Abstract
Deep sequencing of the gut microbiomes of 1,135 participants from a Dutch population-based cohort shows relations between the microbiome and 126 exogenous and intrinsic host factors, including 31 intrinsic factors, 12 diseases, 19 drug groups, 4 smoking categories, and 60 dietary factors. These factors collectively explain 18.7% of the variation seen in the inter-individual distance of microbial composition. We could associate 110 factors to 125 species and observed that fecal Chromogranin A (CgA), a protein secreted by enteroendocrine cells, was exclusively associated with 61 microbial species whose abundance collectively accounted for 53% of microbial composition. Low CgA levels were seen in individuals with a more diverse microbiome. These results are an important step towards better understanding of environment-diet-microbe-host interactions.
One Sentence Summary
126 factors collectively explain 18.7% of the variation in human gut microbiome composition, with strongest effect observed for the fecal level of Chromogranin A.
The human gut microbiome plays a major role in the production of vitamins, enzymes, and other compounds that digest and metabolize food and regulate our immune system (1). It can be considered as an extra organ, with remarkable dynamics and a major impact on our physiology. The composition of the gut microbiome can be considered as a complex trait, with the quantitative variation in the microbiome affected by a large number of host and environmental factors, each of which may have only a small additive effect, making it difficult to identify the association for each separate item. In this study, we present a systematic metagenomic association analysis on 207 intrinsic and exogenous factors from the LifeLines-DEEP cohort, a Dutch population-based study (2, 3). Our study reveals covariates in the microbiome and, more importantly, provides a list of factors that correlate with shifts in the microbiome composition and functionality.
This study includes stool samples from 1,179 LifeLines-DEEP participants from the general population of the northern part of the Netherlands (2). The cohort comprised predominantly Dutch participants; 93.7% had both parents born in the Netherlands. The gut microbiome was analyzed using paired-end metagenomic shotgun sequencing (MGS) on a HiSeq2000, generating an average of 3.0 Gb of data (about 32.3 million reads) per sample (4). After excluding 44 samples with low read counts, 1,135 participants (474 males and 661 females) remained for further analysis. We tested 207 factors with respect to the microbiomes of these participants: 41 intrinsic factors of various physiological and biomedical measures, 39 self-reported diseases, 44 categories of drugs, 5 categories of smoking status and 78 dietary factors (fig. S1 and table S1). These factors cover dietary habits, life-style, medication use, and health parameters. Most of the factors showed a low or modest inter-correlation (table S2A–C, 2A–D); many are highly variable, including, as expected in the Dutch population, the high consumption of milk products and low use of antibiotics. Antibiotic use in the Netherlands is the lowest in the Europe, at a level half that of the UK and one-third that of Belgium. To cover health-domain factors relevant to the host immune system and gut health, we collected cell counts for eight different blood cell types, measured blood cytokine levels, assessed stool frequency and stool type by Bristol Stool Score, and measured fecal levels of several secreted proteins including calprotectin as a marker for the immune system activation, human-β-defensin-2 (HBD-2) as a marker for defense against invading microbes and chromogranin A (CgA) as a marker for neuroendocrine system activation.
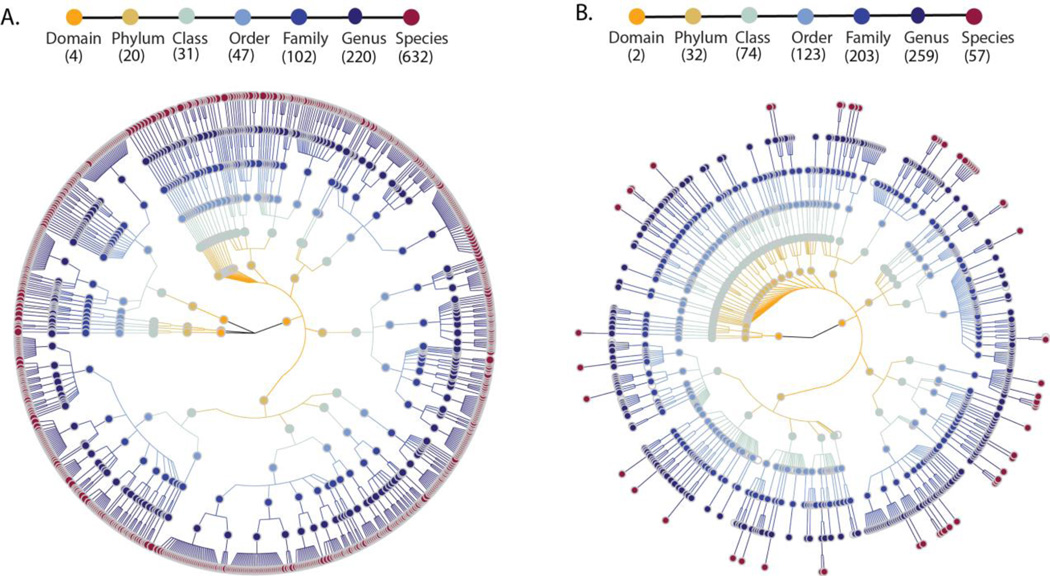
After quality control and removal of sequence reads mapping to the human genome, the microbiome sequence reads were mapped to approximately 1 million microbial-taxonomy-specific marker genes using MetaPhlAn 2.0 (5) to predict the abundance of microorganisms (fig. S3A). For each participant, we predicted the abundance levels for 1,649 microbial taxonomic clades ranging from four different domains to 632 species (Fig 1A). The majority of the reads (97.6%) came from Bacteria, 2.2% from Archaea, 0.2% from Viruses and <0.01% from Eukaryotes. Comparison to previous taxonomic profiles of the same subjects by 16S rRNA gene sequencing (Fig. 1B) showed MGS predicted more microbial species but fewer families and genera. At the phylum level, the abundance of dominant bacterial phyla Firmicutes (63.7%) and Bacteroidetes (8.1%) were similar to estimates based on 16S rRNA gene sequencing, but the abundance of Actinobacteria was higher in MGS (22.3%) than 16S (12.3%) (fig. S4). The microbiome quality control project has recently suggested that microbial composition estimates may not be comparable between studies if sample preparation and data analysis are not done in the same way (6). For instance, compared to the composition reported in other studies of a similar size that used different methods (7, 8), our study detected a higher abundance of Actinobacteria but a lower abundance of Bacteroidetes. Importantly, all samples in our study were isolated and processed using the same pipeline, ensuring low technical variation and high analysis power to access the association of multiple factors with the microbiome.
Fig. 1. The taxonomic tree of microbial taxonomies predicted by MGS and 16S rRNA gene sequencing.
(A) Taxonomic tree based on MGS shotgun sequencing data. (B) Taxonomic tree based on16S rRNA gene sequencing data. Each dot represents a taxonomic entity. From the inner to outer circles, the taxonomic levels range from domains to species. Different colors of dots indicate different taxonomy levels according to the color key shown. Numbers in brackets indicate the total number of unique taxonomies detected at each level.
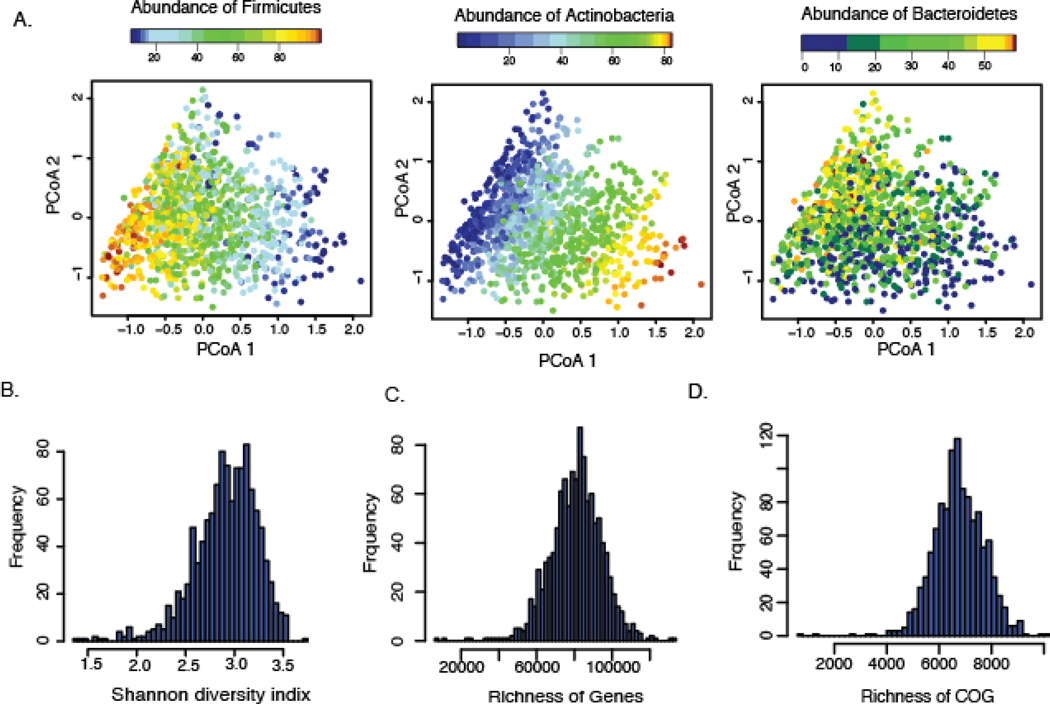
The high inter-individual variation reflects the community composition (fig. S5) and is clearly driven by the abundance of the dominant phyla (fig. 2A). Our further analysis of microbial composition was confined to the 632 unique species (table S3). For the functional profiling, the abundance of 568,874 UniRef gene families were grouped into clusters of orthologous groups (COG) based on the EggNOG database and MetaCyc pathways (fig. S3A). Although the distribution of diversity, genes, and COG richness showed high inter-variability (Fig. 2B–D), functional profiles based on 23 non-redundant, gene ontology molecular function categories remained stable (fig. S6) within our cohort, similar to previous reports (9).
Fig. 2. The inter-individual variation of microbial composition and function profile.
(A) Principal coordinates plots of Bray-Curtis distance of microbial composition. The composition was driven by the most dominant phyla: Firmicutes, Actinobacteria, and Bacteroidetes. Each dot represents one individual. Color indicates the relative abundance of each phylum. (B) Distribution of Shannon’s diversity index. (C) Distribution of the gene richness. (D) Distribution of the clusters of orthologous groups (COG) richness.
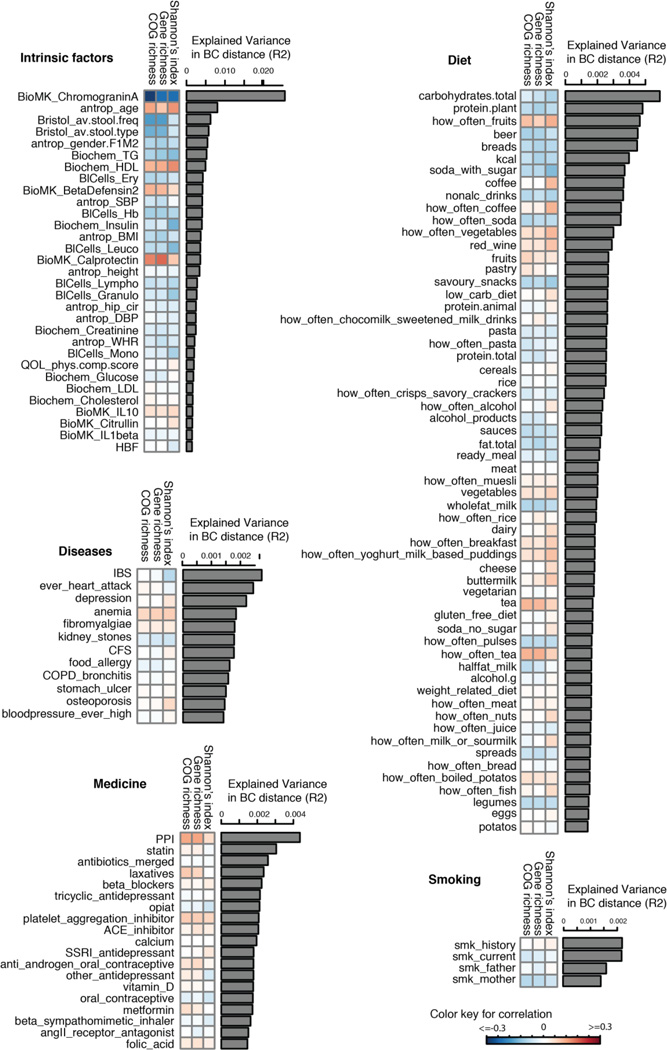
We correlated 207 factors to the inter-individual variation in microbial composition, diversity, richness of genes, and COGs (fig. S3B). At false discovery rate (FDR) <0.1 level, 126 factors were associated with inter-individual distance of microbial composition (Bray-Curtis distance) (Fig 3, table S4), of which 90% could be replicated in 16S rRNA data from the same subjects (table S5, fig. S7), together explaining 18.7% of the variation in composition distance (fig. S8A). A total of 35 factors were associated with Shannon’s diversity index of microbial composition (together explaining 13.7% variation, table S6, fig. S8A), of which 80% were replicated in 16S rRNA data from the same subjects (table S7); 31 factors were associated with gene richness (together explaining 16.7% variation, table S8) and 34 factors with COG richness (explaining 18.8% variation) (table S9, fig. S8A, for replication rates see table S10). We saw a large overlap between different diversity and richness analyses, and most of them were also associated with composition distance (fig. S8B).
Fig. 3. Factors associated with inter-individual variation of gut microbiome.
A total of 126 factors (FDR<0.1) were associated with inter-individual variation of the gut microbiome. The bar plot indicates the explained variation of each factor in the inter-individual variation of microbial composition (Bray-Curtis distance). The heatmap under the bar plot shows the correlation coefficients of each factor with Shannon’s index of diversity, gene richness, and COG richness, respectively. Color key for correlation is shown.
We performed multivariate association analyses between each factor with 170 abundant species (>0.01% of total microbial composition and present in at least 10 individuals) and 215 MetaCyc pathways (fig. S3C). When corrected for age, gender, and sequence depth, we found 485 associations at FDR<0.1 between 110 factors and 125 species (table S11) and 524 associations between 71 factors and 176 MetaCyc pathways (table S12). By correcting the correlation structures among all 207 factors, the number of associations was reduced to 128 independent associations with species (table S13) and 215 associations with pathways (table S14).
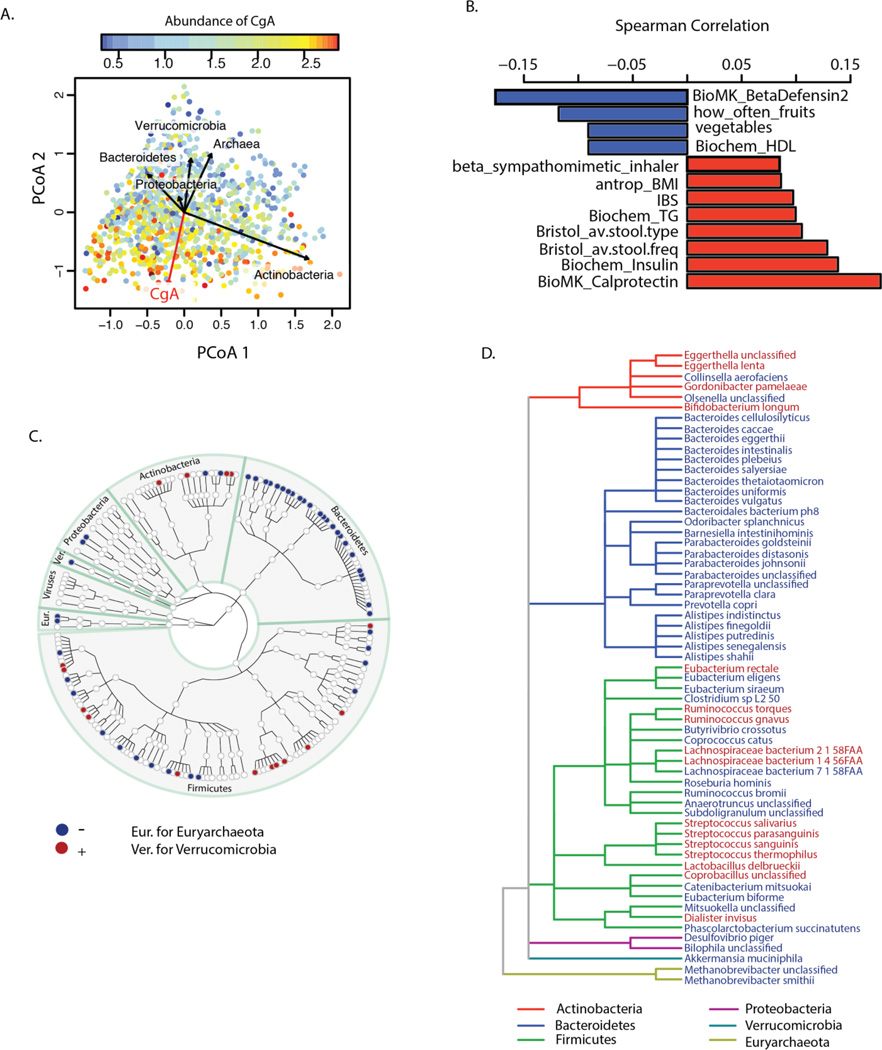
Our data confirmed some previous findings and also yielded novel associations. In our study, age and gender were correlated not only with microbial composition distance and diversity but also with functional richness. Women showed higher COG richness than men (adjusted P=0.03), and COG richness increased with age (adjusted P=0.002) (fig. S9). Multiple intrinsic parameters, such as blood cell counts and lipid levels, were associated to composition and function levels as well. For example, a higher level of hemoglobin was consistently associated with lower diversity and functional richness (Fig 3, table S6–S9). The strongest associations we found were for the fecal levels of several secreted proteins, including human-β-defensin-2 (HBD-2), calprotectin (10, 11), and chromogranin A (CgA), with microbial composition, diversity, and functional richness (Fig. 3, Fig. 4A), as well as with specific species (table S11) and pathways (table S12). Among these associations, CgA showed the strongest association with composition distance (adonis R2=0.03, adjusted P=0.0006), microbial diversity (Spearman r=−0.22, adjusted P=1.49×10−12), gene richness (Spearman r=−0.23, adjusted P=9.4×10−13), and COG richness (Spearman r=−0.285, adjusted P=2.53×10−20) (tables S4–S9). The association of CgA with composition distance was then validated in an independent cohort of 19 individuals for whom 16S rRNA gene sequencing data was available (P=0.0065) (fig. S10). A lower CgA level was associated with higher diversity, with functional richness, with high levels of high-density lipoprotein (HDL), and with intake of fruits and vegetables. In contrast, elevated fecal CgA was associated with high fecal levels of calprotectin, high blood levels of triglycerides, high stool frequency, soft stool type, and self-reported irritable bowel syndrome (IBS) (Fig. 4B). After correcting for the confounding effect of all other factors, our analysis revealed 61 species exclusively associated with CgA (Fig. 4C–D, table S13) whose abundance levels collectively accounted for 53% of the total abundance of the microbiome on average, and with 40 MetaCyc pathways (table S14) that accounted for 34.6% of the pathway profiles. The strongest association to CgA was observed for the Archaea species Methanobrevibacter smithii (fig. S11A), which plays an important role in the digestion of polysaccharides by consuming the end products of bacterial fermentation and methanogenesis (12)(fig. S11B). A negative association with CgA abundance was observed for 24 out of 36 species from phylum Bacteroidetes (Fig. 4C–D).
Fig. 4. The association of fecal level of Chromogranin A.
(A) Principal coordinate plots of Bray-Curtis distance of microbial composition. Each dot represents one individual and its color is based on the abundance level of CgA: warm colors indicate high abundance and cool colors low abundance. The red arrow indicates the association direction of CgA, while the directions of the CgA-associated phyla are shown as black arrows. (B) Correlation between CgA and other factors at FDR<0.1. (C) Taxonomic tree of 170 species, of which 61 species were exclusively associated with CgA level. Each dot represents a taxonomic entity. Red dots indicate positively associated species. Blue dots indicate negatively associated species. (D) Taxonomic tree of the 61 species exclusively associated with CgA level. The branches are colored to show phylum levels as shown in the color key. Species in red show increased abundance associated with higher CgA levels. Species in blue show lower abundance associated with higher CgA levels.
CgA is a member of the granine peptides, which are secreted in nervous, endocrine and immune cells under stress (13), and during active periods of gut-related diseases such as IBS and inflammatory bowel disease, although some findings are contradictory (14–16). Many different functions have been proposed for CgA and other granine peptides, including roles in neurological pathways, pain regulation, and antimicrobial activity against bacteria, fungi, and yeasts (17, 18). However, their mechanism of action and physiological importance need further detailed investigation. To test whether genetic variants that influence CHGA gene expression (encoding CgA) can affect fecal CgA level and the gut microbiome, we tested the effect of six SNPs known to regulate gene expression of CHGA on fecal CgA and abundances of species (table S15). No significant association was observed, suggesting that genetic variation in CHGA expression does not explain the variation observed in the fecal CgA levels and microbiome composition (table S16–S17). Our observation that CgA strongly correlates with microbiome composition, especially with a large number of species from Bacteroidetes phylum, and with diversity will hopefully encourage studies to unravel the role of CgA in gut health.
We also observed associations (FDR<0.1) between 63 dietary factors and inter-individual distances in microbiota composition, including energy (kcal), intake of carbohydrates, proteins and fats, and of specific food items such as bread and soft drinks (Fig. 3, table S4). Drinking buttermilk (sour milk with a low fat content) was associated with high diversity, while drinking high-fat (whole) milk (3.5% fat content) was associated with lower diversity (table S6). Two of the species most strongly associated with drinking buttermilk are Leuconostoc mesenteroides (q=9.1×10−46) and Lactococcus lactis (q=2.5×10−8), both used as a starter culture for industrial fermentation (table S11). The abundance of dairy-fermentation-related bacteria increased with increasing dairy consumption, indicating potential for the use of probiotic drinks to augment and alter the gut microbiome composition. Consumption of alcohol-containing products, coffee, tea, and sugar-sweetened drinks were also correlated with microbial composition. Consumption of sugar-sweetened soda had a negative effect on microbial diversity (adjusted P=5×10−4), whereas consumption of coffee, tea and red wine, which all have a high polyphenol content, was associated with increased diversity (19–21). Red wine consumption correlated with F. prausnitzii abundance, which has anti-inflammatory properties, correlates negatively with inflammatory bowel disease (22), and shows higher abundance in high-richness microbiota (23). Apart from the negative associations between sugar-sweetened soda and bacterial diversity, other features of a Western-style diet, such as higher intake of total energy, snacking, and high-fat (whole) milk, were also associated with lower microbiota diversity (Fig. 3). A higher amount of carbohydrates in the diet was associated with lower microbiome diversity. Total carbohydrate intake was positively associated with Bifidobacteria, but negatively with Lactobacillus, Streptococcus, and Roseburia species. A low carbohydrate diet consistently showed opposite directions for these species. We did not observe an association of carbohydrate intake to prevotella species, as has been described previously (24).
As expected, the use of antibiotics was significantly associated with microbiome composition, in particular with strong and significant decreases in two species from the genus Bifidobacterium (Actinobacteria phylum) (table S11), in line with previous studies (25). Several other drug categories, such as proton pump inhibitors (PPI) (95 users), metformin (15 users), statins (56 users), and laxatives (21 users) also had a strong effect on the gut microbiome. PPI users were found to have profound changes in 33 bacterial pathways (table S12). The most significant positive correlation of PPIs was observed with the pathway of 2,3-butanediol biosynthesis (q=5.3×10−14). We also observed overlap between species and pathways associated to PPI and with calprotectin levels, particularly for bacteria typical of the oral microbiome (table S2A–C, table S11, fig. S12). This is in line with the correlations of PPI with calprotectin levels reported in the literature (26). Even after excluding the 95 PPI users from our analysis, the positive correlation of calprotectin to most oral bacteria remained significant, indicating this association is not due to the confounding effect of PPI (fig. S12). Furthermore, the levels of calprotectin were positively correlated with age and metabolic phenotypes (body mass index (BMI), diabetes, use of statins and metformin, HBAc1, and systolic blood pressure), but negatively correlated with the consumption of vegetables, plant proteins, chocolate, and breads. Multivariate analysis correcting for all factors revealed 14 species (table S13) and 114 bacterial metabolic pathways (table S14) exclusively associated with calprotectin, suggesting calprotectin is robustly associated with gut microbiome.
Metformin is commonly used to control blood sugar levels for treating type 2 diabetes, but can cause gastrointestinal intolerance (27). In 15 metformin users, we observed an increased abundance of Escherichia coli (E. coli) and a positive correlation with specific pathways, including the degradation and utilization of D-glucarate and D-galactarate and pyruvate fermentation pathways. Previous studies in C. elegans indicated the specific drug-bacteria interaction of metformin and E. coli (28). Our results are in line with recent observations in humans (29) that suggest that metformin can impact the microbiome through short-chain fatty acid (SCFA) production. To confirm this observation, we profiled acetate, propionate and butyrate in 24 type 2 diabetes patients in our cohort: 9 non-metformin users and 15 users (4), and found that SCFA levels were consistently higher in metformin-users, especially for propionate (Wilcoxon test P=0.035) (fig. S13).
We assessed the effect of current smoking status, smoking history, parental smoking, and maternal smoking during pregnancy on the gut microbiome. These parameters were associated with Bray-Curtis distance, albeit with very modest effect. We did not detect significant associations for individual species or at pathways. In this study we included 39 self-reported diseases, for which participants had reported at least five cases. IBS was reported by 9.9% of participants (n=112, table S1) and was associated with changes in the gut microbiome and a lower microbial diversity (adjusted P=0.05) (table S6). Species from the Eggerthella and Coprobacillus genera were positively associated with medication and food allergies, respectively. Individuals who had suffered a heart attack (n=10) in the past had a significantly lower abundance of Eubacterium eligens bacterium, even after correcting for all other factors (q=4.6×10−4).
Linking the deep-sequenced MGS data to various intrinsic and exogenous factors from the same individual not only allowed us to detect associations at species level, but also provided new insights into the interaction between the host, microbiota, and environmental factors, including diet. For instance, we have replicated and expanded our association of BMI and blood lipid levels with the gut microbiota based on 16S rRNA gene sequencing data (30) by showing associations with four specific species of the family Rikenellaceae. We previously associated this family with BMI and triglycerides in 16S rRNA data. In the current study we observed higher BMI was associated with lower level of two species from the family Rikenellaceae, Alistipes finegoldii, and Alistipes senegalensis, while blood lipids were associated with other two species, Alistipes shahii and Alistipes putredinis (table S11). Strikingly, these species were also associated to certain dietary factors and drugs. For instance, a high level of Alistipes shahii, which was associated to low TG levels, was linked to higher fruit intake (q= 0.00027). Individuals with a higher abundance level of Alistipes shahii had a higher number of different species in the gut (species richness) (Spearman r=0.2, adjusted p=3.96×10−11), suggesting a beneficial effect on the microbial ecosystem (table S18). Correlations with the number of different species were also found for other bacteria including Roseburia hominis, Coprococcus catus, and Barnesiella intestinihominis and unclassified species from genus Anaerotruncus that also showed correlation both with fruit, vegetable, and nut consumption and with intrinsic phenotypes like HDL, triglycerides and quality of life. Based on this data, it would be interesting to explore the potential to modulate disease-associated species through medication or diet, although we still need to address the causality and underlying mechanism.
Conclusions
Our study revealed significant associations between the gut microbiome and various intrinsic, environmental, dietary and medication parameters, and disease phenotypes, with a high replication rate between MGS and 16S rRNA gene sequencing data from the same subjects. Moreover, our study provides many new intrinsic and exogenous factors that correlate with shifts in the microbiome composition and functionality that can be potentially be manipulated to improve microbiome-related health and we hope our results will inspire further experiments to explore the biological relevance of associated factors. While most of the factors we assessed exerted a very modest effect, fecal levels of Chromogranin A showed a high potential as a biomarker for gut health.
Supplementary Material
Acknowledgments
We thank the LifeLines-DEEP participants and the Groningen LifeLines staff for their collaboration. We thank Jackie Dekens, Mathieu Platteel, and Astrid Maatman for management and technical support. We thank Jackie Senior and Kate Mc Intyre for editing the manuscript.
This project was funded by grants from the Top Institute Food and Nutrition, Wageningen, to C.W. (TiFN GH001), the Netherlands Organization for Scientific Research to J.F. (NWO-VIDI 864.13.013), L.F. (ZonMW-VIDI 917.14.374), and R.W. (ZonMW-VIDI 016.136.308), CardioVasculair Onderzoek Nederland to M.H. and A.Z. (CVON 2012-03). A.Z. holds a Rosalind Franklin Fellowship (University of Groningen) and M.C.C. holds a postdoctoral fellowship from the Fundación Alfonso Martín Escudero. This research received funding from the European Research Council under the European Union’s Seventh Framework Program: C.W. is supported by FP7/2007-2013)/ERC advanced Grant Agreement no. 2012-322698. M.G.N. is supported by an ERC Consolidator Grant (#310372). L.F. is supported by FP7/2007-2013, grant agreement 259867, and by an ERC Starting Grant, grant agreement 637640 (ImmRisk). J.R. and G.F. are supported by FP7 METACARDIS HEALTH-F4-2012-305312, VIB, FWO, IWT, the Rega institute for Medical Research, and KU Leuven. S.V.S. and M.J. are supported by postdoctoral fellowships from FWO.
Footnotes
A.Z., C.W. and J.F. designed the study. A.Z., E.F.T., L.F., and C.W. initiated the cohort and collected cohort data. A.Z., E.F.T., Z.M., S.A.J., M.C.C., and D.K. generated data. A.Z., A.K., M.J.B., E.F.T., M.S., T.V., A.V.V., G.F., S.V.S, J.W., F.I., P.D., M.A.S., C.H., R.J.X., and J.F. analyzed data. G.F, S.V.S., J.W. E.B., M.J., R.K.W., E.J.M.F., M.G.N., D.G., D.J., L.F., Y.S.A., C.H., J.R., R.J.X., and M.H.H. participated in integral discussions. A.Z., A.K., M.J.B., R.J.X., C.W., and J.F. wrote the manuscript.
The authors have no conflicts of interest to report.
Data: The data is currently being uploaded to the European Genotyping Agency (https://www.ebi.ac.uk/ega/) (ega-box-423).
Informed consent: The study was approved by the institutional review board of UMCG, ref.M12.113965.
Supplementary Materials:
Materials and Methods
Figures S1–S13
Tables S1–S19
References (31– 54)
References and Notes
- 1.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tigchelaar EF, et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scholtens S, et al. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int. J. Epidemiol. 2014;44:1172–1180. doi: 10.1093/ije/dyu229. [DOI] [PubMed] [Google Scholar]
- 4.Information on materials and methods is available at the Science Web site
- 5.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodrich JK, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falcony G, et al. Population-level analysis of gut microbiome variation. 2016 doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 9.Huttenhower C, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrand F, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedin C, et al. Siblings of patients with Crohn’s disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut. 2015 doi: 10.1136/gutjnl-2014-308896. [DOI] [PubMed] [Google Scholar]
- 12.Miller TL, Wolin MJ, Conway de Macario E, Macario AJ. Isolation of Methanobrevibacter smithii from human feces. Appl. Environ. Microbiol. 1982;43:227–232. doi: 10.1128/aem.43.1.227-232.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T, et al. Evaluation of psychosomatic stress in children by measuring salivary chromogranin A. Acta Paediatr. 2006;95:935–939. doi: 10.1080/08035250500538940. [DOI] [PubMed] [Google Scholar]
- 14.Ohman L, Stridsberg M, Isaksson S, Jerlstad P, Simrén M. Altered levels of fecal chromogranins and secretogranins in IBS: relevance for pathophysiology and symptoms? Am. J. Gastroenterol. 2012;107:440–447. doi: 10.1038/ajg.2011.458. [DOI] [PubMed] [Google Scholar]
- 15.Sciola V, et al. Plasma chromogranin a in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 2009;15:867–871. doi: 10.1002/ibd.20851. [DOI] [PubMed] [Google Scholar]
- 16.Wagner M, et al. Increased fecal levels of chromogranin A, chromogranin B, and secretoneurin in collagenous colitis. Inflammation. 2013;36:855–861. doi: 10.1007/s10753-013-9612-4. [DOI] [PubMed] [Google Scholar]
- 17.Aslam R, et al. Chromogranin A-derived peptides are involved in innate immunity. Curr. Med. Chem. 2012;19:4115–4123. doi: 10.2174/092986712802430063. [DOI] [PubMed] [Google Scholar]
- 18.Bartolomucci A, et al. The extended granin family: structure, function, and biomedical implications. Endocr. Rev. 2011;32:755–797. doi: 10.1210/er.2010-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Queipo-Ortuño MI, et al. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am. J. Clin. Nutr. 2012;95:1323–1334. doi: 10.3945/ajcn.111.027847. [DOI] [PubMed] [Google Scholar]
- 20.Duda-Chodak A, Tarko T, Satora P, Sroka P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: a review. Eur. J. Nutr. 2015;54:325–341. doi: 10.1007/s00394-015-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills CE, et al. In vitro colonic metabolism of coffee and chlorogenic acid results in selective changes in human faecal microbiota growth. Br. J. Nutr. 2015;113:1220–1227. doi: 10.1017/S0007114514003948. [DOI] [PubMed] [Google Scholar]
- 22.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Chatelier E, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 24.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korpela K, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016;7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poullis A, Foster R, Mendall MA, Shreeve D, Wiener K. Proton pump inhibitors are associated with elevation of faecal calprotectin and may affect specificity. Eur. J. Gastroenterol. Hepatol. 2003;15:573–574. doi: 10.1097/00042737-200305000-00021. [DOI] [PubMed] [Google Scholar]
- 27.Burton JH, et al. Addition of a Gastrointestinal Microbiome Modulator to Metformin Improves Metformin Tolerance and Fasting Glucose Levels. J. Diabetes Sci. Technol. 2015;9:808–814. doi: 10.1177/1932296815577425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabreiro F, et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu J, et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ. Res. 2015;117:817–824. doi: 10.1161/CIRCRESAHA.115.306807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibble JA, et al. High prevalence of NSAID enteropathy as shown by a simple faecal test. Gut. 1999;45:362–366. doi: 10.1136/gut.45.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi S, Lewis SJ, Creanor S, Ayling RM. Age-related faecal calprotectin, lactoferrin and tumour M2-PK concentrations in healthy volunteers. Ann. Clin. Biochem. 2010;47:259–263. doi: 10.1258/acb.2009.009061. [DOI] [PubMed] [Google Scholar]
- 33.Harder J, Bartels J, Christophers E, Schröder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 34.Langhorst J, et al. Elevated human beta-defensin-2 levels indicate an activation of the innate immune system in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2009;104:404–410. doi: 10.1038/ajg.2008.86. [DOI] [PubMed] [Google Scholar]
- 35.El-Salhy M, Lomholt-Beck B, Hausken T. Chromogranin A as a possible tool in the diagnosis of irritable bowel syndrome. Scand. J. Gastroenterol. 2010;45:1435–1439. doi: 10.3109/00365521.2010.503965. [DOI] [PubMed] [Google Scholar]
- 36.Sidhu R, Drew K, McAlindon ME, Lobo AJ, Sanders DS. Elevated serum chromogranin A in irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD): a shared model for pathogenesis? Inflamm. Bowel Dis. 2010;16:361. doi: 10.1002/ibd.20982. [DOI] [PubMed] [Google Scholar]
- 37.Windmueller HG, Spaeth AE. Source and fate of circulating citrulline. Am. J. Physiol. 1981;241:E473–E480. doi: 10.1152/ajpendo.1981.241.6.E473. [DOI] [PubMed] [Google Scholar]
- 38.Crenn P, Messing B, Cynober L. Citrulline as a biomarker of intestinal failure due to enterocyte mass reduction. Clin. Nutr. 2008;27:328–339. doi: 10.1016/j.clnu.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Farzi A, et al. Synergistic effects of NOD1 or NOD2 and TLR4 activation on mouse sickness behavior in relation to immune and brain activity markers. Brain. Behav. Immun. 2015;44:106–120. doi: 10.1016/j.bbi.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whelan RA, et al. A transgenic probiotic secreting a parasite immunomodulator for site-directed treatment of gut inflammation. Mol. Ther. 2014;22:1730–1740. doi: 10.1038/mt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 44.UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimmer EC, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gene Ontology Consortium: going forward. Nucleic Acids Res. 2014;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oksanen J, et al. vegan: Community Ecology Package. R package version 2.3-0. 2015 [Google Scholar]
- 48.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 51.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brandt BW, Bonder MJ, Huse SM, Zaura E. TaxMan: A server to trim rRNA reference databases and inspect taxonomic coverage. Nucleic Acids Res. 2012;40:W82–W87. doi: 10.1093/nar/gks418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta RK, Pant CS, Singh AK, Behl P. Real time ultrasonography in the evaluation of hydrocephalus and associated abnormalities. Indian Pediatr. 1986;23:249–254. [PubMed] [Google Scholar]
- 54.Neoptolemos JP, et al. Study of common bile duct exploration and endoscopic sphincterotomy in a consecutive series of 438 patients. Br. J. Surg. 1987;74:916–921. doi: 10.1002/bjs.1800741014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.