Abstract
Purpose: EGFRvIII as the most common mutant variant of the epidermal growth factor receptor is resulting from deletion of exons 2–7 in the coding sequence and junction of exons 1 and 8 through a novel glycine residue. EGFRvIII is highly expressed in glioblastoma, carcinoma of the breast, ovary, and lung but not in normal cells. The aim of the present study was identification of a novel single chain antibody against EGFRvIII as a promising target for cancer therapy.
Methods: In this study, a synthetic peptide corresponding to EGFRvIII protein was used for screening a naive human scFv phage library. A novel five-round selection strategy was used for enrichment of rare specific clones.
Results: After five rounds of screening, six positive scFv clones against EGFRvIII were selected using monoclonal phage ELISA, among them, only three clones had expected size in PCR reaction. The specific interaction of two of the scFv clones with EGFRvIII was confirmed by indirect ELISA. One phage clone with higher affinity in scFv ELISA was purified for further analysis. The purity of the produced scFv antibody was confirmed using SDS-PAGE and Western blotting analyses.
Conclusion: In the present study, a human anti- EGFRvIII scFv with high affinity was first identified from a scFv phage library. This study can be the groundwork for developing more effective diagnostic and therapeutic agents against EGFRvIII expressing cancers.
Keywords: Human single chain antibody, Cancer, EGFRvIII, Phage display
Introduction
Epidermal growth factor receptor (EGFR) as a transmembrane glycoprotein from tyrosine kinases family is a crucial regulator of normal cellular growth in epithelial tissues.1
Dysregulated EGFR following overexpression or mutation of EGFR is one of the main factors involved in epithelial malignancies.2 EGF receptor variant III )EGFRvIII( is the most common mutation in EGFR and is expressed mostly in glioblastoma, and carcinoma of the breast, ovary, and lung, whereas normal tissues lacking EGFRvIII.3,4 This variant receptor with molecular mass of 145kDa is resulting from deletion in exons 2–7 in the coding sequence and junction of exons 1 and 8 through a novel glycine residue.5,6 This deletion creates a tumor specific immunogenic epitope and leads to unregulated growth, invasion, and angiogenesis.7
In recent years, several murine antibodies have been developed against EGFRvIII, including L8A4, MR1, MR1-1, 3C10; most of them have cross reactivity with wild type EGFR or have lower affinity to EGFRvIII. On the other hand, unfavorable HAMA responses induced by the murine antibodies limit their therapeutic applications.4,5-8
Since, the large size of full length antibodies limits their efficiency for treatment of solid tumors due to weak penetration, small fragments of antibodies like scFvs with higher tissue penetration rate are considered as attractive therapeutic or diagnostic alternatives.9
In contrast to the conventional methods for antibodies generating, scFv phage libraries with high diversity of gene repertoires provide a rich source of scFvs to almost any antigen.10,11 The present study aimed to isolate a novel human scFv against EGFRvIII using phage display technology as a potential candidate for treatment of EGFRvIII expressing cancers.
Materials and Methods
ScFv-Phage Library, Bacteria, and Reagents
The Human scFv phage libraries I + J (Tomlinson I+ J), HB2151and KM13 helper phage,12 (The Medical Research Council (MRC), Cambridge, UK) and, E. coli TG1were used for isolation of specific antibody clones and production of scFvs.12-14
Synthetic Peptide
The synthetic peptide LEEKKGNYVVTDHSGGK was selected from the N terminal region of EGFRvIII in which the first 13 residues contained the tumor-specific deletion junction sequence and the SGG sequence served as a flexible spacer.5 This peptide was synthesized with 97.2% purity (Biomatik, Life science).
Biopanning of scFv phage library
The stock libraries I & J were initially mixed together then were amplified and titrated as described by the Medical Research Council (MRC) protocol.15 Biopanning process was performed with 50μg/mL EGFRvIII peptide on a Maxisorp 96-well plate (Nunc Thermo Scientific Inc., Rochester, NY) in 50 mM carbonate buffer (pH 9.6). To avoid loss of rare specific clones, the concentration of EGFRvIII peptide was kept constant during biopanning. Since, a significant part of antigen is often desorbed from solid phase during blocking and washing processes, low antigen concentration can lead to reduction of phage yield.15,16 Hence, in this study in order to increase the output of screening steps; we didn’t reduce peptide concentration during biopanning rounds. However, for increasing the screening efficiency, incubation time of the pool phages with antigen were decreased while washing numbers were increased between screening rounds (Table 1).
Table 1. Details of five rounds Biopanning including EGFRvIII peptide concentration, blocking buffers, Tween 20 percentage, washing numbers and incubation period of phage.
| Round | I | II | III | IV | V |
| Peptide (μg/ml) | 50 | 50 | 50 | 50 | 50 |
| Blocking buffers | 2% BSA | 3%BSA | 2% BSA | 3%BSA | 2%MPBS |
| %Tween 20 | 0.1% | 0.1% | 0.15% | 0.2% | 0.2% |
| Washing numbers | 3 | 5 | 10 | 10 | 10 |
| Incubation period of phage(hrs) | 2 | 1.5 | 1 | 1 | 1 |
After washing EGFRvIII- immobilized plate with PBS and blocking with 3% Bovine Serum Albumin (BSA) for 2hrs, 1012-13pfu phages diluted with blocking solution were added. The plate was incubated on a platform shaker (Heidolph Titramax 1000) with speed of 150 rpm for 30 min at room temperature (RT) and then stood for further 30 min at RT. Afterward, plate was washed with PBS containing Tween 20 (PBST), the bound phages were eluted by treatment with 100μl/well of trypsin-PBS (100μl of 10mg/ml trypsin stock solution in10ml PBS) and incubating for 15 min at RT on a platform shaker (150rpm). Details related to the phage incubation period and washing number is presented in Table 1. All the titration, amplification and purification processes of phages were carried out as described elsewhere.15
Totally, five rounds of biopanning were carried out to select EGFRvIII -specific phage clones. In round 4, before starting screening an invert biopanning against BSA was performed to deplete sublibrary from specific clones to BSA, then the depleted library from BSA binders was added to the EGFRvIII- immobilized wells.17
To perform invert biopanning, in a 96-well Maxisorp plate, 16 wells were coated with 3% BSA, and incubated overnight at 4°C. After washing plate with PBS, the eluted phages from 3th round (diluted in PBS) were added to BSA-immobilized wells and incubated for 1hr at RT. The depleted library from BSA binders was then added to the EGFRvIII- immobilized wells and incubated for 1hr at RT. After washing with PBST, the bound phages were eluted by trypsin as detailed above.
Specificity analysis of selected phages by polyclonal phage ELISA
After five rounds of biopanning, a polyclonal phage ELISA was performed to examine the specificity of selected phages from each round of panning against EGFRvIII peptide. For this purpose, EGFRvIII peptide with concentration of 25µg/ml and BSA were coated into the plates separately and incubated overnight at 4°C. After blocking with 3% BSA for 2hrs and washing with PBST, the eluted phages (1:10 dilution in1% BSA–PBS) were added to plates and incubated for 1hr. After washing, the plates were incubated with anti-M13- HRP (1:2000 dilutions in 1% BSA–PBS) for 1hr and the reaction was developed with TMB substrate. The optical density (OD) was read at 450nm.
Selection of scFvs clones against EGFRvIII peptide by monoclonal phage ELISA
Single colonies from the 5th round of panning were selected randomly, and their affinity was determined against EGFRvIII peptide using monoclonal phage ELISA. For this purpose individual colonies from 5th round were inoculated into 2xTY medium (1.6% [w/v] tryptone, 1% [w/v] yeast extract, and 0.5% [w/v] sodium chloride) containing100 µg/ml ampicillin and 4% glucose in a 96 well plate at 37°C for 2 hrs. After 2hrs incubation, 109 helper phages were added to each wells and incubated for 1hr at 37°C without shaking. Then the plate was spined in 3000 g for 10 min, aspirated off the supernatants and bacterial pellets were resuspended in 2xTY containing 100 µg/ml ampicillin and 50 µg/ml kanamycin. Cultures were continued overnight at 30°C with shaking (250 rpm) and culture supernatants (1:2 dilution in1% BSA–PBS) were used in phage ELISA. For this, EGFRvIII peptide with concentration of 25µg/ml was coated into the plates overnight at 4°C. After blocking with 3% BSA for 2hrs and washing with PBST, the eluted phages (1:2 dilution in1% BSA–PBS) were added to the plates and incubated for 1hr. After washing, the plates were incubated with anti-M13- HRP (1:2000 dilutions in 1% BSA–PBS) for 1hr and the reaction was developed with TMB substrate. The optical density (OD) was read at 450nm.
PCR amplification and sequence analysis
To confirm the presence of VH and VL fragments, plasmid DNA were extracted from positive phages clones identified and PCR amplification was performed using pIT2-vector specific primers: Forward: 5´-CAGGAAACAGCTATGAC-3´, Reverse: 5´-CTATGCGGCCCCATTCA-3´ in a 25ml reaction for 31 cycles (94°C for 30s, 58°C for 30s, 72°C for 45s) after an initial denaturation at 94°C for 4 min. Products were analyzed on %1 (w/v) agarose gel. The positive clones with confirmed size were sequenced using pIT2-vector specific primers and sequences were then analyzed with V-BASE (http:// vbase. mrc- cpe. cam. ac. uk/ ) and Ig BLAST (www. ncbi. nlm. nih. gov/ blast/ Blast. cgi) using the Kabat numbering system to identify the complementarity determining regions (CDRs) in heavy chain (VH) and light chain (VL) variable regions.18
Screening of clones by soluble fragment ELISA
The binding specificity of positive phage clones against EGFRvIII was examined by scFv ELISA. The positive clones confirmed by PCR were grown up in E. coli TG1 suppressor strain for producing soluble scFv in which all selected clones can produce scFv even clones containing TAG stop codon.
For this, the positive clones were grown up in 2xTY/ ampicillin/ 0.1 % glucose media at 37°C until the OD600 was approximately 0.9. At this stage, scFv expression was induced with 1mM IPTG and shaking was continued at 200 rpm overnight at 30°C. Soluble scFvs obtained from the periplasmic/osmotic fractions were then retested by the EGFRvIII -captured ELISA. Accordingly, EGFRvIII peptide with concentration of 25µg/ml was coated into the plate overnight at 4°C. After blocking with 3% BSA for 2hrs and washing with PBST, soluble scFvs were added to the plate in different dilutions and incubated for 1hr. After washing with PBS-T, the plate was incubated with HRP conjugated Protein L (1:2000 dilution in 1% BSA–PBS) for 1hr, and then washed. The reaction was developed with TMB substrate and the OD was read at 450nm.
Purification of soluble scFv fragments and western blotting analysis
Among positive clones detected by scFv ELISA, scFv clone with higher OD was selected for production of soluble scFv. The selected positive clone was grown up in 2xTY/ampicillin/0.1 % glucose media at 37°C until the OD600 was approximately 0.9. At this stage, scFv expression was induced with 1mM IPTG and shaking was continued at 200 rpm overnight at 30 °C. Soluble scFvs enriched in the periplasmic/osmotic fractions were then purified by Ni-NTA column.16 The purity of expressed and extracted soluble scFv was evaluated by 12% SDS– PAGE.
For western blot analysis, the purified scFv (30ug/lane) was resolved by 12% SDS-PAGE and transferred onto PVDF membrane (Roche, Germany) and blocked with 5% skimmed milk overnight at 4°C. For detection of scFv, membrane was incubated with HRP conjugated Protein L (HRP) as secondary antibody for 1hr in RT and after washing with PBS-T, reaction was developed by staining with DAB.
Results and Discussion
Enrichment of specific phage clones by five rounds of biopanning
For selecting specific phages against EGFRvIII, five rounds of biopanning was performed also an invert biopanning in fourth round was carried for elimination of BSA binding clones. Since BSA is used as blocking agent during biopanning, an invert biopanning in the fourth round was also carried for elimination of BSA binding clones. As expected, in the first round of screening the yield of EGFRvIII -specific phages was very low. During the first three rounds, output phages were increased from 2.07× l05 in the first round to 3.79×108 in the 3rd round. However, with implementation of invert biopanning method in fourth round, yield of output phages was reduced due to removing BSA- binding clones but in the fifth round again the titer of phages raised due to enrichment of EGFRvIII-specific phage clones. Totally, the eluted phages were increased from 2.07× l05 in the first round to 0.7× l08 in the latest round with enrichment about 338-fold (Table2). These results were displaying a successful screening to obtain specific phages against EGFRvIII peptide.
Table 2. Enrichment of specific phages during five rounds of biopanning.
| Round | input(pfu) | Output(pfu) | Enrichment(output/input) |
| 1 | 1.72×l012 | 2.07× l05 | 1.20×l0-7 |
| 2 | 1.88×l012 | 5.21×106 | 2.77× l0-6 |
| 3 | 2.08×l012 | 3.79×108 | 1.82×l0-4 |
| 4 | 2×l012 | 6.3×107 | 3.15×l0-5 |
| 5 | 2.5×l012 | 0.7×108 | 0.028×l0-4 |
Specificity of scFv phages against EGFRvIII
Specificity of eluted phages after each round of biopanning was verified by the polyclonal phage ELISA against EGFRvIII peptide and BSA. For this, polyclonal phages in two dilutions (1:10 and 1:20) were added to wells coated with EGFRvIII peptide and BSA separately. The wild type phages were used as negative control which revealed only weak signal in ELISA (Figure 1). After five round of panning, the eluted phages from 2nd and 3rdrounds showed an enhancement in binding activity to EGFRvIII and BSA. Therefore, for removing BSA binding clones, invert biopanning was carried in the beginning of the fourth round. Although invert biopanning led to reducing optical density in the fourth round but BSA-binding phages were removed. So that, in the fifth round, the binding affinity to EGFRvIII was again increased (OD450= 3.63). So, the 5th round with highest affinity against EGFRvIII was selected for further screening processes (Figure 1).
Figure 1.
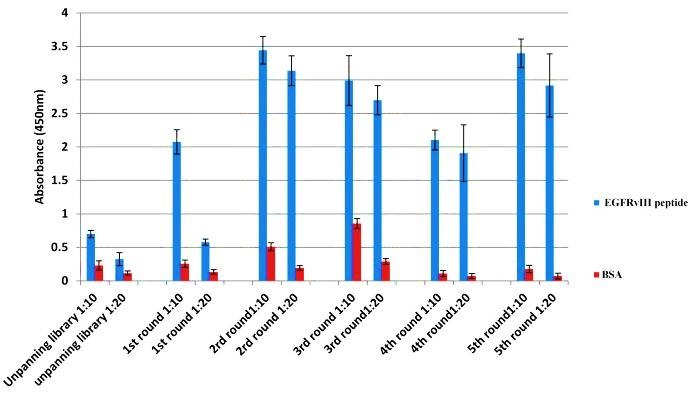
Binding of selected phage-scFv clones from 5 rounds panning to EGFRvIII peptide by polyclonal phage ELISA. ELISA plates were coated with 25μg/ml EGFRvIII peptide and BSA separately and binding of the precipitated phages of each round was detected with anti-M13 -HRP conjugate at 1:2000 dilution. Absorbance values are represented as the mean ± standard deviation (SD) of three independent determinations. Error bars show the standard deviation for each set of data.
Identification of scFv phage clones with high binding to EGFRvIII
Approximately 120 individual clones from the fifth round were examined by monoclonal phage ELISA for isolation of specific phage clones against EGFRvIII. Almost 30 specific phage clones were identified with a range of optical densities from 0.2 to 3.334 indicating significant differences in the binding affinity of selected phage clones. Among positive clones, 6 phage clones (2, 10, 13, 14, 15 and 27) with higher binding affinity to EGFRvIII (OD range 0.8 - 3.3) were selected for further analysis (Figure 2).
Figure 2.
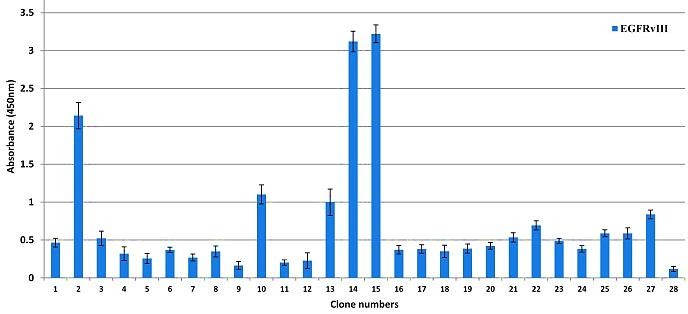
Characterization of the selected phage clones for binding to EGFRvIII by ELISA. The EGFRvIII peptide with concentration of 25μg/ml was coated and 50μl supernatant of each phage was separately added per well. Clone number 1 was as negative control related to wild type phage. The reactivity was determined using anti-M13 -HRP conjugate at 1:2000 dilution. Absorbance values are represented as the mean ± SD for three independent determinations. Error bars show the standard deviation for each set of data.
Identification of antigen-specific scFv clones by DNA sequencing and PCR reaction
Results of PCR reaction on DNA coding sequences of the six phage clones with higher reactivity displayed that only clones 2, 10 and 15 had expected size (930bp) indicating the presence of both VL and VH fragments (Figure 3). The clone 13 with two bands and clone 27 with a single band of 700bp were considered as false positive clones (Figure 3). To confirm integrity of VH–VL fragments in positive phage clones, sequencing was performed on three positive phage clones (2, 10, and 15) selected by ELISA. Sequencing results showed that scFv clones 2 and 15 were bearing an amber stop codon (TAG) and clone number 10 showed a frame shift mutation. Multiple alignments of the amino acid sequences related to three scFvs indicated a diverse panel within CDRs regions. So that, highest diversity was related to the CDR2 and CDR3 regions from heavy chain and CDR3 region of light chain (Table 3).
Figure 3.
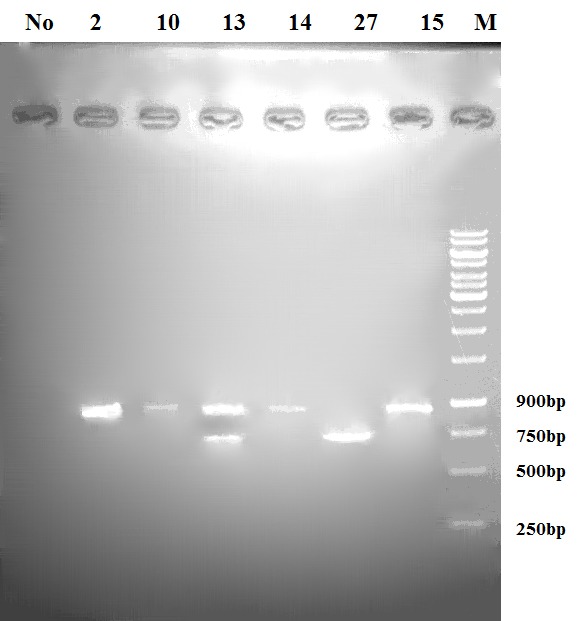
Identification of antigen-specific scFv clones by PCR reaction
Table 3. Amino acid residues related to CDR regions of positive scFv clones to EGFRvIII .
| Clones | CDR1 | CDR2 | CDR3 |
| VH10 | SYAMS | SIAANGCTTYYADSVKG | TNGSFDY |
| VH2 | SYAMS | SISK@GGRTTYADSVKG | RGAKFDY |
| VH15 | SYAMS | TIFK@GEHT NYADSVKG | MGRRFDY |
| VL10 | RASQSISSYLN | SASYLQS | QQAGSTPAT |
| VL2 | RASQSISSYLN | SASYLQS | QQTAKRPIT |
| VL15 | RASQSISSYLN | RASSLQS | QQVRSTPVT |
Results of PCR reaction on DNA coding sequences of the six selected phage clones with higher reactivity displayed that only clones 2, 10 and 15 had expected size (930bp) indicating the presence of both VL and VH fragments. The clone 13 with two bands and clone 27 with a band in a size of 700bp were found as false positive clones.
Specificity of scFv clones
Results of EGFRvIII -captured ELISA showed that among three reactive clones in phage ELISA, the scFv clones 2 and 15 were able to recognize and bind to EGFRvIII peptide but the scFv clone 10 indicated a negative signal. The positive clones displayed a range of optical densities dependent on scFv concentrations. The maximum OD value observed in EGFRvIII -captured ELISA was 3.27 related to scFv clone 2 (Figure 4). So, the EGFRvIII-scFv clone 2 was selected for correction of amber stop codon and further studies.
Figure 4.
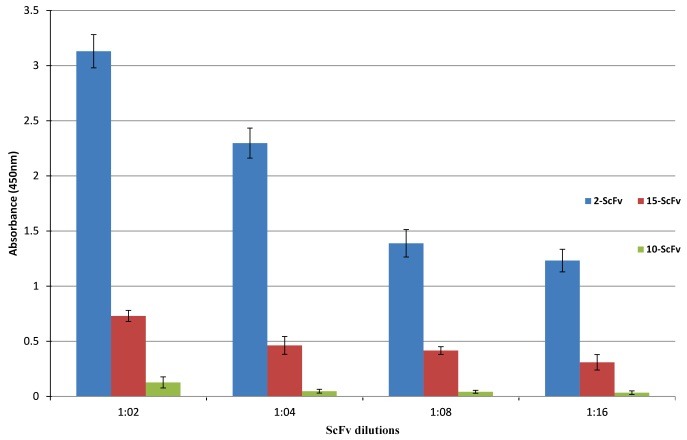
Binding of three selected scFvs clones with high reactivity to EGFRvIII. The reactivity of different dilutions of scFv-2, scFv-15, and scFv- 10 with EGFRvIII was assessed by indirect ELISA. The EGFRvIII peptide with concentration of 25μg/ml was coated on ELISA plates and binding of the produced scFvs in TG1 were detected with Protein L conjugated to HRP at 1:2000 dilutions. Absorbance values are represented as the mean ± SD for three independent determinations. Error bars show the standard deviation for each set of data.
Purification of anti- EGFRvIII scFv and western blot analysis
For production of soluble scFv fragments, scFv clone 2 with higher affinity in ELISA was grown in E. coli TG1 suppressor strain. The scFv fragments were then purified from soluble fraction with high purity. SDS-PAGE analysis illustrated the purified anti- EGFRvIII scFv as fused to PIII with molecular weight of about 52KDa which was consistent with results previously reported from Tomlinson library (Figure 5A).19 Also, the presence of the purified scFv antibody was also confirmed using western blot (Figure 5B).
Figure 5.
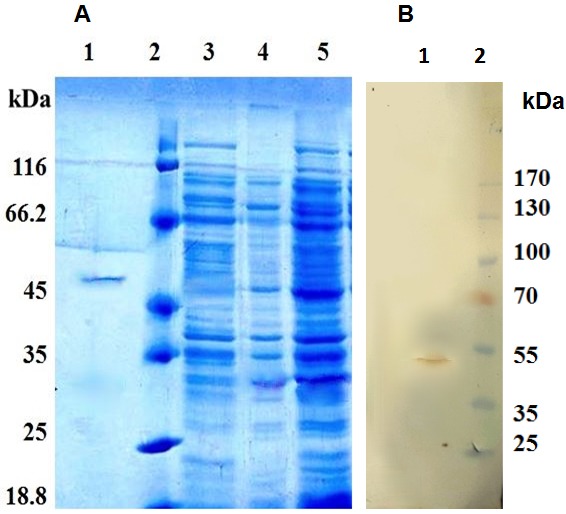
SDS-PAGE and western blot analyses anti–EGFRvIII scFv. (A) After induction with 1mM IPTG at 30°C, anti- EGFRvIII scFv was mainly expressed in TG1 E. coli as a band around 52 kDa indicating pIII - scFv fusion. Lane (1) Purified scFv, Lane (2) size marker, Lane (3) before IPTG induction, Lane (4) the expression of pIII-scFv in soluble fraction, Lane (5) the bacterial lysate. (B) The presence of scFv was confirmed by Western blotting analysis. The purified scFv was resolved (5ug/lane) by 12% SDS-PAGE and transferred onto a PVDF membrane. Lane (1) the purified scFv was recognized as a 52kDa band corresponds to the size of scFv fused to pIII. Lane (2) size marker.
The versatile application of monoclonal antibodies in diagnosis and therapy of cancer types, have been led to notable progresses in the field of cancer target therapy.
In this respect, phage display technology as a selection based strategy plays a crucial role in the development of specific antibodies against cancer types. Unique ability of phage libraries for displaying proteins on bacteriophages surfaces offers rich repertoires for isolation of specific antibodies with high affinity for the vast majority of antigens. In the present study, we aimed to isolate high affinity scFvs from a human phage library against EGFRvIII as a tumor specific antigen. Recently, EGFRvIII has been emerged as an attractive candidate for antibody based therapy of several cancers. Studies have showed that EGFRvIII is expressed in 57% of high grade and 86% of low grade glioma and almost 78% of breast carcinomas.20 So far, several antibodies have been developed against EGFRvIII, but none of them have been approved for clinical use, yet.4,5,21 In this respect, Lorimer et al., constructed a scFv phage library from the spleen of mouse immunized with EGFRvIII and then isolated an anti EGFRvIII-scFv (MR1) by screening this library.5 Gupta et al., improved the specificity of MR1 scFv by mutagenesis to eliminate cross reaction with wild type EGFRGupta.21 Recently, Safdari et al., humanized the MR1 antibody by CDR grafting method for reducing its immunogenicity.3 However, this antibody is still not fully human and there is a potential risk of immunogenicity for human that limit their clinical application.
In this study, we first identified a fully human anti- EGFRvIII scFv from a scFv phage library. For effective screening of scFv phage library, we used a novel five rounds strategy for enrichment of rare specific clones against EGFRvIII with a negative selection in the 4th round for elimination of BSA binding clones.17
Results of polyclonal phage ELISA and monitoring phage titers in different rounds were indicating a successful enrichment toward increasing specificity (338-fold) to the EGFRvIII peptide. Results of PCR and DNA sequencing showed that, only three clones (2, 10, and 15) from six positive clones had expected size (930bp) representing presence of both VL and VH fragments. However, two of them (2 and15clones) had amber stop codon and clone 10 had a frame shift mutation in its coding sequences which these defects limited production of soluble scFv in non-suppressor strains e.g. HB2151. Studies performed by Xia et al., and Hung et al., were indicating the presence of extra amber (TAG) stop codon in scFvs isolated from semi-synthetic phage antibody libraries (e.g., Tomlinson I & J).22,23 Results of scFv- ELISA showed specific interaction of two scFv clones 2 and 15 with EGFRvIII. In this study, the presence of the amber stop codons in two scFvs (2 and15) obtained from Tomlinson I & J phage library led to production of scFvs fused to the pIII in E. coli TG1 (amber stop codon suppressor strain) among which the scFv clone 2 with higher binding affinity to EGFRvIII was produced in E. coli TG1 as a fusion protein with pIII. The quality of the purified scFv was confirmed by SDS-PAGE and western blotting analyses.
Conclusion
In this study, a scFv clone with high reactivity to EGFRvIII was selected using a novel selection strategy. The isolated svFv could be used for developing more effective diagnostic and therapeutic agents against EGFRvIII expressing cancers.
Acknowledgments
This study was supported by Drug Applied Research Center, Tabriz University of Medical Science, Tabriz, Iran. The results presented in this work are based on data set of PhD thesis, registered in Tabriz University of Medical Sciences.
Ethical Issues
Not applicable.
Conflict of Interest
The authors declare no conflict of interests.
Abbreviations
Mabs: Monoclonal antibodies, Tomlinson I+ J: The Human scFv phage libraries I + J, scFv: single-chain variable fragment, MPBS: Marvel milk powder in PBS, IPTG: β-D thiogalactopyranoside, SDS–PAGE: sodium dodecyl sulphate–polyacrylamide gel electrophoresis, DAB: chromomeric substrate - 3, 3’diaminobenzidine, MW: molecular weight, PVDF: poly vinylidine difluoride membrane.
References
- 1.Yarden Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37 Suppl 4:S3–8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 2.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): where wild things are altered. FEBS J. 2013;280(21):5350–70. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 3.Safdari Y, Farajnia S, Asgharzadeh M, Omidfar K, Khalili M. humMR1, a highly specific humanized single chain antibody for targeting EGFRvIII. Int Immunopharmacol. 2014;18(2):304–10. doi: 10.1016/j.intimp.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Jungbluth AA, Stockert E, Huang HJ, Collins VP, Coplan K, Iversen K. et al. A monoclonal antibody recognizing human cancers with amplification/overexpression of the human epidermal growth factor receptor. Proc Natl Acad Sci U S A. 2003;100(2):639–44. doi: 10.1073/pnas.232686499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lorimer IA, Keppler-Hafkemeyer A, Beers RA, Pegram CN, Bigner DD, Pastan I. Recombinant immunotoxins specific for a mutant epidermal growth factor receptor: targeting with a single chain antibody variable domain isolated by phage display. Proc Natl Acad Sci U S A. 1996;93(25):14815–20. doi: 10.1073/pnas.93.25.14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS. et al. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 1995;6(10):1251–9. [PubMed] [Google Scholar]
- 7.Grandis JR, Sok JC. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol Ther. 2004;102(1):37–46. doi: 10.1016/j.pharmthera.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto S, Yoshikawa K, Obata Y, Shibuya M, Aoki S, Yoshida J. et al. Monoclonal antibody against the fusion junction of a deletion-mutant epidermal growth factor receptor. Br J Cancer. 1996;73(11):1366–72. doi: 10.1038/bjc.1996.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotný J, Margolies MN. et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85(16):5879–83. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL. et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13(14):3245–60. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;15:624–8. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- 12.Kristensen P, Winter G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des. 1998;3(5):321–8. doi: 10.1016/S1359-0278(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 13.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19(15):4133–7. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18(9):989–94. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- 15.Harrison JL, Williams SC, Winter G, Nissim A. Screening of phage antibody libraries. Methods Enzymol. 1996;267:83–109. doi: 10.1016/s0076-6879(96)67007-4. [DOI] [PubMed] [Google Scholar]
- 16.Eteshola E. Isolation of scFv fragments specific for monokine induced by interferon-gamma (MIG) using phage display. J Immunol Methods. 2010;358(1-2):104–10. doi: 10.1016/j.jim.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rahbarnia L, Farajnia S, Babaei H, Majidi J, Veisi K, Tanomand A, et al. Invert biopanning: A novel method for efficient and rapid isolation of scFvs by phage display technology. Biologicals 2016. [DOI] [PubMed]
- 18.Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147(5):1709–19. [PubMed] [Google Scholar]
- 19.Su YC, Lim KP, Nathan S. Bacterial expression of the scFv fragment of a recombinant antibody specific for Burkholderia pseudomallei exotoxin. J Biochem Mol Biol. 2003;36(5):493–8. doi: 10.5483/bmbrep.2003.36.5.493. [DOI] [PubMed] [Google Scholar]
- 20.Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW. et al. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55(23):5536–9. [PubMed] [Google Scholar]
- 21.Gupta P, Han SY, Holgado-Madruga M, Mitra SS, Li G, Nitta RT. et al. Development of an EGFRvIII specific recombinant antibody. BMC Biotechnol. 2010;10:72. doi: 10.1186/1472-6750-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia J, Zhang Y, Qian J, Zhu X, Zhang Y, Zhang J. et al. Isolation, identification and expression of specific human CD133 antibodies. Sci Rep. 2013;3:3320. doi: 10.1038/srep03320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, Samanta M, Crawford SE, Estes MK, Neill FH, Atmar RL. et al. Identification of human single-chain antibodies with broad reactivity for noroviruses. Protein Eng Des Sel. 2014;27(10):339–49. doi: 10.1093/protein/gzu023. [DOI] [PMC free article] [PubMed] [Google Scholar]


