Abstract
Purpose of review
Asthma is a complex and heterogeneous disease with strong genetic and environmental components that manifests within a variety of clinical features and diverse patterns of immune responses. Asthma prevalence has dramatically increased over the last decade in Westernize societies, thereby suggesting a key function of environmental factors in disease promotion and development.
Recent Findings
“Early-life” microbial exposures and bacterial colonization are crucial for the maturation and the education of the immune system. The commensal flora is also critical in order to maintain immune homeostasis at the mucosal surfaces and may consequently play an important function in allergic disease development. Recent evidences demonstrate that asthma influences and is also impacted by the composition and function of the human intestinal and respiratory microbiome.
Summary
In this review, we will summarize the most recent findings on how asthma development is connected with respiratory and intestinal microbial dysbiosis. We will highlight and discuss the recent research that reveal the existence of a “gut-lung” microbial axis and its impact on asthma development. We will also analyze how “early-life” microbial exposure affects the immune response and their consequences on asthma development.
Keywords: Asthma, Microbiome, Mycobiome, Lower airways, Gut, Metabolites
Introduction
Asthma is a chronic inflammatory disease of the respiratory airways characterized by an inappropriate immune response resulting in reversible airflow obstruction, airway hyper-responsiveness (AHR), mucus overproduction, tissue eosinophilia, and intense airway wall remodeling [1,2]. It is a complex and heterogeneous disease which exists under different phenotypes, known as allergic and non-allergic or intrinsic asthma. Allergic asthma affects mostly children and is triggered by aeroallergens, such as house dust mite (HDM), pollen, and fungal spores. Predominantly developing during the first years of life, allergic asthma involves the production of allergen-specific immunoglobulin type E (IgE) and the participation of an adaptive T helper cells Type 2 (Th2) immune response. On the contrary, non-allergic intrinsic asthma occurs later in life independently of aeroallergen sensitization, results from air pollution, chronic or recurrent bacterial and viral infections of the bronchi and sinuses, is more severe and predominant in women [3]. Compared with non-allergic patients, allergic asthmatic subjects respond better to a medical treatment combining β2-adrenergic receptor agonists and inhaled corticosteroids. However, 5 to 10% of bronchial asthmatic subjects are unresponsive to this treatment and considered as refractory or steroid resistant asthmatic [4,5].
Asthma and allergic diseases have become a major health issue worldwide with increased prevalence over the last 50 years. In the USA, subjects affected with asthma rose from 20.3 millions in 2001 to 25.7 million in 2010 [6]. Although genetic polymorphisms have been associated with asthma development [7], this steep increase in asthma prevalence over the last decades implicates the existence of environmental factors which promote disease on genetically predisposed hosts. Among them, early-life sensitization to aeroallergen [8] and microbial exposure [9] as well as infections with respiratory viruses [10] and changes in the host microbiome composition [11] have been associated with increased risk of asthma development.
In the present review we will highlight and discuss major findings demonstrating a connection between the gut and lung microbiome with asthma development and how host-microorganism interactions in early-life affect the immune response and their consequences on allergic diseases development.
Immunology of asthma
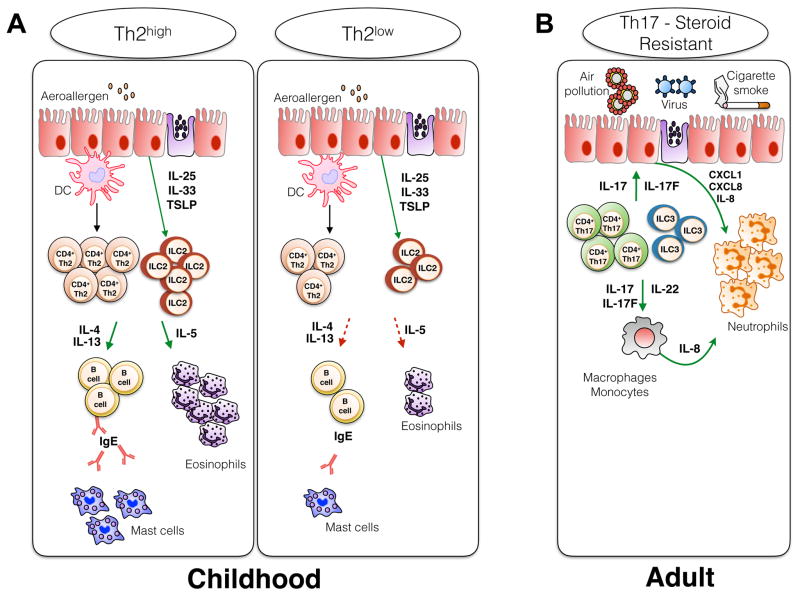
Asthma is a complex and heterogeneous disease initially thought to be mediated by eosinophils and Th2 immune cells as evidenced by elevated numbers of interleukin 4 (IL-4) and IL-5 producing CD4+ T cells in bronchoalveolar lavage (BAL) fluid collected from allergic asthmatic patients [12]. Asthma heterogeneity was further characterized by gene expression analysis and the identification of two human endotypes, Th2high and Th2low, which differ mainly by their Th2 cytokine expression pattern and their response to medical treatment [13] (Figure 1). The Th2high endotype is associated with increased activation of dendritic cells (DCs), CD4+ Th2 T cells, innate lymphoid cells type 2 (ILC2) and B cells resulting in heightened allergen-specific IgE and Th2 cytokine production as well as elevated numbers of lung infiltrating and circulating eosinophils. On the other hand, the Th2low endotype display reduced levels of Th2 cells activation, Th2 cytokine production and eosinophil accumulation in the lungs (Figure 1). Strategies aiming to block the α chain of the IL-4 receptor and thereby disrupting both IL-4 and IL-13 signaling (Dupilumab) [14] or targeting directly IL-13 (Lebrikizumab) [15] improve asthma clinical aspects for the Th2high phenotype. However, those therapeutical approaches appear to be less successful for patients with a Th2low profile. Recent studies demonstrate that intrinsic non-allergic, non-IgE dependent, late-onset asthma triggered by environmental factors such as respiratory airway infection, air pollution and/or smoking, involves a Th1/Th17 immune response and intense neutrophils recruitment in the lungs [16–18] (Figure 1). The secretion of IL-8, IL-17, IL-21, IL-22, and monocyte colony stimulatory factor by either CD4+ Th17 T cells or ILC3s will promote the recruitment of neutrophils and airway inflammation [19,20]. This Th17-driven asthma phenotype is linked to a more severe pathology and a poor response to corticosteroid treatment [21–23] (Figure 1). Asthma endotype classification and disease heterogeneity are not fixed and exclusive. While each endotype is characterized by specific immune responses and pathological patterns, the possibility that those pathways concur and act synergistically in asthmatic patients cannot be excluded.
Figure 1. Asthma heterogeneity and immunological patterns involved in disease pathology.
(A) The Th2high endotype is characterized by increased activation of DCs, Th2 CD4+ T cells, ILC2s and B cells which result in heightened allergen-specific IgE and Th2 cytokine production as well as elevated numbers of lung infiltrating and circulating eosinophils. On the other hand, the Th2low endotype display reduced activation of Th2 cells and Th2 cytokine production as well as decreased numbers of eosinophils. (B) Intrinsic non-allergic and steroid resistant asthma is IgE and Th2 independent, associated with a Th1/Th17 immune response and the participation of ILC3s as well as intense neutrophils recruitment in the lungs.
The microbiome
The advancement and the development of affordable next generation high-throughput sequencing techniques has expanded our knowledge on the composition of the human microbiome as well as its relation to disease [24]. Recently, it has been estimated that the human body is composed of 3x1013 eukaryotic cells and colonized by 4 x1013 bacteria, roughly representing a 1:1 ratio [25]. The microbiome is constituted of bacteria, fungi, and viruses that colonize body surfaces exposed or opened to the outside environment such as the skin [26], the lung [27,28], the oral cavity [29] and the gastrointestinal tract [30]. Most of the current studies focus on the characterization of the bacterial microbiota and omit the presence of the virome and the mycobiome. Fungi are often considered as pathogenic, however many fungal species do not trigger disease and coexist with the bacterial microbiota. Compared to the bacterial microbiome, the characterization of the mycobiome composition is rendered more complicated by the lack of a complete fungal database; it is currently estimated that only 1% of fungal species are present in the NCBI GenBank database and among those sequences a large part are not classified correctly or of uncharacterized origin [31]. Currently, up to 75 different fungal genera have been described and are present at different body sites such as the oral cavity [32], intestine [33], skin [34] and lungs [35]. Commensal fungi also regulate immune responses and host physiology as fungal dysbiosis is associated with changes in the commensal bacteria composition that could potentially fuel further inflammation and disease [31,36,37].
Early microbial exposure influences the atopic status
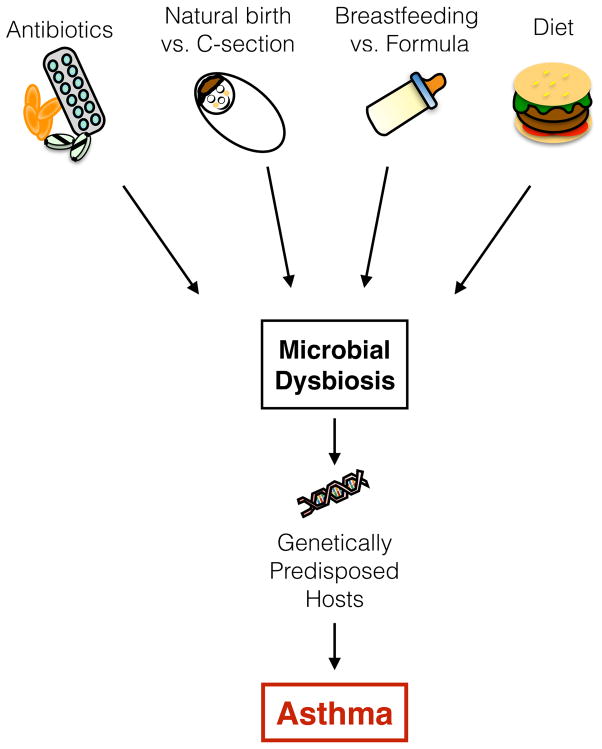
The continuous rise in asthma incidence in industrialized societies cannot be attributed to genetic factors alone and implies that some environmental factors resulting from the modern lifestyle promotes asthma [38] (Figure 2). The “hygiene hypothesis” states that personal hygiene improvement, declining family size and decreased infection burden result in reduced early-life microbial exposure and promotion of atopic diseases [39]. The current dogma is that microbial colonization begins at birth from exposure to the mother’s vaginal and fecal microbial flora, although recent studies demonstrate that microbial exposition may start earlier, in utero, as indicated by the presence of bacterial DNA in the placenta and meconium of pre-term babies [40,41]. Maternal farming microbial exposure during pregnancy is correlated with elevated levels of regulatory T cells (TReg) and decreased Th2 cytokine secretion [42]. Those studies suggest the possibility that in utero bacterial exposure could initiate colonization at different fetal surfaces during pregnancy. Alternatively, it is the maternal exposure that directly influences the fetal immune development. The mode of delivery is also known to strongly influence the infant gut bacterial colonization [43]. Infants born by Caesarian section (C-section) are at higher risks of developing atopic diseases [44,45] and are principally colonized by microbial communities similar to the mother’s skin, such as Staphylococcus species [43,46]. Early colonization patterns of the neonatal gut, but not the airways, is also affected by the mode of delivery; C-section promotes the emergence of Citrobacter, Klebsiella, Enterobacter, and Enterococcus species whereas E. coli colonization was associated with natural delivery [47]. Right after birth, inhalation of environmental microorganisms will result in lung and airways colonization [48,49]. In mice neonates, the main bacterial phyla of the lung microbiome, Proteobacteria and Firmicutes, will stabilize during the first month of life and aging is correlated with an outgrowth of Bacteroidetes [49]. Similar bacterial clusters were detected in the gut and respiratory airways of young infants diagnosed with cystic fibrosis, thereby suggesting the possibility of dynamic crosstalk and interactions between the intestinal and the respiratory microbiota [48].
Figure 2. Environmental factors influencing asthma development.
The continuous rise in asthma incidence in “westernized” societies cannot be only attributed to a genetic component alone; environmental factors resulting from a modern lifestyle are involved in this increased prevalence. The composition of the microbial flora is constantly fluctuating and strongly influenced by environmental factors. Microbial dysbiosis can be triggered by stress, the modern-lifestyle diet, antibiotic use, birth and feeding mode. By promoting the growth of pathogenic bacteria, microbial dysbiosis will also prevent early exposure to health-promoting bacteria. In genetically predisposed hosts, this alteration in microbial communities will promote the development of asthma.
Breast- versus formula-feeding is another factor that influences the infant microbiome composition and development. Breast milk shapes the infant gut microbial communities and promotes intestinal homeostasis through diverse mechanisms; it is a source of bacterial communities transmitted to the infant [50], it provides maternal antibodies, such as secretory IgA, that further promotes gut homeostasis [51], and supplies nutrients that shape the microbial flora composition during the first year of life [46]. Consequently, compared to formula-fed infants, breast-fed infants harbor a more uniform commensal flora composition associated with higher abundance of Bifidobacteria and Lactobacillus, whereas formula-fed infants display higher abundances of Bacteroides, Clostridium, Streptococcus, Enterobacteria, and Veillonella species (reviewed in [52]). Compared to formula-fed infants, breast-feeding is also associated with decreased risks of developing asthma and this trend is even higher in infants with family history of asthma [53–55]. However, whether infant feeding mode can be correlated with increased risks to develop atopic diseases still remains uncertain and will require further studies in order to identify the specific microbial communities present in breast-versus formula-fed infants.
Early microbial exposure and bacterial colonization are important for the maturation and the generation of a normal immune system. Compared to specific pathogen-free (SPF) mice, Germ-free (GF) mice which are devoid of any microbiota harbor distinct physical differences such as increased size cecum, decreased gastrointestinal motility, abnormal morphology, and smaller Peyer’s patches and mesenteric lymph nodes [56]. Absence of microbial flora in GF mice is linked to a Th2-skewed immune response and elevated levels of serum IgE [57,58]. Mono-colonization of GF mice with either Bacteroides fragilis or segmented filamentous bacteria will redirect and balance the T cell immune response towards stable Th1/Th2 or Th17 phenotypes respectively [59,60]. There is a critical “time window of opportunity” in early-life for an appropriate education of the immune system by the microbiota [56]. Therefore, early-life events able to alter the microbiota composition or disrupt microbiota-immune interactions can be deleterious and promote immune deviation towards atopy. In mice, interruption of microbial sensing by the immune system promotes the development of allergic inflammation and food allergy development has been associated with the emergence of a specific microbial signature [61–63]. In humans, the use of antibiotics during the pre- or post-natal period is associated with increased development of atopic dermatitis and asthma [64,65]. Early-life intestinal microbial flora dysbiosis in children is correlated with increased asthma development and a reduction of four specific bacterial genera: Faecalibacterium, Lachnospira, Veillonella and Rothia [66]. Supplementation of these 4 bacterial species in GF mice reduced asthma incidence in their adult offspring by decreasing neutrophil recruitment in the lungs and reducing the Th1/Th17 immune response associated with severe human asthma [66]. Whether early-life microbial dysbiosis triggered by environmental factors elicit allergic disease or is a consequence of allergic disease development remains unknown and will require further investigations.
Asthma and the lung microbiome
Until recently, healthy lungs and respiratory airways were considered sterile. The use of 16S rDNA sequencing techniques demonstrated the presence of a lung microbiota in healthy subjects as well as its specific topographical distribution since bacterial biomass decreases from upper to lower respiratory airways [27]. The characterization of the lung microbiome is rendered more difficult due to its low microbial load compared to other organs such as the skin, the oral cavity and the gut, and the possible presence of bacterial contaminants from either the upper respiratory airways during sample collection by bronchoscopy or contaminants present in DNA extraction kit [67].
In mice, absence of lung microbial colonization in OVA-sensitized and intranasally challenged GF mice results in enhanced allergic airway inflammation with increased levels of Th2 cytokines and serum IgE as well as augmented lymphocytes and eosinophils infiltrations. Colonization of GF mice by SPF flora before allergen exposure protects from allergic airway inflammation development [68]. Furthermore, SPF mice intra-nasally treated with either Staphylococcus sciuri or Escherichia coli were protected from allergic airway development [69,70].
In humans, 16s rDNA sequencing of various type of specimens, including bronchial brushing [71,72], BAL [73], and induced sputum samples [74] allowed the detection in the lower airways of bacterial species belonging to the 5 major phyla (Proteobacteria, Firmicutes, Actinobacteria, Fusobacterium and Bacteroidetes). The asthmatic airway microbiota composition differs from healthy subjects and is characterized by a higher bacterial load and diversity as well as increased abundance of species belonging to the Proteobacteria phylum such as Comamonadaceae, Nitrosomonadaceae, Oxalobacteraceae, Pastereurellaceae and Pseudomonadaceae families [71,72,74]. On the other hand, members from the Bacteroidetes and Firmicutes phyla have been found more abundantly in the airways of healthy controls subjects [71,72,74]. The microbial airway composition also differs based on clinical features and the type of immune response triggered by the aeroallergens. For instance, the airways of corticosteroid-resistant and severe asthmatic patients are predominantly colonized by pathogenic organisms such as M. catarrhalis or members of the Haemophilus or Streptococcus genera [75]. Th17 immune-mediated asthma exhibit a predominance of Proteobacteria belonging to the Pasteurellaceae, Enterobacteriaceae, and Bacillaceae families [76]. This specific bacterial signature was only observed in Th17-driven inflammation and did not overlap with a Th2-driven airway inflammation [76]. Studies linking the metabolic activities of those dysbiotic bacteria to asthma severity and treatment prognosis are further required. For example, steroid-resistance treatment could be related to the presence and expansion in the airways of bacterial species that are capable of degrading steroids.
Fungi are also present in the sputum of allergic and healthy subjects [77]. Malassezia pachydermatis; a fungus already associated with the development of atopic dermatitis, was present in the sputum of asthmatic patients and absent from healthy controls [77]. The last couple of years have seen huge improvement in high-throughput fungal rDNA sequencing techniques and refinement of fungal sequences databases [31]. Therefore, in order to gain a better understanding of the lung mycobiome function during asthma, further studies taking advantage of those advances are required to define the role of fungal colonization in asthma.
Asthma and the gut microbiome
The intestinal microbiota is composed of hundreds to thousands of bacterial species belonging mostly to the Firmicutes and Bacteroidetes phyla [24]. Its composition varies within the distinct section of the gastrointestinal tract and their physiological characteristics, resulting in different intestinal micro-habitats such as the gut lumen, colonic mucus and colonic crypts and their capacities to promote differently the growth of bacterial species (Reviewed in [78]). The bacterial composition of the intestinal microbiota is known to affect the development and the phenotype of immune responses [79]. Defined bacteria or their derived microbial products will trigger and modulate distinctively either a regulatory or an effector immune response. Commensal flora derived products, such as polysaccharide A of Bacteroides fragilis [80], several short-chain fatty acids (SCFAs) generated by fermentation of dietary fibers [81–83], or the colonization of GF mice by a defined mix of Clostridium strains [84,85], prevent intestinal pathology by promoting the differentiation of suppressor TReg cells.
On the other hand, the gut microbiota is a potent stimulus for the generation of pro-inflammatory and harmful autoimmune responses. Alterations in the intestinal flora composition are associated with disease development, however how intestinal dysbiosis affects the host systemic immune response remains uncertain. Diet is known to modulate the gut microbiota composition in humans and mice [48,86]. Increased asthma incidence in developed countries could be related to recent lifestyle changes characterized by reduced dietary fibers consumption and higher fat intake. Intestinal microbiota as well as their derived metabolites production can be influenced by those dietary components. Bacterial-derived metabolites are not specifically confined to the intestinal tract but can enter the blood circulation and affect immune cells and responses at distant sites. Trompette et al. demonstrated in a murine model of allergic airway inflammation that dietary fibers influence the intestinal microbial flora composition and increase the circulating levels of SCFAs [86]. Treatment with the SCFA propionate promotes the hematopoiesis of DCs and convert those cells into poor drivers of lung Th2 immune responses resulting in reduced airway inflammation [86].
The strong influence of diet on intestinal flora composition and bacterial metabolite production on atopic disease development has also been highlighted in human studies. In young children, development of allergic disease is often preceded by alteration of the intestinal microbial flora composition [11,87]. The intestinal human microbiota coevolves with the diet as demonstrated by a study of the intestinal microbiota composition of 2 different children cohorts; one from Europe and the other one from Burkina-Faso. The fiber rich diet from Burkinabe children is associated with intestinal microbiome compositional changes and the emergence of bacterial species producing high levels of SCFAs and specialized in energy intake from polysaccharides [88]. On the other hand, the western diet with reduced fiber and increased animal protein, sugar and starch content consumed by European children is correlated to a complete lack or under representation of those SCFAs bacterial producers and consequently with a significant decrease in SCFAs production [88].
How the commensal fungi within the gastrointestinal tract influences the systemic immune response and affects the development of allergic airway inflammation is still under investigation. Recently, in a murine model of allergic airway inflammation Wheeler et al. demonstrated that commensal fungi dysbiosis, obtained by means of oral antifungal treatment, is associated with exacerbated allergic airway inflammation and the growth of Aspergillus, Wallemia, and Epicoccum species [37]. Oral gavage of these 3 fungi into HDM-sensitized mice was sufficient to recapitulate similar and exaggerated levels of allergic airway inflammation as the ones observed in mice treated with antifungal drugs [37]. Importantly, fungal dysbiosis was also associated with a complete restructuring of the commensal microbial communities, including a decrease in abundance of Bacteroides, Clostridium, and Desulfovibrio and an increase of Anaerostipes, Coprococcus, and Streptococcus in mice with allergic airway inflammation [37]. On the other hand, gut bacterial dysbiosis induced by antibiotics treatment also promotes allergic airway inflammation by boosting the overgrowth of intestinal Candida fungal species and the polarization of lung M2 macrophages [89]. These data demonstrate that commensal fungi and microbial communities are deeply connected and dependent on one another thereby suggesting that the impact of the mycobiome on atopic disease development in underestimated and should be further investigated.
Conclusion
Asthma is an heterogeneous and complex disease that can manifest within a variety of different clinical features and patterns of immune responses. A growing number of studies demonstrate that asthma influences and is also impacted by the composition of the intestinal and respiratory microbiota. As discussed above, evidences generated from human and murine investigations indicate that lung microbial as well as intestinal microbial and fungi communities diverge between healthy and allergic airway inflammation. Additionally, the composition of those communities is also impacted by a wide variety of environmental factors such as diet and microbial exposures in “early-life”. The specific mechanisms by which this “gut-lung” microbiota axis impact health and disease requires further investigation. However, modulation of systemic immune responses by the intestinal microbial flora can occur through the release in the circulation of bacterial-derived metabolites with immuno-modulatory properties. Deciphering further the mechanisms and mediators of this “gut-lung” axis could potentially lead to a new therapeutic strategies aiming to modulate the composition of the respiratory and/or intestinal microbiome in order to promote resistance to allergic airway disease.
Key points.
Asthma is a complex and heterogeneous disease with strong genetic and environmental components that manifests within a variety of clinical features and immune responses.
Lung tissues are not sterile; fungi and bacterial species have been detected in various type of airway specimens collected from both healthy and asthmatic subjects.
The lower airway and intestinal microbial composition is different between healthy and asthmatic subjects and early-life intestinal microbial flora dysbiosis is correlated with increased asthma incidence.
Existence of a “gut-lung” axis that could potentially be used for therapeutics purposes.
Acknowledgments
Financial support and sponsorship.
This work was supported by the following grants: R01AI117968-01A1 (to M.A.) and R21AI112826-02 (to T.R.C.).
Abbreviation
- AHR
Airway hyper-responsiveness
- HDM
House Dust Mite
- IgE
Immunoglobulin type E
- Th2
T helper cells Type 2
- IL-
Interleukin
- BAL
Bronchoalveolar lavage
- ILC
Innate lymphoid cells
- DCs
Dendritic cells
- TReg
Regulatory T cells
- SPF
Specific pathogen free
- ICSs
Inhaled corticosteroids
- SCFA
short-chain fatty acid
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Locksley RM. Asthma and allergic inflammation. Cell. 2010;140:777–83. doi: 10.1016/j.cell.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambrecht BN, Hammad H. The immunology of asthma. Nat Immunol. 2014;16:45–56. doi: 10.1038/ni.3049. [DOI] [PubMed] [Google Scholar]
- 3.Romanet-Manent S, Charpin D, Magnan A, et al. Allergic vs nonallergic asthma: what makes the difference? Allergy. 2002;57:607–13. doi: 10.1034/j.1398-9995.2002.23504.x. [DOI] [PubMed] [Google Scholar]
- 4.Bush A, Zar HJ. WHO universal definition of severe asthma. Curr Opin Allergy Clin Immunol. 2011;11:115–21. doi: 10.1097/ACI.0b013e32834487ae. [DOI] [PubMed] [Google Scholar]
- 5.Wenzel S. Mechanisms of severe asthma. Clin Exp Allergy. 2003;33:1622–8. doi: 10.1111/j.1365-2222.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- 6.Moorman JE, Akinbami LJ, Bailey CM, et al. National surveillance of asthma: United States, 2001–2010. Vital Health Stat. 2012;3:1–58. [PubMed] [Google Scholar]
- 7.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sly PD, Boner AL, Björkstén B, et al. Early identification of atopy in the prediction of persistent asthma in children. Lancet. 2008;372:1100–6. doi: 10.1016/S0140-6736(08)61451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ege Markus J, Mayer M, Normand A-C, et al. Exposure to Environmental Microorganisms and Childhood Asthma. N Engl J Med. 2011;364:701–9. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 10.James KM, Gebretsadik T, Escobar GJ, et al. Risk of childhood asthma following infant bronchiolitis during the respiratory syncytial virus season. J Allergy Clin Immunol. 2013;132:227–9. doi: 10.1016/j.jaci.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalliomäki M, Kirjavainen P, Eerola E, et al. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–34. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 12.Robinson DS, Hamid Q, Ying S, et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992;326:298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 13.Woodruff PG, Modrek B, Choy DF, et al. T-helper Type 2–driven Inflammation Defines Major Subphenotypes of Asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel S, Ford L, Pearlman D, et al. Dupilumab in Persistent Asthma with Elevated Eosinophil Levels. N Engl J Med. 2013;368:2455–66. doi: 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 15.Corren J, Lemanske RF, Hanania NA, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 16.Woodruff PG, Khashayar R, Lazarus SC, et al. Relationship between airway inflammation, hyperresponsiveness, and obstruction in asthma. J Allergy Clin Immunol. 2001;108:753–8. doi: 10.1067/mai.2001.119411. [DOI] [PubMed] [Google Scholar]
- 17.Molet S, Hamid Q, Davoine F, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108:430–8. doi: 10.1067/mai.2001.117929. [DOI] [PubMed] [Google Scholar]
- 18.Brandt EB, Kovacic MB, Lee GB, et al. Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol. 2013;132:1194–1204. e2. doi: 10.1016/j.jaci.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Annunziato F, Romagnani C, Romagnani S. The 3 major types of innate and adaptive cell-mediated effector immunity. J Allergy Clin Immunol. 2015;135:626–35. doi: 10.1016/j.jaci.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Kim HY, Lee HJ, Chang Y-J, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nature Med. 2014;20:54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKinley L, Alcorn JF, Peterson A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181:4089–97. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ano S, Morishima Y, Ishii Y, et al. Transcription factors GATA-3 and RORγt are important for determining the phenotype of allergic airway inflammation in a murine model of asthma. J Immunol. 2013;190:1056–65. doi: 10.4049/jimmunol.1202386. [DOI] [PubMed] [Google Scholar]
- 23.Shaw DE, Berry MA, Hargadon B, et al. Association between neutrophilic airway inflammation and airflow limitation in adults with asthma. Chest. 2007;132:1871–5. doi: 10.1378/chest.07-1047. [DOI] [PubMed] [Google Scholar]
- 24.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164:337–40. doi: 10.1016/j.cell.2016.01.013. In this study, the authors revisit the common dogma that in the human body bacteria outnumber human cells. Until recently, most of the microbiome studies refer to a 10:1 bacteria to human cells ratio based on a crude assessment published in a study from 1977. By revisiting how the number of human cells and bacteria are counted, the authors estimate that the ratio of human cells to bacteria is 1:1. [DOI] [PubMed] [Google Scholar]
- 26.Grice EA, Segre JA. The skin microbiome. Nat Rev Micro. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–63. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–16. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghannoum MA, Jurevic RJ, Mukherjee PK, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scupham AJ, Presley LL, Wei B, et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Findley K, Oh J, Yang J, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–70. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charlson ES, Diamond JM, Bittinger K, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–45. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iliev ID, Funari VA, Taylor KD, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–7. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37***.Wheeler ML, Limon JJ, Bar AS, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016;19:865–73. doi: 10.1016/j.chom.2016.05.003. This study demonstrates that fungi dysbiosis induced by oral treatment with an antifungal drug exacerbates the severity of intestinal colitis and allergic airway inflammation. Fungal dysbiosis is associated with compositional change in both the mycobiome but also with dysbiosis in commensal bacterial flora. The authors also identified 3 fungal species which adoptive transfer increases the severity of allergic airway disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eder W, Ege MJ, Mutius von E. The asthma epidemic. N Engl J Med. 2006;355:2226–35. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 39.Strachan DP. Family size, infection and atopy: the first decade of the “hygiene hypothesis”. Thorax. 2000;55(Suppl 1):S2–10. doi: 10.1136/thorax.55.suppl_1.s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65–5. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moles L, Gómez M, Heilig H, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE. 2013;8:e66986. doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaub B, Liu J, Höppler S, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123:774–5. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 43.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renz-Polster H, David MR, Buist AS, et al. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy. 2005;35:1466–72. doi: 10.1111/j.1365-2222.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 45.Pistiner M, Gold DR, Abdulkerim H, et al. Birth by cesarean section, allergic rhinitis, and allergic sensitization among children with a parental history of atopy. J Allergy Clin Immunol. 2008;122:274–9. doi: 10.1016/j.jaci.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46***.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. This study analyzed the fecal microbiome of mothers and their infant during the first year of life in order to determine how the gut microbiota initiates and maturate in early life. The authors show that compared to vaginally delivered infants, the microbiome of infants born by C-section is more dissimilar with their respective mother. They also demonstrate that change in nutrients and cessation of breastfeeding was required for the infant flora to maturate into a more adult one. [DOI] [PubMed] [Google Scholar]
- 47***.Stokholm J, Thorsen J, Chawes BL, et al. Cesarean section changes neonatal gut colonization. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.01.028. [Epub ahead of print] In this study, the authors follow a cohort of Danish children and collect their fecal as well as hypopharyngeal microbiomes at different time-point in order to determine if the mode of delivery, vaginally versus C-section, influences intestinal and airways bacterial early colonization patterns. Intestinal microbiome composition differences, but not for the airways, were observed between delivery modes at 1 week. However with aging, those differences became less apparent. [DOI] [PubMed] [Google Scholar]
- 48.Madan JC, Koestler DC, Stanton BA, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio. 2012;3(4) doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nature Med. 2014;20:642–7. doi: 10.1038/nm.3568. [DOI] [PubMed] [Google Scholar]
- 50.Hunt KM, Foster JA, Forney LJ, et al. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE. 2011;6:e21313–8. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogier EW, Frantz AL, Bruno MEC, et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. PNAS. 2014;111:3074–9. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arrieta M-C, Stiemsma LT, Amenyogbe N, et al. The intestinal microbiome in early life: health and disease. Front Immunol Frontiers. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oddy WH, Holt PG, Sly PD, et al. Association between breast feeding and asthma in 6 year old children: findings of a prospective birth cohort study. BMJ. 1999;319:815–9. doi: 10.1136/bmj.319.7213.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gdalevich M, Mimouni D, Mimouni M. Breast-feeding and the risk of bronchial asthma in childhood: A systematic review with meta-analysis of prospective studies. J Pediatr. 2001;139:261–6. doi: 10.1067/mpd.2001.117006. [DOI] [PubMed] [Google Scholar]
- 55.van Odijk J, Kull I, Borres MP, et al. Breastfeeding and allergic disease: a multidisciplinary review of the literature (1966–2001) on the mode of early feeding in infancy and its impact on later atopic manifestations. Allergy. 2003;58:833–43. doi: 10.1034/j.1398-9995.2003.00264.x. [DOI] [PubMed] [Google Scholar]
- 56***.Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539–44. doi: 10.1126/science.aad9378. This review highlight recent evidences demonstrating the importance of early-life microbial colonization for the immune system maturation and how early-life microbial exposure can either promote tolerance or disease development later in life. The authors also review new data demonstrating the existence of a “window of opportunity” for early-life microbial colonization leading to either improper or proper immune education. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sudo N, Sawamura S, Tanaka K, et al. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–45. [PubMed] [Google Scholar]
- 58.Cahenzli J, Köller Y, Wyss M, et al. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–70. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 60.Ivanov II, Atarashi K, Manel N, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nature Med. 2012;18:538–46. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bashir MEH, Louie S, Shi HN, Nagler-Anderson C. Toll-Like Receptor 4 Signaling by Intestinal Microbes Influences Susceptibility to Food Allergy. J Immunol. 2004;172:6978–87. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 63.Noval Rivas M, Burton OT, Wise P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–12. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee S-Y, Yu J, Ahn K-M, et al. Additive effect between IL-13 polymorphism and cesarean section delivery/prenatal antibiotics use on atopic dermatitis: a birth cohort study (COCOA) PLoS ONE. 2014;9:e96603. doi: 10.1371/journal.pone.0096603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Metsälä J, Lundqvist A, Virta LJ, et al. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy. 2015;45:137–45. doi: 10.1111/cea.12356. [DOI] [PubMed] [Google Scholar]
- 66***.Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152–2. doi: 10.1126/scitranslmed.aab2271. This study demonstrate that change in the intestinal microbiome during the critical “window” of early colonization could be associated with asthma development. By characterizing the microbiome of children during their first 100 days of life, the authors of this study demonstrate that infants at higher risks of developing asthma exhibit intestinal dysbiosis associated decreased abundance of 4 bacterial genera. [DOI] [PubMed] [Google Scholar]
- 67.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herbst T, Sichelstiel A, Schär C, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 69.Hagner S, Harb H, Zhao M, et al. Farm-derived Gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy. 2013;68:322–9. doi: 10.1111/all.12094. [DOI] [PubMed] [Google Scholar]
- 70.Nembrini C, Sichelstiel A, Kisielow J, et al. Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax. 2011;66:755–63. doi: 10.1136/thx.2010.152512. [DOI] [PubMed] [Google Scholar]
- 71.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. In: Neyrolles O, editor. PLoS ONE. Vol. 5. 2010. p. e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. e1–3. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goleva E, Jackson LP, Harris JK, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188:1193–201. doi: 10.1164/rccm.201304-0775OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marri PR, Stern DA, Wright AL, et al. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–52. e1–3. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–84. doi: 10.1016/j.jaci.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Woerden HC, Gregory C, Brown R, et al. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Micro. 2015;14:1–13. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molloy MJ, Bouladoux N, Belkaid Y. Intestinal microbiota: shaping local and systemic immune responses. Seminars in Immunology. 2012;24:58–66. doi: 10.1016/j.smim.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 84.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Atarashi K, Tanoue T, Oshima K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 86.Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Med. 2014;20:159–66. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 87.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56:661–7. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. PNAS. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim Y-G, Udayanga KGS, Totsuka N, et al. Gut Dysbiosis Promotes M2 Macrophage Polarization and Allergic Airway Inflammation via Fungi-Induced PGE2. Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]