Abstract
Vibrio cholerae has become a model organism for studies connecting virulence, pathogen evolution and infectious disease ecology. The coordinate expression of motility, virulence and biofilm enhances its pathogenicity, environmental fitness and fecal-oral transmission. The histone-like nucleoid structuring protein negatively regulates gene expression at multiple phases of the V. cholerae life cycle. Here we discuss: (i) the regulatory and structural implications of H-NS chromatin-binding in the two-chromosome cholera bacterium; (ii) the factors that counteract H-NS repression; and (iii) a model for the regulation of the V. cholerae life cycle that integrates H-NS repression, cyclic diguanylic acid signaling and the general stress response.
Keywords: H-NS, Vibrio cholerae, Life cycle
1. Introduction
Cholera is an acute, water-borne diarrheal disease caused by the Gram-negative and motile bacterium Vibrio cholerae of serogroup O1 of the classical and El Tor biotypes and serogroup O139. Mankind has experienced seven recorded cholera pandemics. The seventh and current pandemic is characterized by the predominance of O1 strains of the El Tor biotype with sporadic emergence of serogroup O139. Endemic cholera continues to be a major public health problem in vast regions of South Asia and Africa. In these areas, the occurrence of cholera exhibits a seasonal pattern that correlates with climatic changes. Introduction of virulent V. cholerae O1 in non-endemic areas with poor sanitation commonly results in rapidly spreading outbreaks.
Cholera vibrios gain access to the human small intestine by oral ingestion of contaminated water and food. In the gastrointestinal tract, vibrios are exposed to low pH, bile acids, elevated osmolarity, iron limitation and antimicrobial peptides. Nevertheless, V. cholerae can grow to high titers in the human gut, and cholera patients commonly shed 107–109 virulent vibrios per mL back to the environment in the rice-watery stool. In the aquatic environment, vibrios withstand diverse physical, chemical and biological stresses that include nutrient limitation, extreme temperatures, oxidative stress, bacteriophage predation and protozoan grazing. V. cholerae employs multiple strategies to cause infection and persist outside the human host. These include: (i) the activation of general and specific stress responses; (ii) expression of chemotaxis and motility; (iii) surface attachment; (iv) formation of biofilms; and (v) detachment. In this review we examine the critical contribution of the histone-like nucleoid structuring protein (H-NS) to V. cholerae adaptation to its diverse and dynamic environments. The emerging theme is a regulatory pattern in which H-NS silences large gene clusters required for vibrio adaptation to specific environments, including its ability to switch between alternative lifestyles. Genes within these clusters are turned on in response to environmental changes by the action of transcription factors that counteract H-NS repression.
2. The nucleoid-associated protein family
The nucleoid-associated proteins comprise a group of basic, low molecular weight DNA binding proteins that participate in chromatin organization, restraining of DNA supercoiling and transcription regulation. The family includes H-NS, the factor for inversion stimulation (Fis), integration host factor (IHF), the heat-stable protein HU, and the leucine-responsive protein (Lrp). A vast literature has accumulated on the structure and function of these proteins in Escherichia coli and Salmonella (reviewed in [1]). In contrast to E. coli and Salmonella, V. cholerae contains two circular chromosomes [2] and has evolved to colonize disparate ecological niches such as the chitinous exoskeleton of crustacean [3], chironomid egg masses [4], the contractile vacuole of Acanthamoeba castellanii [5] and the mucosal side of the human small intestine [6]. These differences may demand significant variation in the mode through which nucleoid-associated proteins execute their interrelated architectural and regulatory functions. Amongst the nucleoid-associated protein family, Fis was shown to indirectly affect V. cholerae virulence by modulating quorum sensing [7]. The IHF enhances expression of the ctxAB operon encoding the A and B subunits of cholera toxin (CT) [8]. However, most studies related to the function of nucleoid-associated proteins in pathogenic vibrios have centered on H-NS. Here we focus on: (i) the regulatory and structural implications of H-NS DNA binding in the two-chromosome cholera bacterium; (ii) the factors that counteract H-NS repression; and (iii) we discuss a model for the regulation of V. cholerae behavior that integrates quorum sensing, the general stress response, cyclic diguanylic acid (c-di-GMP) signaling and H-NS antirepression.
3. The histone-like nucleoid-structuring protein
H-NS is a highly abundant protein that functions as a nucleoid organizer and a transcriptional silencer at promoters exhibiting AT-rich highly-curved DNA. Several excellent reviews have been published covering the structure and function of H-NS in E. coli and Salmonella [9,10]. In this review, we examine the regulatory function of H-NS in V. cholera, highlighting similarities and differences found between vibrios and the above Enterobacteriaceae. The E. coli 15 kDa and 137 amino acid H-NS protein consists of an N-terminal coiled-coil oligomerization domain connected by a flexible linker to a nucleic acid binding domain. The N-terminal oligomerization domain exhibits two dimerization interfaces located within residues 1–46 and 57–80. The functional H-NS subunit binds DNA as a dimer that associates through the N-terminal dimerization interface. Simultaneous dimerization through the N-terminal and central interfaces results in the formation of high order oligomers or fibers. H-NS fibers can associate with a single stretch of a DNA molecule (stiffening mode) or bring together distant regions of the DNA duplex (bridging mode) to create topological domains. The C-terminal DNA-binding domain of H-NS is highly conserved among H-NS orthologs, whereas the N-terminal oligomerization region appears more variable. Both DNA binding and oligomerization are required for the biological activities of H-NS [11]. H-NS has been suggested to exploit a variety of mechanisms to silence transcription. Reported mechanisms include: (i) direct occlusion of promoter −10 and −35 elements (promoter occlusion); (ii) entrapment of RNA polymerase within a repression loop; (iii) binding to DNA upstream from the promoter followed by oligomerization to contact and stall RNA polymerase; and (iv) constraining DNA supercoiling [12–14]. Promoters repressed by H-NS may also contain several H-NS binding sites to which H-NS can bind in a positive cooperative manner [15].
The 135 amino acid V. cholerae H-NS protein shares 69 % similarity and 55 % identity with E. coli H-NS, with most differences located outside the DNA binding domain. Site-directed mutagenesis of V. cholerae H-NS suggested that more extensive regions of the V. cholerae protein contribute to oligomerization compared to E. coli and Salmonella H-NS [16]. The biological significance of this difference is unclear. However, a 2.5Å crystal structure of V. cholerae H-NS showed that the vibrio and E. coli H-NS N-terminal oligomerization domains adopt a similar fold and dimeric assembly [17].
The vicH (hns) gene encoding V. cholerae H-NS was cloned by complementation of an E. coli hns mutant [18]. The gene was shown to be expressed from two promoters arranged in tandem and enhanced by cold shock [18]. Cold shock induction of hns transcription was consistent with the presence of a Y-box for binding of cold shock protein A in its promoter region [18]. Chromatin immunoprecipitation (ChiP) and parallel DNA sequencing (ChIP-Seq) showed that H-NS binds to its own promoter, suggesting that, similarly to E. coli, H-NS can negatively regulate its own expression in V. cholerae [19,20]. The regulation of H-NS expression could be more complex, as one study showed that E. coli H-NS oligomerization and activity can be modulated by temperature and osmolarity [21]. Analogous studies have not been conducted in vibrios, which nevertheless are subject to similar stressors in the aquatic environment and the human host.
In E. coli, truncated H-NS proteins lacking the DNA binding domain act as H-NS antagonists [22]. Similarly, overexpression of a C-terminal truncated hns allele in V. cholerae of the classical biotype induced pleiotropic effects such as the formation of mucoid colonies and reduced swarming in semisolid agar [18]. El Tor biotype V. cholerae hns mutants formed small colonies in agar plates, exhibited diminished growth rate in broth, elongated cell morphology and enhanced resistance to low pH and hydrogen peroxide [23,24]. Although El Tor biotype hns mutants appeared flagellated and expressed elevated flagellin A, they showed reduced motility in semisolid agar [24]. Finally, deletion of hns induced an endogenous envelope stress response resulting in elevated expression of rpoE encoding the extracytoplasmic sigma factor E (σE) [25].
4. H-NS global regulation and chromatin binding in V. cholerae
Transcription profiling of a V. cholerae hns mutant of the El Tor biotype by RNA-Seq showed that H-NS affects the expression of 18% of all predicted genes in a growth-phase-dependent manner. In exponentially growing cells cultivated in rich medium, 84% of differentially expressed genes were repressed. However, in cells harvested from the same medium in stationary phase, only 48% of differentially expressed genes were repressed by H-NS [25]. The change in the percentage of repressed genes in stationary phase was explained by the expression of nucleoid-associated proteins, transcription factors and the RNA polymerase σS subunit (RpoS) reported to antagonize H-NS repression in stationary phase [25]. Similar to other bacteria, positive regulation by H-NS was found to be mostly indirect and included multiple genes predicted to affect chemotaxis, explaining the diminished motility of El Tor biotype hns mutants in semisolid agar in spite of being flagellated [24,25].
It has been suggested that H-NS acts to silence the expression of genes acquired by horizontal gene transfer [26,27]. The V. cholerae genome exhibits 17 putative genomic islands encompassing 446 genes [28]. Of these genes, 66 (15%) were found to be repressed by H-NS [25]. This percentage is higher than the percent of genes repressed within the entire genome (7%), suggesting a statistical bias for repression of genes located within genomic islands. The majority of genes predicted to be laterally acquired and repressed by H-NS were located within the 40 kb Vibrio pathogenicity island (VPI) in chromosome I and the 125 kb superintegron located on chromosome II [19,20]. In addition, H-NS silenced transcription of the rtx cluster, encoding a member of the repeat toxin superfamily and its transport system [25]. This cluster, located adjacent to the CTX prophage in chromosome I, exhibits features suggesting recent acquisition by current pandemic V. cholerae strains [29]. H-NS, however, represses many genes outside predicted genomic islands, suggesting a much broader function.
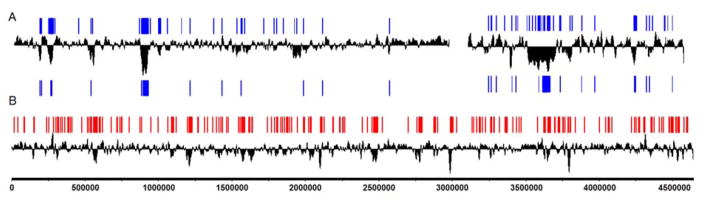
ChIP-Seq showed that H-NS associates with 6.3% of the V. cholerae genome [19,20]. Similarly to enterobacteria, H-NS was found preferentially associated with regions in the genome exhibiting low GC content. However, this study revealed important differences in the manner in which H-NS associates with the chromatin between vibrios and E. coli. As shown in Fig. 1, the association of H-NS with V. cholerae chromosomes appeared significantly uneven compared to the more uniform distribution observed along the E. coli chromosome [19,20,30]. A similar pattern was observed in independent ChiP-Seq analysis using a different El Tor biotype V. cholerae strain (Fig. 1) [31]. H-NS bound to fewer sites along V. cholerae chromosomes, but occupied longer stretches of DNA compared to E. coli. This pattern could be due to the presence of an additional oligomerization domain in V. cholerae H-NS [16], which may allow the protein to form oligomers of higher order in V. cholerae. Binding of H-NS in V. cholerae showed higher bias toward regions located upstream from open reading frames (85%) compared to E. coli (67%) [19,20,30]. These differences raise the question of whether binding of H-NS to chromatin plays a similar architectural role in two-chromosome versus single-chromosome bacteria [32]. The uneven binding pattern of H-NS observed in V. cholerae suggests that the H-NS bridging activity may play a more regional role in chromatin organization. It remains unclear as to whether the differences observed between V. cholerae and E. coli result from adaptation to different ecological niches, chromosome number or other undetermined factors. It is noteworthy that more of chromosome 2 (8.3%) is bound by H-NS compared to chromosome 1 (5.6%) [19,20]. Chromosome conformation capture revealed architectural differences between V. cholerae chromosomes [33]. Chromosome 1, which exhibits less H-NS binding, was found to adopt a more open structure [33]. However, the potential role of H-NS binding in this difference remains to be determined.
Fig. 1. Comparison of the distribution of H-NS clusters in the V. cholerae and E. coli genomes.
A. H-NS occupancy along V. cholerae chromosomes I (left) and II (right) is represented by blue bars. Top blue bars correspond to ChIP-Seq peaks in V. cholerae C7258 [19,20]. Bottom blue bars correspond to ChIP-Seq peaks in V. cholerae C6706 [31]. B. H-NS occupancy along the chromosome of E. coli K-12 strain MG1655 is represented by red bars [30]. Deviation from average GC content is plotted along chromosomes in black.
Recent studies have led to the view that organization of the chromatin by H-NS and transcriptional silencing are interrelated functions of this protein in which gene regulation drives nucleoid organization [10,34,35]. In V. cholerae, significant clustering of H-NS is observed at large operons controlling surface attachment and pathogenicity such as VPI, the ctxAB operon; the vas (virulence associated secretion) operon encoding a type VI secretion system, the vps-rbm cluster encoding components of the biofilm matrix and the chitin utilization program [19,20]. Clustering of H-NS at these sites of the chromatin could bring these regions into proximity, rendering their coordinate regulation more effective. Further, H-NS clustering at these sites could function to synchronize expression of coregulated genes in response to environmental conditions that affect DNA supercoiling, such as temperature, pH, osmotic shifts, transitions from aerobiosis to anaerobiosis and starvation. The following examples illustrate how the DNA bridging activity of H-NS could enhance coordinate gene expression. For instance, bridging could bring into closer proximity the VPI and ctxAB operon to facilitate communication between their common regulatory gene toxT and its target promoters. Another example is provided by the bitopic regulator ToxR. The cytoplasmic domain of the transmembrane protein ToxR directly activates the transcription of toxT, ompU and ompT (encoding outer membrane proteins) and the vpsL-Q operon while remaining anchored to the inner membrane [31,36]. The bridging activity of H-NS could facilitate chromosome-inner membrane interactions at several sites, allowing ToxR molecules to simultaneously engage multiple target promoters. The V. cholerae chitin utilization program provides an additional example of how H-NS chromatin organization could contribute to coordinate gene expression. Significant binding of H-NS can be detected at loci encoding the chitin-regulated pilus (ChiRP, VC2423) and chitoporin (ChiP, VC0972) [19,20]. Both genes are under positive regulation by the sensor kinase ChiS and are required for colonization of chitin and its utilization as sole carbon source [37]. The mechanism of this regulation is not fully understood, but similarly to VPI and ctxAB, a nucleoid structure placing these genes in close proximity to their regulator may well favor their coordinated expression.
5. H-NS transcriptional silencing and anti-silencing
Vibrio transition from the aquatic environment to the human host requires significant changes in gene expression patterns. These changes ensure effective dispersal of infecting biofilms, colonization of the distal small intestine, expression of virulence factors, detachment from the microvilli and in vivo biofilm formation [6]. Adaptive gene expression is further required upon vibrio transmission to a secondary host or shedding of bacteria back to the aquatic environment. Recent studies suggest that H-NS acts at multiple stages of the V. cholerae life cycle as a transcription gatekeeper to prevent gratuitous gene expression that could negatively impact fitness. Commonly, genes repressed by H-NS are expressed in response to environmental cues that induce expression of transcription factors that antagonize H-NS binding, bridging or oligomerization [9].
5.1. H-NS silencing and anti-silencing of the ToxR regulon
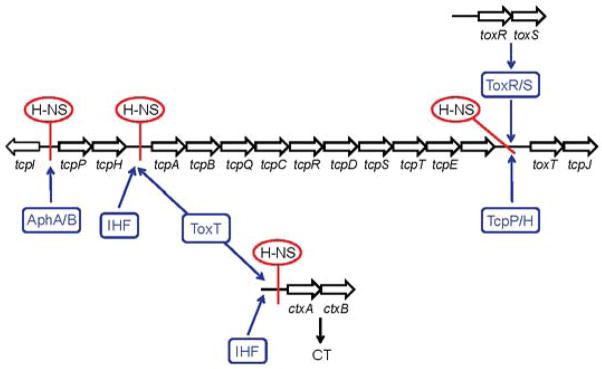
CT and the toxin co-regulated pilus (TCP) are the major virulence factors produced by cholera vibrios responsible for intestinal colonization and the rice-watery diarrhea typical of this illness. The expression of CT and TCP is regulated by a complex regulatory network (Fig. 2). At the top of the regulatory cascade, transcription factors AphA and AphB enhance the expression of the transmembrane regulators TcpP and TcpH. TcpP/H, in concert with transmembrane regulators ToxR/S, activates the expression of the soluble AraC-family regulator ToxT. Finally, ToxT interacts with the ctxA and tcpA promoters to activate production of CT and TCP, respectively (reviewed in [38]). The genes regulated by ToxR and ToxT are collectively known as the ToxR regulon. In Fig. 2, we show that H-NS represses the ToxR regulon at multiple levels, which include transcription initiation at the tcpPH promoter [25,39], the toxT promoter [40] and the tcpA and ctxAB promoters [40]. ChIP-Seq showed that H-NS occupies a large stretch of DNA encompassing the tcpPH promoter [19,20]. AphA and AphB are the only transcription factors known to activate tcpPH transcription in a cooperative manner [39]. DNAse I footprinting showed that AphB binds more proximally to the promoter, suggesting that it acts as the primary activator contacting RNA polymerase [39]. The specific mechanism by which H-NS represses tcpPH transcription is not clear, but it could entail cooperative binding to multiple sites followed by oligomerization. It is possible that binding of AphA and/or AphB could alleviate H-NS repression at the tcpPH promoter by displacing H-NS from occupied regions or preventing its oligomerization along the promoter. ToxR recruits TcpP to the toxT promoter, which interacts with RNA polymerase to enhance its transcription [41]. An antagonistic effect between the activators of toxT transcription and H-NS has not been confirmed by experiments. However, there are significant overlaps between ToxR-protected regions at the toxT promoter and H-NS binding motifs, suggesting that ToxR could function as an antirepressor. In the case of the tcpA promoter, ToxT was shown to function as both an H-NS antirepressor and RNA polymerase activator [42]. The ctxAB promoter contains four H-NS binding regions, two of which overlap ToxT binding sites [43]. As expected, ToxT was shown to displace H-NS from the promoter [43]. Transcription regulation at the tcpA promoter differed from ctxAB in that H-NS showed higher affinity for the ctxAB promoter, whose activation by ToxT did not involve interaction with RNA polymerase [43]. The transcription of ctxAB and tcpA is positively regulated by a second nucleoid-associated protein, IHF [8]. DNAse I footprinting and competitive DNA binding assays showed that H-NS and IHF associate with overlapping DNA sequences at the tcpA promoter, suggesting an antagonistic effect between these nucleoid-associated proteins at this locus [8].
Fig. 2. Transcriptional silencing by H-NS and anti-silencing of the ToxR regulon.
H-NS repression is represented by a red line through the intergenic regions. The blue arrows indicate transcription activation at the corresponding promoter. Abbreviations: CT, cholera holotoxin; IHF, integration host factor.
A recent study showed that the ToxR regulon is significantly larger than previously recognized and encompasses multiple genes repressed by H-NS, impacting intestinal colonization and biofilm development [31]. ChiP-Seq showed a 39% overlap between regions bound by ToxR and H-NS in the V. cholerae genome. A double mutant analysis showed that the colonization defect of a toxRS mutant was reversed by deletion of hns [31]. The authors reported that ToxR antirepression at newly found loci did not require partnership with TcpP [31].
The ToxR regulon exemplifies the role of H-NS in promoting fitness by silencing gratuitous gene expression. Classical biotype strains, which have been largely supplanted by the El Tor biotype, express CT and TCP in LB medium at 37°C, whereas El Tor biotype strains require special culture conditions that mimic the environment of the small intestine. The different requirement for toxin production is due to the differential regulation of tcpPH transcription in V. cholerae biotypes [44]. El Tor biotype hns mutants that overexpress tcpPH produce CT and TCP in LB medium, similarly to classical biotype strains [40]. Although the causes of the V. cholerae O1 population shift from classical to El Tor biotypes are not clearly understood, loss of H-NS in El Tor biotype V. cholerae mimics an evolutionary backward change. A recent study of wave 3 El Tor O1 strains isolated during the 2010 cholera outbreak in Haiti revealed that these isolates, which express elevated CT and hemolysin compared to earlier wave 1 strains, harbored mutations in the hns gene [45]. This finding highlights the importance of H-NS transcriptional silencing and antirepression in pathogen evolution.
5.2. H-NS silencing of the transcription of ancillary toxins
V. cholerae produces additional toxic factors outside the ToxR regulon, such as hemolysin (HlyA) and the repeat toxin (RTX) with confirmed noxious activities in cell culture and animal models [46]. Expression of these toxins could modulate the course of an infection from asymptomatic to severe in a strain-specific manner. Transcription profiling of a V. cholerae O1 El Tor hns mutant showed that H-NS silences transcription of numerous potential virulence factors outside the ToxR regulon [19,20,25] (Table 1).
Table 1.
Selected virulence traits outside the ToxR regulon silenced by H-NS.
| Locus | Gene product |
|---|---|
| VC1415 | Hcp-1, secreted hemolysin (HlyA)-co-regulated protein |
| VC1446 | RtxE, repeat toxin transporter |
| VC1447 | RtxD, repeat toxin transporter |
| VC1448 | RtxB, repeat toxin transporter |
| VC1450 | RtxC, repeat toxin-activating protein |
| VC1451 | RtxA, repeat toxin |
| VCA0017 | Hcp-2, secreted hemolysin (HlyA)-coregulated protein |
| VCA0107 - VCA0123 | Virulence-associated secretion (vas) operon |
| VCA0218 | Thermolabile hemolysin |
| VCA0219 | HlyA, hemolysin |
| VCA0220 | HlyB, hemolysin secretion protein |
5.2.1. The El Tor hemolysin/cytolysin
RNA-Seq analysis and DNA binding assays showed that H-NS binds to the hlyA promoter to silence the expression of the El Tor pore-forming hemolysin/cytolysin (HlyA) [25]. Transcriptional silencing of hlyA by H-NS was also reported in V. vulnificus [47] and V. anguillarum [48]. Expression of hlyA in V. cholerae is regulated by iron [49] and HlyU, a member of the SmtB/ArsR family of metalloregulators [50]. Deletion of hlyU diminished hlyA expression in the absence of H-NS, suggesting that HlyU does not act solely as an antirepressor. ChIP, however, showed that ectopic expression of HlyU diminishes H-NS occupancy at the hlyA promoter [25]. Taken together, the above data suggests that activation of hlyA transcription could involve both antirepression and activation of RNA polymerase. The regulation of hlyA expression in V. cholerae provides another example of the contribution of H-NS transcriptional silencing and anti-silencing to fitness. In wild type V. cholerae, expression of hlyA is induced by iron limitation, a well-recognized host signal [49]. The hns mutant, though overexpressing hlyA under laboratory conditions, was unresponsive to iron limitation [25].
5.2.2. The repeat toxin cluster
V. cholerae produces a multifunctional toxin, a member of the RTX family that causes actin depolymerization [29]. The RTX gene cluster in V. cholerae encodes the presumptive cytotoxin (rtxA), an acyltransferase (rtxC) and an associated ATP-binding cassette transporter system (rtxB and rtxD). H-NS binds to the intergenic region located between the divergently transcribed rtxAC and rtxBD genes to repress their transcription [25]. In V. vulnificus, HlyU enhances the transcription of rtx genes by antagonizing H-NS repression [51]. HlyU also enhances the transcription of rtx genes in V. cholera, but is still required for maximal expression of rtxCA and rtxBD in the absence of H-NS [25]. Further, ectopic expression of HlyU did not significantly influence H-NS occupancy at the intergenic region between rtxCA and rtxBD, suggesting that the interplay between H-NS and HlyU at this promoter differs in V. cholerae and V. vulnificus.
5.3. H-NS regulation of biofilm development
V. cholerae transition between motile and sessile (biofilm) lifestyles is regulated by the intracellular concentration of c-di-GMP [52]. Three major regulators sense the intracellular concentration of c-di-GMP: the σ54-dependent activator FlrA required for the expression of flagellar motility [53] and the biofilm activators VpsR [54] and VpsT [55].
The genes responsible for making the V. cholerae exopolysaccharide matrix are located in two clusters (vpsU, vpsA-K) and vpsL-Q on V. cholerae chromosome I [56]. These clusters comprise two operons in which vpsA and vpsL are the first genes of operon I and II, respectively [56]. A third gene cluster, rbmA-F, located between the vpsA-K and vpsL-Q operons and bap1, encodes protein components of the biofilm matrix [57]. When the intracellular concentration of c-di-GMP is elevated, VpsR and VpsT enhance the expression of vps and rbm genes required to make the biofilm matrix [52].
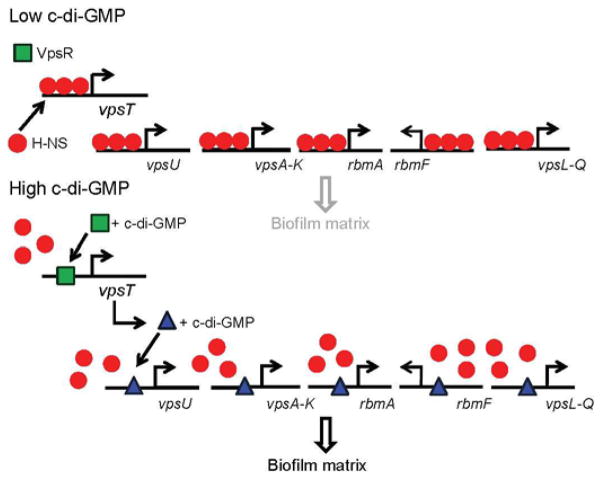
In V. cholerae, H-NS represses biofilm formation [58] by binding to the promoters of genes vpsU, vpsA-K, rbmA, rbmF-D, vpsL-Q and vpsT [25,58,59]. The promoters of vpsU, vpsA, rbmA, rbmF and vpsL contain overlapping H-NS and VpsT binding motifs [19,20]. Further, DNase I footprinting showed that H-NS and VpsT protect overlapping DNA sequences at these promoters [19,20]. In the presence of elevated c-di-GMP, VpsT displaces H-NS from promoters to activate vps and rbm gene expression, whereas at low c-di-GMP levels, H-NS displaces VpsT from promoters to reset gene expression to silent [20]. Deletion of vpsT enhanced H-NS occupancy at the vpsU, vpsA, rbmA, rbmF and vpsL promoters, while artificially increasing the c-di-GMP pool had the opposite effect [20]. Although VpsT associates with its own promoter [60], its binding site is located upstream and does not overlap with the H-NS binding site [20]. Consistently, deletion of the vpsT open reading frame does not diminish H-NS occupancy at its own promoter [20]. However, the vpsT promoter contains a VpsR binding motif immediately downstream from the H-NS binding site [59]. This arrangement suggests that binding of VpsR to the vpsT promoter could debilitate H-NS binding or block its oligomerization along the promoter. It was shown that artificially increasing the c-di-GMP pool diminished H-NS occupancy at the vpsT promoter and that this effect required VpsR [20]. Inactivation of AphA, which binds to the vpsT promoter, increased H-NS occupancy at this site. However, AphA was not required for c-di-GMP-induced H-NS antirepression. We note that the capacity of AphA to diminish H-NS occupancy could contribute to antirepression at other loci such as the tcpP promoter discussed previously. The above results suggest that the biosynthesis of the biofilm extracellular matrix is regulated by an H-NS antirepression cascade that responds to environmentally induced fluctuations in the c-di-GMP pool (Fig. 3). A recent study proposing that ToxR binds to the vpsL promoter to antagonize H-NS repression [31] suggests that additional signals are channelled through this transmembrane regulator to release biofilm development from H-NS repression.
Fig. 3. Model for the regulation of vps and rbm gene expression by H-NS transcriptional silencing and anti-silencing.
H-NS binds to the promoters of vps and rbm genes to silence their expression when the c-di-GMP concentration is low. When the c-di-GMP pool is elevated, VpsR and VpsT sequentially displace H-NS from promoters to turn on gene expression. Diminished biosynthesis of the biofilm matrix at low c-di-GMP due to transcriptional silencing is indicated by a light gray font.
We note that, similarly to ToxR, VpsT could have a wider role as an H-NS antirepressor. Overlapping H-NS and VpsT binding motifs are present upstream from the open reading frames encoding the RTX toxin transporter, hemolysin, the ankyrin-like protein AnkB, catalase KatB and the thermolabile hemolysin [20]. The finding that artificially increasing the c-di-GMP pool enhanced expression of rtxC, hlyA, ankB and katB supports a more global role for VpsT as an H-NS antirepressor [61]. These results suggest an unrecognized role for VpsT and c-di-GMP in the anti-silencing of V. cholerae ancillary toxins that could modulate the course of an infection.
5.4. H-NS regulation of mucosal escape
The expression of motility requires a hierarchical regulatory cascade that begins with the alternative RNA polymerase subunit σ54 (RpoN) and the σ54-dependent transcriptional activator FlrA [62]. Binding of c-di-GMP to FlrA results in downregulation of the entire flagellar system [53]. Both the housekeeping RNA polymerase σ70 and σS (RpoS) subunits contribute to transcription initiation at the rpoN and flrA promoters [24]. Consequently, V. cholerae rpoS mutants exhibit diminished flagellar motility compared to wild type strains [23,24,63].
Detachment of vibrios from abiotic or biotic surfaces and mature biofilms is a critical step in the bacterium’s life cycle. The reversal of a cell population from a sessile to a motile lifestyle is favored by environmentally-induced downshifts in the c-di-GMP pool. Expression of the quorum sensing regulator HapR at high cell density and RpoS in stationary phase diminish the c-di-GMP pool [64,65]. Lowering of the c-di-GMP pool enhances motility by increasing the activity of FlrA [53]. Experiments using the rabbit ileal loop model showed that V. cholerae cells detached from the intestinal epithelium into the lumen in stationary phase and that this process, known as “mucosal escape”, was dependent on RpoS activation of motility [63]. Analysis of rpoN-lacZ and flrA-lacZ promoter fusions and DNA binding assays demonstrated that H-NS binds to the rpoN and flrA promoters to diminish their transcription [24]. Expression of RpoS in the stationary phase enhanced transcription of rpoN and flrA by two distinct mechanisms: transcription initiation resistant to H-NS and displacement of H-NS from the rpoN and flrA promoters [24]. The first mechanism is consistent with studies in E. coli and Salmonella suggesting that H-NS contributes to σS promoter specificity [66]. Open initiation complex formation by RNA polymerase containing σ70 induces topological changes that facilitate lateral oligomerization of H-NS by cooperative recruitment of H-NS molecules to form a repression loop [67]. In contrast, the complex formed by RNA polymerase containing σS does not promote H-NS oligomerization and exhibits H-NS-resistant transcription initiation [67]. The second mechanism of RpoS-mediated antirepression is indirect and entails the induction in stationary phase of another nucleoid-associated protein, IHF [24]. A comparison of H-NS occupancy at the flrA and rpoN promoters between wild type V. cholerae and its isogenic ΔihfA mutant using a ChiP assay showed that IHF acts to diminish H-NS occupancy and repression at these loci [24]. Consistent with the occurrence of both antirepression mechanisms, a double deletion mutant lacking the A subunit of IHF and RpoS exhibited the highest (approximately additive) H-NS occupancy at the flrA promoter [24].
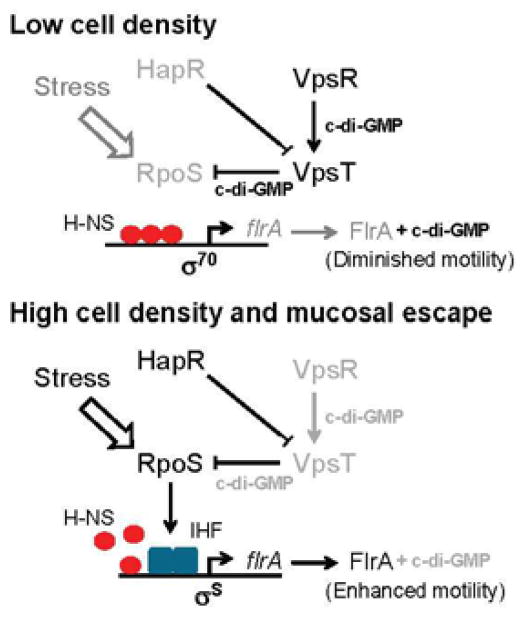
The biofilm activator VpsT represses the transcription of rpoS in a c-di-GMP-dependent manner by binding to its promoter at a region that overlaps a transcription initiation site [68]. This findings is integrated into the model shown in Fig. 4 for detachment or mucosal escape involving quorum sensing, VpsT and RpoS. In a low-cell-density population, HapR is not expressed, the c-di-GMP content is high and VpsT silences the transcription of rpoS [68]. In a high-cell-density population, HapR is expressed to lower the c-di-GMP pool and terminate the transcription of vpsT [64]. In the absence of VpsT, RpoS is expressed to further lower the c-di-GMP pool and relieve flrA and rpoN from H-NS repression [24,65,68]. The end result is activation of the flagellar system so that vibrios can detach and swim towards an unspent substratum. We note that HapR and RpoS also activates the expression of hemagglutinin/protease, a mucinase shown to promote detachment from the mucus gel lining the small intestine [69]. It is worth mentioning that, during infection, vibrios detach from the villi into the bile-rich luminal compartment prior to exiting the host. The elevated bile concentration in this compartment enhances the c-di-GMP pool and favors biofilm formation [70]. This finding is consistent with the notion that V. cholerae increases its c-di-GMP pool late in infection, an event that could enhance the fitness of vibrios returning to the aquatic environment [71]. Since V. cholerae biofilms exhibit a lower infective dose, this late event could also increase the probability of vibrio transmission to a secondary host [72]. In summary, the results discussed above show that the reversible transition between planktonic and biofilm lifestyle is regulated by c-di-GMP through the action of VpsR-, VpsT- and RpoS-dependent H-NS antirepression switches.
Fig. 4. Regulatory pathway controlling RpoS-dependent activation of motility and detachment.
In a low-cell-density community, the c-di-GMP content is elevated, resulting in expression and activation of VpsR and VpsT. Motility is diminished by binding of H-NS to the flrA promoter and inhibition of FlrA by c-di-GMP. In a high-cell-density community, HapR is expressed to lower the c-di-GMP pool and terminate transcription of vpsT. Environmental stress and the absence of VpsT favor expression of RpoS. RpoS further diminishes the c-di-GMP pool to enhance the activity of FlrA and activates expression of IHF that lessens H-NS occupancy at the flrA promoter. In addition, initiation of flrA transcription by σS is more resistant to H-NS remaining bound to the promoter. Downregulated or inactive transcription factors, as well as disabled regulatory conections, are represented in light graey font. Symbols: ↓, positive regulation; ⊥, negative regulation.
6. Conclusion
In V. cholerae, multiple signal transduction pathways participate in the continuous reprogramming of gene expression to cope with environmental and host-specific stresses. In this review, we highlight the overarching role of H-NS repression and antirepression in this process. At the gene level, H-NS acts as a transcription gatekeeper to prevent gratuitous gene expression that could result in poor fitness. Transcriptional silencing can be reversed by antagonistic regulators such as ToxR, ToxT, VpsR and VpsT in response to environmental cues and fluctuations in the c-di-GMP pool. At the genome level, H-NS binding to the chromatin and bridging could contribute to vibrio adaptation by bringing clusters of coregulated genes into proximity or coordinating their expression in response to environmental stresses that affect DNA topology.
Acknowledgments
Research in the author’s laboratory has been funded by the National Institute of Allergy and Infectious Disease, National Institutes of Health, Bethesda, Maryland, USA. We are grateful for the technical support provided by the Morehouse School of Medicine Research Core Facility and the University of Alabama at Birmingham Heflin Center for Genomic Sciences and the Targeted Metabolomics and Proteomics Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dorman CJ. Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation. J Mol Microbiol Biotechnol. 2014;24:316–331. doi: 10.1159/000368850. [DOI] [PubMed] [Google Scholar]
- 2.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14:1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 4.Broza M, Gancz H, Kashi Y. The association between non-biting midges and Vibrio cholerae. Environ Microbiol. 2008;10:3193–3200. doi: 10.1111/j.1462-2920.2008.01714.x. [DOI] [PubMed] [Google Scholar]
- 5.Van der Henst C, Scrignari T, Maclachlan C, Blokesch M. An intracellular replication niche for Vibrio cholerae in the amoeba Acanthamoeba castellanii. The ISME journal. 2016;10:897–910. doi: 10.1038/ismej.2015.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva AJ, Benitez JA. Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl Trop Dis. 2016;10:e0004330. doi: 10.1371/journal.pntd.0004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenz DH, Bassler BL. The small nucleoid protein Fis is involved in Vibrio cholerae quorum sensing. Mol Microbiol. 2007;63:859–871. doi: 10.1111/j.1365-2958.2006.05545.x. [DOI] [PubMed] [Google Scholar]
- 8.Stonehouse E, Kovacikova G, Taylor RK, Skorupski K. Integration host factor positively regulates virulence gene expression in Vibrio cholerae. J Bacteriol. 2008;190:4736–4748. doi: 10.1128/JB.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman CJ, Kane KA. DNA bridging and antibridging: A role for bacterial nucleoid-associated proteins in regulating the expression of laterally acquired genes. FEMS Microbiol Rev. 2009;33:587–592. doi: 10.1111/j.1574-6976.2008.00155.x. [DOI] [PubMed] [Google Scholar]
- 10.Winardhi RS, Yan J, Kenney LJ. H-NS regulates gene expression and compacts the nucleoid: Insights from single-molecule experiments. Biophys J. 2015;109:1321–1329. doi: 10.1016/j.bpj.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin M, Lagda AC, Lee JW, Bhat A, Rhee JH, Kim JS, Takeyasu K, Choy HE. Gene silencing by H-NS from distal DNA site. Mol Microbiol. 2012;86:707–719. doi: 10.1111/mmi.12012. [DOI] [PubMed] [Google Scholar]
- 14.Dorman CJ. Co-operative roles for DNA supercoiling and nucleoid-associated proteins in the regulation of bacterial transcription. Biochem Soc Trans. 2013;41:542–547. doi: 10.1042/BST20120222. [DOI] [PubMed] [Google Scholar]
- 15.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nature Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 16.Nye MB, Taylor RK. Vibrio cholerae H-NS domain structure and function with respect to transcriptional repression of toxR regulon genes reveals differences among H-NS family members. Mol Microbiol. 2003;50:427–444. doi: 10.1046/j.1365-2958.2003.03701.x. [DOI] [PubMed] [Google Scholar]
- 17.Cerdan R, Bloch V, Yang Y, Bertin P, Dumas C, Rimsky S, Kochoyan M, Arold ST. Crystal structure of the N-terminal dimerisation domain of VicH, the H-NS-like protein of Vibrio cholerae. J Mol Biol. 2003;334:179–185. doi: 10.1016/j.jmb.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 18.Tendeng C, Badaut C, Krin E, Gounon P, Ngo S, Danchin A, Rimsky S, Bertin P. Isolation and characterization of vicH, encoding a new pleiotropic regulator in Vibrio cholerae. J Bacteriol. 2000;182:2026–2032. doi: 10.1128/jb.182.7.2026-2032.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayala JC, Wang H, Benitez JA, Silva AJ. RNA-Seq analysis and whole genome DNA-binding profile of the histone-like nucleoid structuring protein (H-NS) Genomics data. 2015;5:147–150. doi: 10.1016/j.gdata.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayala JC, Wang H, Silva AJ, Benitez JA. Repression by H-NS of genes required for the biosynthesis of the Vibrio cholerae biofilm matrix is modulated by the second messenger cyclic diguanylic acid. Mol Microbiol. 2015;97:630–645. doi: 10.1111/mmi.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stella S, Falconi M, Lammi M, Gualerzi CO, Pon CL. Environmental control of the in vivo oligomerization of nucleoid protein H-NS. J Mol Biol. 2006;355:169–174. doi: 10.1016/j.jmb.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Williamson HS, Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- 23.Silva AJ, Sultan SZ, Liang W, Benitez JA. Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae. J Bacteriol. 2008;190:7335–7345. doi: 10.1128/JB.00360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Ayala JC, Benitez JA, Silva AJ. Interaction of the histone-like nucleoid structuring protein and the general stress response regulator RpoS at Vibrio cholerae promoters that regulate motility and hemagglutinin/protease expression. J Bacteriol. 2012;194:1205–1215. doi: 10.1128/JB.05900-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Ayala JC, Benitez JA, Silva AJ. RNA-Seq analysis identifies new genes regulated by the histone-like nucleoid structuring protein (H-NS) affecting Vibrio cholerae virulence, stress response and chemotaxis. PLoS One. 2015;10:e0118295. doi: 10.1371/journal.pone.0118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 28.Dhillon BK, Laird MR, Shay JA, Winsor GL, Lo R, Nizam F, Pereira SK, Waglechner N, McArthur AG, Langille MG, et al. Islandviewer 3: More flexible, interactive genomic island discovery, visualization and analysis. Nucl Acids Res. 2015;43:W104–108. doi: 10.1093/nar/gkv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci USA. 1999;96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucl Acids Res. 2011;39:2073–2091. doi: 10.1093/nar/gkq934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazi MI, Conrado AR, Mey AR, Payne SM, Davies BW. ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog. 2016;12:e1005570. doi: 10.1371/journal.ppat.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorman CJ. Integrating small molecule signalling and H-NS antagonism in Vibrio cholerae, a bacterium with two chromosomes. Mol Microbiol. 2015;97:612–615. doi: 10.1111/mmi.13063. [DOI] [PubMed] [Google Scholar]
- 33.Val ME, Marbouty M, de Lemos Martins F, Kennedy SP, Kemble H, Bland MJ, Possoz C, Koszul R, Skovgaard O, Mazel D. A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae. Science Adv. 2016;2:e1501914. doi: 10.1126/sciadv.1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorman CJ. Genome architecture and global gene regulation in bacteria: Making progress towards a unified model? Nature Rev Microbiol. 2013;11:349–355. doi: 10.1038/nrmicro3007. [DOI] [PubMed] [Google Scholar]
- 35.Peeters E, Driessen RP, Werner F, Dame RT. The interplay between nucleoid organization and transcription in archaeal genomes. Nature Rev Microbiol. 2015;13:333–341. doi: 10.1038/nrmicro3467. [DOI] [PubMed] [Google Scholar]
- 36.Crawford JA, Krukonis ES, DiRita VJ. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol Microbiol. 2003;47:1459–1473. doi: 10.1046/j.1365-2958.2003.03398.x. [DOI] [PubMed] [Google Scholar]
- 37.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matson JS, Withey JH, DiRita VJ. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infec Immun. 2007;75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacikova G, Skorupski K. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol. 2001;41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 40.Nye MB, Pfau JD, Skorupski K, Taylor RK. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol. 2000;182:4295–4303. doi: 10.1128/jb.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krukonis ES, DiRita VJ. DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae. Mol Cell. 2003;12:157–165. doi: 10.1016/s1097-2765(03)00222-3. [DOI] [PubMed] [Google Scholar]
- 42.Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol Microbiol. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]
- 43.Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK. H-NS binding and repression of the ctx promoter in Vibrio cholerae. J Bacteriol. 2011;193:979–988. doi: 10.1128/JB.01343-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murley YM, Carroll PA, Skorupski K, Taylor RK, Calderwood SB. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satchell KJ, Jones CJ, Wong J, Queen J, Agarwal S, Yildiz FH. Phenotypic analysis reveals Haiti cholera epidemic linked to hypervirulent strain. Infect Immun. 2016 doi: 10.1128/IAI.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olivier V, Haines GK, 3rd, Tan Y, Satchell KJ. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elgaml A, Miyoshi S. Role of the histone-like nucleoid structuring protein (H-NS) in the regulation of virulence factor expression and stress response in Vibrio vulnificus. Biocontrol Sci. 2015;20:263–274. doi: 10.4265/bio.20.263. [DOI] [PubMed] [Google Scholar]
- 48.Mou X, Spinard EJ, Driscoll MV, Zhao W, Nelson DR. H-NS is a negative regulator of the two hemolysin/cytotoxin gene clusters in Vibrio anguillarum. Infect Immun. 2013;81:3566–3576. doi: 10.1128/IAI.00506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoebner JA, Payne SM. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect Immun. 1988;56:2891–2895. doi: 10.1128/iai.56.11.2891-2895.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams SG, Manning PA. Transcription of the Vibrio cholerae haemolysin gene, hlyA, and cloning of a positive regulatory locus, hlyU. Mol Microbiol. 1991;5:2031–2038. doi: 10.1111/j.1365-2958.1991.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu M, Naka H, Crosa JH. HlyU acts as an H-NS antirepressor in the regulation of the rtx toxin gene essential for the virulence of the human pathogen Vibrio vulnificus cmcp6. Mol Microbiol. 2009;72:491–505. doi: 10.1111/j.1365-2958.2009.06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. Living in the matrix: Assembly and control of Vibrio cholerae biofilms. Nature Rev Microbiol. 2015;13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol. 2013;90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSEtr-associated phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fong JC, Yildiz FH. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol. 2007;189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Ayala JC, Silva AJ, Benitez JA. The histone-like nucleoid structuring protein (H-NS) is a repressor of Vibrio cholerae exopolysaccharide biosynthesis (vps) genes. Appl Environ Microbiol. 2012;78:2482–2488. doi: 10.1128/AEM.07629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zamorano-Sanchez D, Fong JC, Kilic S, Erill I, Yildiz FH. Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J Bacteriol. 2015;197:1221–1235. doi: 10.1128/JB.02439-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Srivastava D, Harris RC, Waters CM. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol. 2011;193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyhan S, Tischler AD, Camilli A, Yildiz FH. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J Bacteriol. 2006;188:3600–3613. doi: 10.1128/JB.188.10.3600-3613.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prouty MG, Correa NE, Klose KE. The novel sigma54- and sigma28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol Microbiol. 2001;39:1595–1609. doi: 10.1046/j.1365-2958.2001.02348.x. [DOI] [PubMed] [Google Scholar]
- 63.Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Wu JH, Ayala JC, Benitez JA, Silva AJ. Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. J Bacteriol. 2011;193:6529–6538. doi: 10.1128/JB.05166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hengge-Aronis R. Stationary phase gene regulation: What makes an Escherichia coli promoter sigmas-selective? Curr Opin Microbiol. 2002;5:591–595. doi: 10.1016/s1369-5274(02)00372-7. [DOI] [PubMed] [Google Scholar]
- 67.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: Specific requirement of Eσ70 as a cofactor for looping. Genes Develop. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang H, Ayala JC, Benitez JA, Silva AJ. The LuxR-type regulator VpsT negatively controls the transcription of rpoS, encoding the general stress response regulator, in Vibrio cholerae biofilms. J Bacteriol. 2014;196:1020–1030. doi: 10.1128/JB.00993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benitez JA, Spelbrink RG, Silva A, Phillips TE, Stanley CM, Boesman-Finkelstein M, Finkelstein RA. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells: An in vitro colonization model. Infect Immun. 1997;65:3474–3477. doi: 10.1128/iai.65.8.3474-3477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koestler BJ, Waters CM. Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect Immun. 2014;82:3002–3014. doi: 10.1128/IAI.01664-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe. 2007;2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamayo R, Patimalla B, Camilli A. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun. 2010;78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]