Abstract
DnaK is a major heat shock protein of Escherichia coli and the homolog of hsp70 in eukaryotes. We demonstrate the mechanism by which DnaK and another heat shock protein, DnaJ, render the plasmid P1 initiator RepA 100-fold more active for binding to the P1 origin of replication. Activation is the conversion of RepA dimers into monomers in an ATP-dependent reaction and the monomer form binds with high affinity to oriP1 DNA. Reversible chemical denaturants also convert RepA dimers to monomers and simultaneously activate oriP1 DNA binding. Increasing protein concentration converts monomers to dimers and deactivates RepA. Based on our data and previous work, we present a model for heat shock protein action under normal and stress conditions.
Full text
PDF
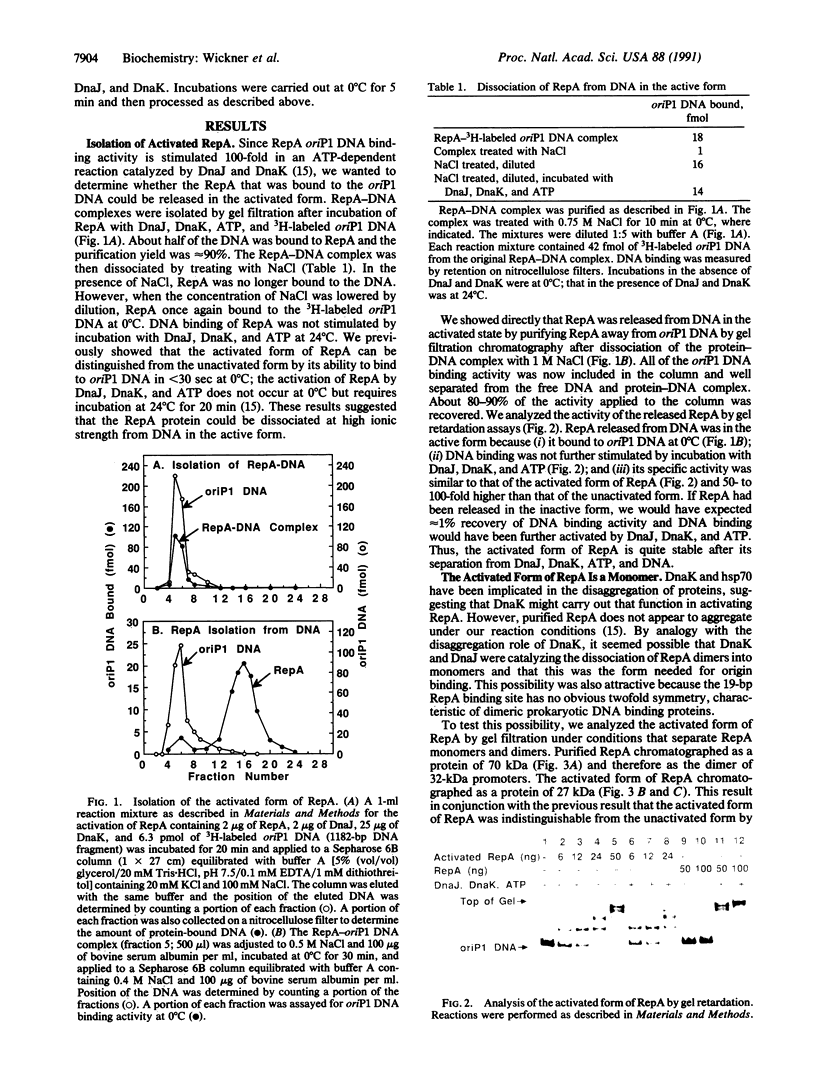



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles A. L., Snyder K. M., Chattoraj D. K. P1 plasmid replication: replicon structure. J Mol Biol. 1984 Mar 5;173(3):307–324. doi: 10.1016/0022-2836(84)90123-2. [DOI] [PubMed] [Google Scholar]
- Alfano C., McMacken R. Heat shock protein-mediated disassembly of nucleoprotein structures is required for the initiation of bacteriophage lambda DNA replication. J Biol Chem. 1989 Jun 25;264(18):10709–10718. [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989 May;171(5):2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Delta dnaK52 mutants of Escherichia coli have defects in chromosome segregation and plasmid maintenance at normal growth temperatures. J Bacteriol. 1989 Nov;171(11):6030–6038. doi: 10.1128/jb.171.11.6030-6038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D. K., Snyder K. M., Abeles A. L. P1 plasmid replication: multiple functions of RepA protein at the origin. Proc Natl Acad Sci U S A. 1985 May;82(9):2588–2592. doi: 10.1073/pnas.82.9.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J. D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: localized unwinding of duplex DNA by a six-protein reaction. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Catalytic RNA: a Nobel Prize for small village science. New Biol. 1990 Jan;2(1):1–4. [PubMed] [Google Scholar]
- Pelham H. R. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell. 1986 Sep 26;46(7):959–961. doi: 10.1016/0092-8674(86)90693-8. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Georgopoulos C., Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990 Sep 7;62(5):939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- Swack J. A., Pal S. K., Mason R. J., Abeles A. L., Chattoraj D. K. P1 plasmid replication: measurement of initiator protein concentration in vivo. J Bacteriol. 1987 Aug;169(8):3737–3742. doi: 10.1128/jb.169.8.3737-3742.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- Tilly K., Yarmolinsky M. Participation of Escherichia coli heat shock proteins DnaJ, DnaK, and GrpE in P1 plasmid replication. J Bacteriol. 1989 Nov;171(11):6025–6029. doi: 10.1128/jb.171.11.6025-6029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H., Chattoraj D. K. Replication of mini-P1 plasmid DNA in vitro requires two initiation proteins, encoded by the repA gene of phage P1 and the dnaA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3668–3672. doi: 10.1073/pnas.84.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S. H. Three Escherichia coli heat shock proteins are required for P1 plasmid DNA replication: formation of an active complex between E. coli DnaJ protein and the P1 initiator protein. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2690–2694. doi: 10.1073/pnas.87.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Hoskins J., McKenney K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature. 1991 Mar 14;350(6314):165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- Zylicz M., Ang D., Liberek K., Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: the role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989 May;8(5):1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]




