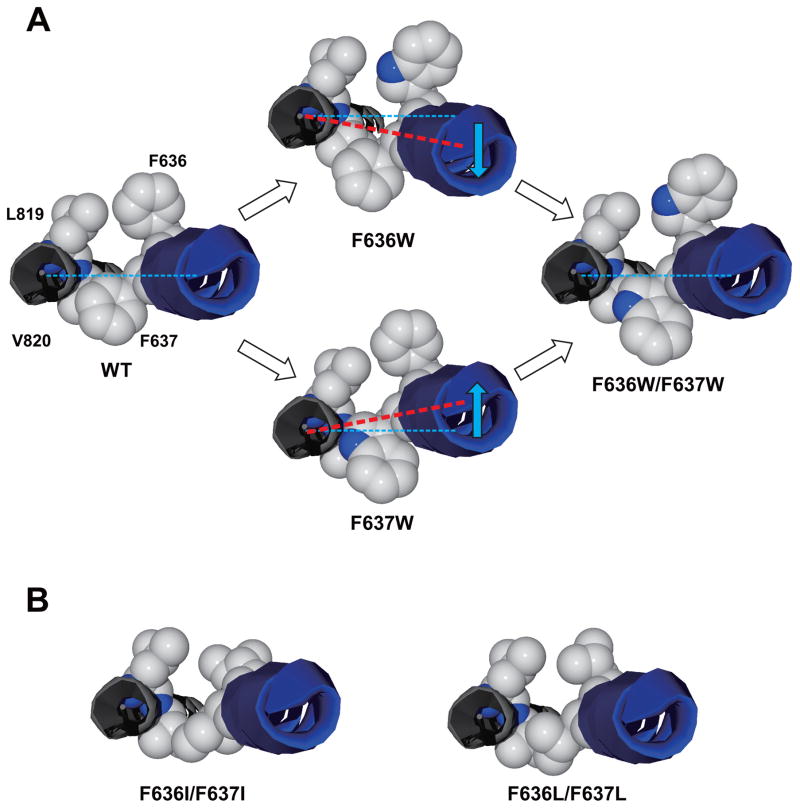
Fig. 10. A simple conceptual model for the effects of mutations at F636 and F637 in the GluN2A M3 domain.
A, The GluN1 M4 domain and GluN2A M3 domain helices are shown from the extracellular side; side chains of GluN1 residues M818 and L819 and GluN2A residues F636 and F637 are shown. In the wild-type ion channel (left), the normal balance of forces at this level of the M3 and M4 helices allows for normal gating. Tryptophan substitution at either position 636 or 637 (middle) could alter ion channel gating in opposite directions by shifting the M3 helix (blue arrows) in this region relative to the GluN1 M4 domain (or by altering the forces on M3 in this region) due to interactions with adjacent residues, whereas tryptophan substitution at both positions (right) may restore the balance, and with it, normal ion channel gating. The wild-type structure is that of the rat GluN1/GluN2B receptor from Karakas and Furukawa (2014). The differences in the relative positions of M3 and M4 in the single mutants were made by manually shifting the M3 helix relative to M4; these are for the purpose of illustration and are not intended to represent actual molecular distances. B, The model shown in A with dual isoleucine (left) or leucine (right) substitutions to illustrate the subtle differences in structure between these mutants.
Although the small number of substituent side chains that we tested in the present study precludes a detailed analysis of structural features that regulate ion channel gating, the differences observed between the effects of the isomeric substituents leucine and isoleucine suggest that subtle structural changes at these positions may nevertheless have important consequences. In a recent study, we found that isoleucine substitution at F636 did not significantly alter mean open time, whereas a leucine substitution at the same position decreased open time to less than 1 ms (Ren et al., 2013). In contrast, in an earlier study, isoleucine substitution at F637 decreased open time to approximately 1 ms (Ren et al., 2007). Our observation in the present study that the open time in the F636I/F637I mutant was 1.4 ms is consistent with a greater influence of the substituent at 637 in this mutant. The molecular mechanism that accounts for the observed differences between the leucine and isoleucine mutants is not clear, however. Isoleucine and leucine are identical in molecular volume and hydrophobicity, so the differences are not attributable to simple changes in volume occupation or hydrophobicity irrespective of side-chain structure. Multiple hydrophobic side chains from other M domains are adjacent to the phenylalanines at 636 and 637 (Karakas & Furukawa, 2014), which could allow for aromatic interactions with the phenylalanine side-chains (Mahadevi and Sastry, 2013). Either leucine or isoleucine substitution, however, would eliminate the phenyl ring of the phenylalanine side chain in the native protein, disrupting any aromatic interactions. Because the leucine side chain structure more closely resembles that of phenylalanine than does isoleucine, the effect of a leucine substitution on ion channel kinetics would be predicted to be less than that of an isoleucine substitution. That this prediction does not always hold indicates that the structural requirements are more stringent, but the precise explanation for this is not clear at present given the high degree of similarity between the isomers (Fig. 10B). Additional studies will be required to resolve the precise nature of these requirements.
Substitutions at GluN2A positions F636 and F637 also regulated ethanol sensitivity in an interactive manner: introduction of an alanine, cysteine, or tryptophan at either position altered the effect of substituting the same residue at the other position. Although there is no direct evidence that these positions form binding sites for ethanol, the ability of the positions to interactively and strongly regulate ethanol sensitivity is consistent with this view and provides the best evidence for this to date (Ren et al., 2012; Xu et al., 2015). Stronger evidence for this hypothesis awaits future studies demonstrating persistent alteration of NMDA receptor function by covalently-bound alcohol analogs or localization of ethanol or ethanol analogs to these positions using structural methods such as x-ray crystallography (Sauguet et al., 2013). As we observed for regulation of receptor gating, the effect of a single tryptophan substitution at either position could be reversed by substituting a second tryptophan at the other position. Unlike the opposing effects of tryptophan mutations at these positions on mean open time, however, ethanol sensitivity was markedly decreased in both the GluN2(F636W) and GluN2A(F637W) mutants, so that the restoration to normal ethanol sensitivity in the dual tryptophan mutant could not be explained by offsetting shifts in ethanol IC50 values. The effects of combining alanine and tryptophan substitutions on ethanol sensitivity were also unexpected: ethanol sensitivity was unchanged from the wild-type value in the F636W/F637W and F636W/F637A mutants, was decreased in the F636A/F637W mutant, but was increased in the dual alanine mutant. The underlying molecular events responsible for these observations are unclear at present, but may involve both a balancing of forces at this level of the alpha helix and an optimal molecular volume, with smaller side chain molecular volumes in a balanced helix (F636A/F637A, F636C/F637C) generally yielding higher ethanol sensitivities. The marked exception to this was the discrepancy observed between the leucine and isoleucine dual mutants: ethanol IC50 was unchanged in the F636L/F637L mutant, but was over two times greater in the F636I/F637I mutant. Although the magnitude of the difference in ethanol sensitivity between the mutants containing the isomeric amino acids leucine and isoleucine may be larger than expected, studies in both NMDA receptors and glycine receptors (Ye et al., 1998; Yamakura et al., 1999; Ren et al., 2013) have reported similar differences in ethanol sensitivity or agonist potency between leucine and isoleucine mutants at various positions. Because leucine and isoleucine are identical in molecular volume and hydrophobicity, we interpret their disparate effects on ethanol sensitivity as evidence for specific structural requirements of the substituent side chains.
In summary, the results of the present paper demonstrate that the alcohol-sensitive positions F636 and F637 in the M3 domain of the GluN2A subunit interactively regulate ethanol inhibition and NMDA receptor function. This regulation appears to involve counteracting forces on the M3 helix at the level of these positions due to additional molecular interactions with nearby side chains.
Fig. 1. Alignment of M3 domains in GluN1 and GluN2 subunits.
Sequences of the M3 domains in GluN1 and GluN2A-D subunits are shown, with positions F636 and F637 in the GluN2A subunit in bold type and underlined.
Acknowledgments
Funding: These studies were supported by grants AA015203-01A1 and AA015203-06A1 from the National Institutes of Health to R.W.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Colquhoun D. Binding, gating, affinity and efficacy. The interpretation of structure-activity relationships for agonists and of the effects of mutating receptors. Br J Pharmacol. 1998;125:924–47. doi: 10.1038/sj.bjp.0702164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF. Allosteric interaction between zinc and glutamate binding domains on NR2A causes desensitization of NMDA receptors. J Physiol. 2005;569:381–93. doi: 10.1113/jphysiol.2005.095497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms JE, Benveniste M, Kessler M, Stone LM, Arai AC, Partin KM. A charge-inverting mutation in the "linker" region of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors alters agonist binding and gating kinetics independently of allosteric modulators. J Biol Chem. 2014;289:10702–14. doi: 10.1074/jbc.M113.526921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. Ion Channels of Excitable Membranes. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- Honse Y, Ren H, Lipsky RH, Peoples RW. Sites in the fourth membrane-associated domain regulate alcohol sensitivity of the NMDA receptor. Neuropharmacol. 2004;46:647–54. doi: 10.1016/j.neuropharm.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Jones KS, VanDongen HMA, VanDongen AMJ. The NMDA receptor M3 segment is a conserved transduction element coupling ligand binding to channel opening. J Neurosci. 2002;22:2044–53. doi: 10.1523/JNEUROSCI.22-06-02044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–7. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi R, Gan Q, Talukder I, Markowitz M, Salussolia CL, Wollmuth LP. Asynchronous movements prior to pore opening in NMDA receptors. J Neurosci. 2013;33:12052–66. doi: 10.1523/JNEUROSCI.5780-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RM, Howe JR. Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J Neurosci. 2004;24:4941–51. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and δ2 receptor channel properties. Nat Neurosci. 2000;3:315–22. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- Lopez MN, Wilding TJ, Huettner JE. Q/R site interactions with the M3 helix in GluK2 kainate receptor channels revealed by thermodynamic mutant cycles. J Gen Physiol. 2013;142:225–39. doi: 10.1085/jgp.201311000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevi AS, Sastry GN. Cation-π interaction: its role and relevance in chemistry, biology, and material science. Chem Rev. 2013;113:2100–38. doi: 10.1021/cr300222d. [DOI] [PubMed] [Google Scholar]
- Ogden KK, Traynelis SF. Contribution of the M1 transmembrane helix and pre-M1 region to positive allosteric modulation and gating of N-methyl-D-aspartate receptors. Mol Pharmacol. 2013;83:1045–56. doi: 10.1124/mol.113.085209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples RW, Stewart RR. Alcohols inhibit N-methyl-D-aspartate receptors via a site exposed to the extracellular environment. Neuropharmacol. 2000;39:1681–91. doi: 10.1016/s0028-3908(00)00067-8. [DOI] [PubMed] [Google Scholar]
- Peoples RW. Alcohol Actions on Glutamate Receptors. In: Herman BH, Frankenheim J, Litten RZ, Sheridan PH, Weight FF, Zukin SR, editors. Glutamate and Addiction. Humana Press; Totowa, NJ: 2003. pp. 343–56. [Google Scholar]
- Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Honse Y, Karp BJ, Lipsky RH, Peoples RW. A site in the fourth membrane-associated domain of the N-methyl-D-aspartate receptor regulates desensitization and ion channel gating. J Biol Chem. 2003a;278:276–83. doi: 10.1074/jbc.M209486200. [DOI] [PubMed] [Google Scholar]
- Ren H, Honse Y, Peoples RW. A site of alcohol action in the fourth membrane-associated domain of the NMDA receptor. J Biol Chem. 2003b;278:48815–20. doi: 10.1074/jbc.M302097200. [DOI] [PubMed] [Google Scholar]
- Ren H, Salous AK, Paul JM, Lipsky RH, Peoples RW. Mutations at F637 in the NMDA receptor NR2A subunit M3 domain influence agonist potency, ion channel gating and alcohol action. Br J Pharmacol. 2007;151:749–57. doi: 10.1038/sj.bjp.0707254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao Y, Dwyer DS, Peoples RW. Interactions among positions in the third and fourth membrane-associated domains at the intersubunit interface of the N-methyl-D-aspartate receptor forming sites of alcohol action. J Biol Chem. 2012;287:27302–12. doi: 10.1074/jbc.M111.338921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao Y, Wu M, Peoples RW. A novel alcohol-sensitive position in the N-methyl-D-aspartate receptor GluN2A subunit M3 domain regulates agonist affinity and ion channel gating. Mol Pharmacol. 2013;84:501–10. doi: 10.1124/mol.113.085993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald KM, Mirshahi T, Woodward JJ. Ethanol inhibition of N-methyl-D-aspartate receptors is reduced by site- directed mutagenesis of a transmembrane domain phenylalanine residue. J Biol Chem. 2001;276:44729–35. doi: 10.1074/jbc.M102800200. [DOI] [PubMed] [Google Scholar]
- Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, Harris RA, Delarue M. Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun. 2013;4:1697. doi: 10.1038/ncomms2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–8. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegler Retchless B, Gao W, Johnson JW. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci. 2012;15:406–13. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers CT, Woodward JJ. Effects of amino acid substitutions in transmembrane domains of the NR1 subunit on the ethanol inhibition of recombinant N-methyl-D-aspartate receptors. Alcohol Clin Exp Res. 2006;30:523–30. doi: 10.1111/j.1530-0277.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- Smothers CT, Jin C, Woodward JJ. Deletion of the N-terminal domain alters the ethanol inhibition of N-methyl-D-aspartate receptors in a subunit-dependent manner. Alcohol Clin Exp Res. 2013;37:1882–90. doi: 10.1111/acer.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Beck C, Wollmuth LP. Molecular rearrangements of the extracellular vestibule in NMDAR channels during gating. Neuron. 2002;33:75–85. doi: 10.1016/s0896-6273(01)00560-8. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–96. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengeliene V, Bilbao A, Molander A, Spanagel R. Neuropharmacology of alcohol addiction. Br J Pharmacol. 2008;154:299–315. doi: 10.1038/bjp.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalan SP, Czajkowski C. A conserved salt bridge critical for GABAA receptor function and loop C dynamics. Proc Natl Acad Sci USA. 2008;105:13604–9. doi: 10.1073/pnas.0801854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Lopez MN, Huettner JE. Radial symmetry in a chimeric glutamate receptor pore. Nat Commun. 2014;5:3349. doi: 10.1038/ncomms4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ. Ethanol and NMDA receptor signaling. Crit Rev Neurobiol. 2000;14:69–89. doi: 10.1080/08913810008443548. [DOI] [PubMed] [Google Scholar]
- Poznyak V, Rekve D, editors. World Health Organization. Global Status Report on Alcohol and Health. Geneva: WHO Press; 2014. [Google Scholar]
- Wright JM, Peoples RW, Weight FF. Single-channel and whole-cell analysis of ethanol inhibition of NMDA-activated currents in cultured mouse cortical and hippocampal neurons. Brain Res. 1996;748:249–56. doi: 10.1016/s0006-8993(96)00780-9. [DOI] [PubMed] [Google Scholar]
- Xu M, Smothers CT, Woodward JJ. Cysteine substitution of transmembrane domain amino acids alters the ethanol inhibition of GluN1/GluN2A N-methyl-D-aspartate receptors. J Pharmacol Exp Ther. 2015;353:91–101. doi: 10.1124/jpet.114.222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakura T, Mihic SJ, Harris RA. Amino acid volume and hydropathy of a transmembrane site determine glycine and anesthetic sensitivity of glycine receptors. J Biol Chem. 1999;274:23006–12. doi: 10.1074/jbc.274.33.23006. [DOI] [PubMed] [Google Scholar]
- Ye Q, Koltchine VV, Mihic SJ, Mascia MP, Wick MJ, Finn SE, Harrison NL, Harris RA. Enhancement of glycine receptor function by ethanol is inversely correlated with molecular volume at position α267. J Biol Chem. 1998;273:3314–9. doi: 10.1074/jbc.273.6.3314. [DOI] [PubMed] [Google Scholar]
- Yelshansky MV, Sobolevsky AI, Jatzke C, Wollmuth LP. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci. 2004;24:4728–36. doi: 10.1523/JNEUROSCI.0757-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ren H, Dwyer DS, Peoples RW. Different sites of alcohol action in the NMDA receptor GluN2A and GluN2B subunits. Neuropharmacol. 2015;97:240–50. doi: 10.1016/j.neuropharm.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberter Y, Uteshev V, Sokolova S, Khodorov B. Desensitization of N-methyl-D-aspartate receptors in neurons dissociated from adult rat hippocampus. Mol Pharmacol. 1991;40:337–41. [PubMed] [Google Scholar]