Abstract
Although several genes have been implicated in the development of the early-onset autosomal dominant form of Alzheimer's disease (AD), the genetics of late-onset AD (LOAD) is complex. Loci on several chromosomes have been linked to the disease, but so far only the apolipoprotein E gene has been consistently shown to be a risk factor. We have performed a large-scale single-nucleotide polymorphism (SNP)-based association study, across the region of linkage on chromosome 12, in multiple case-control series totaling 1,089 LOAD patients and 1,196 control subjects and report association with SNPs in the glyceraldehyde-3-phosphate dehydrogenase (GAPD) gene. Subsequent analysis of GAPD paralogs on other chromosomes demonstrated association with two other paralogs. A significant association between LOAD and a compound genotype of the three GAPD genes was observed in all three sample sets. Individually, these SNPs make differential contributions to disease risk in each of the casecontrol series, suggesting that variants in functionally similar genes may account for series-to-series heterogeneity of disease risk. Our observations raise the possibility that GAPD genes are AD risk factors, a hypothesis that is consistent with the role of GAPD in neuronal apoptosis.
Alzheimer's disease (AD), the most common form of dementia among the elderly, is a complex neurodegenerative disorder resulting from multiple genetic and nongenetic factors (1, 2, 3). A large body of evidence supports a central role for β-amyloid (Aβ) in AD pathogenesis. Mutations associated with familial AD in Aβ precursor protein and the γ-secretase subunits presenilin-1 and -2 all lead to increased production and/or deposition of Aβ42 (4). Because Aβ42 is known to be neurotoxic and/or neuroinflammatory in various experimental systems, it is believed that increased Aβ42 production leads to synaptic dysfunction and subsequent neuronal cell death, thereby contributing to memory loss and other symptoms (5). It follows that genetic mutations affecting various steps of the Aβ pathway, such as Aβ production, degradation/clearance, deposition, and neuronal apoptosis, could be associated with AD. Indeed, a number of such candidate genes have been reported to be risk factors for late-onset AD (LOAD), although so far only apolipoprotein E (APOE) has been confirmed by multiple independent studies (6). However, only ≈50% of AD cases carry an APOE4 allele,‡‡ and genetic studies have predicted that at least four other genes modify age of disease onset (7). Identification of other genetic risk factors is critical not only to further understanding of the disease mechanism but also to guide development of diagnostic reagents and disease-modifying treatments. The latter may be particularly sensitive to stratification by disease alleles.
Genome-wide linkage screens in affected sibling pairs have identified several candidate gene regions, notably on chromosomes 9, 10, and 12 (8, 9, 10, 11, 12, 13, 14). Association studies have evaluated a small number of biological candidate genes from the linkage regions, but many of these studies have used samples of relatively small size, thus limiting their power to identify disease-risk genes (for review, see ref. 15). Previously, we and others have reported linkage on chromosome 12 in LOAD families (8, 11, 12, 14, 16). The linkage signal in these studies came predominantly from individuals with no APOE4 alleles. Subsequently, association with AD has been reported for polymorphisms in several candidate genes, but none have been consistently replicated (for review, see ref. 17). To follow up our linkage results on chromosome 12, we have genotyped 282 single-nucleotide polymorphisms (SNPs) under our linkage peak in multiple case-control series totaling 1,089 AD subjects and 1,196 nondemented controls. Strong association was observed in a small region that includes a gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPD), which led us to examine this gene and its paralogs on other chromosomes in more depth.
Materials and Methods
General Case-Control Strategy. We first assayed SNPs across a broad region of chromosome 12 in the Washington University case-control sample. Significant markers were then genotyped in other case-control series, and additional fine-mapping markers were added near replicated SNPs. For markers that were replicated [those with similar odds ratios (ORs) and a one-tailed test P value threshold of 0.05], we used marker–marker linkage disequilibrium (LD) information to identify the minimum number of SNPs needed to be genotyped in the sibling pairs from our linkage study to tag the haplotype blocks.
Clinical Samples. Three AD case-control series, independently collected at Washington University (WashU) in St. Louis (419 cases/377 controls), University of California at San Diego (UCSD; 278 cases/412 controls), and a combined set from the Wales College of Medicine and King's College London (UK; 392 cases/407 controls), respectively, were used in the study. Cases from these series have a clinical diagnosis of dementia of the Alzheimer's type according to National Institute of Neurological Disorders and Stroke–Alzheimer's Disease and Related Disorders Association (18) or similar criteria with an age of disease onset of 60 years or more. All individuals are reported to be Caucasians. The three samples combined have 1,089 cases and 1,196 controls. Details of the three series are provided in Table 4, which is published as supporting information on the PNAS web site. The case series from the linkage sample comprises 461 unrelated individuals selected from affected sibling pairs collected by the National Institute of Mental Health–AD Genetic Consortium and the Indiana Alzheimer Disease Center (11). The combined U.S.–sample set controls (WashU + UCSD) were used as controls for the case linkage sample.
Genotyping. The polymorphisms analyzed in this study were taken from either the Celera human genome database that includes publicly available SNP data or the Applera Genome Resequencing Initiative database. SNP genotyping was performed by allele-specific real-time PCR (19). Genotyping accuracy was >99%, as determined by internal comparisons of differentially designed assays for the same marker and comparisons for the same marker across our groups. The case-control sample derived from the linkage sample was genotyped with Pyrosequencing technology (Biotage, Uppsala).
Statistical Analysis. To identify possible genotyping errors, Hardy–Weinberg equilibrium tests were first done in both the case and control samples. Assays with significant deviation from the Hardy–Weinberg equilibrium in controls (P < 0.05, 20 markers) were closely examined for genotyping quality. Allelic association was then examined on nonstratified samples by χ2 tests. ORs and the 95% confidence intervals for an allelic effect were also estimated. Meta analyses that combine P values of both exploratory and confirmatory sample sets were done by using the Cochran–Mantel–Haenszel test (Categorical Data Analysis, ref. 20). The combined ORs were corrected for sample set unless otherwise stated. Evidence for replication, rather than multiple testing corrections, was used to evaluate the significance of associated SNPs. Multiple testing-corrected P values for the unstratified analysis were calculated by permutation tests after performing 1,000 random permutations of disease status for each tested SNP. For the discovery sample, the permutation P values represent the proportion of permutation samples in which at least one SNP had a P value as extreme as or more extreme than the observed P value. For the replication samples, the P value was calculated as the proportion of permutations in which at least one SNP from either of the two replication samples had a P value as extreme as or more extreme than the observed P value. Power for the allelic association test was calculated under a test of binomial proportions, considering the effect size observed in the initially genotyped sample set and the actual sample size of the replication series at a significance level of P = 0.05 (20).
Marker–marker linkage disequilibrium measures, D′ and r2, were calculated by using ldmax (www.sph.umich.edu/csg/abecasis/GOLD/docs/ldmax.html) to elucidate the haplotype block structure. For haplotype analysis, haplotypes were inferred from the genotypic data. Haplotype frequencies were estimated for both the case and control samples, and their association with disease status was then assessed by using a score test with haplotypes coded in an additive fashion (21). In addition to this unstratified analysis, haplotype association tests were also done on the basis of APOE4 strata (presence or absence). The Cochran–Armitage test for trend was used to assess the association of LOAD with the three GAPD genes for the multilocus genotype (22).
Test for Population Stratification. All SNPs tested in the WashU sample, including SNPs located on other chromosomes, were divided into 1,220 bins based on intermarker distances (SNPs within 55 kbp of each other). We randomly selected one SNP from each bin to construct 10 sets of 1,220 markers. All SNPs tested in both the UK and UCSD samples were also divided into bins based on intermarker distances, yielding 183 bins. Ten sets of 183 markers were generated by choosing a single random marker from each bin. An overall test of population stratification was performed by using the method presented by Pritchard and Rosenberg (23). In addition, a structured association method was used to further test each sample set for stratification between cases and controls (24). This method is implemented by the programs structure and strat (25). Studies have shown that ≈100 SNPs are required to accurately resolve population structure (26). Given those estimates, we have sufficient power to resolve population structure and detect stratification.
Results
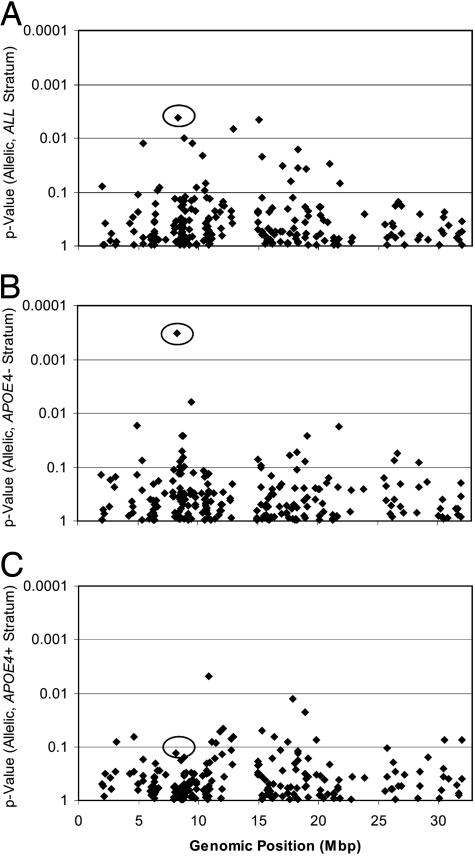
SNP Genotyping Identifies LOAD Gene Region on Chromosome 12. We conducted a SNP-based association screen, targeting the region of chromosome 12p between the telomere and D12S1042 (1.9–32.8 Mbp), an interval that had previously shown modest evidence of linkage to the whole sample and the APOE4-negative subgroup in our family study (11). According to the Celera human genome assembly, this region contains 436 known or predicted genes. One hundred fifty-four of these genes were targeted with 282 polymorphic markers [mean allele frequency ± standard deviation, 24.4 ± 13.2%; mean intermarker distance ± standard deviation, 107 ± 240 kbp; minor allele frequency (MAF) >2%; Fig. 4, which is published as supporting information on the PNAS web site]. A total of 223 exploratory markers (MAF >2%) were screened in the WashU sample (Fig. 1). After testing the first 24 exploratory markers, a strong hit (rs3741916 in GAPD, P = 0.004) was identified that remained significant after correcting for APOE4, gender, and age of onset (AOO). Follow-up by screening the UCSD series with rs3741916 showed that the marker replicated (P = 0.027). Further testing in the UK samples did not replicate the finding, but a combined analysis of the three samples was significant (P = 0.047). Twelve other markers, located in different regions of chromosome 12, were significant in the exploratory screen of the WashU series but did not replicate. As a result, we initiated fine mapping near rs3741916 with 62 additional SNPs (54 SNPs with MAF >2%) in the WashU series, of which 25 were significant (range, P = 0.00002–0.046). One of the 25 markers, also in GAPD, replicated at P < 0.05 in the UCSD series (Table 1). We further assessed these results and applied multiple testing corrections to the combined 277 exploratory and fine-mapping markers tested in the WashU sample (MAF >2%, five other SNPs were tested only in the UK sample). Five SNPs remained significant (rs2079867, rs714774, rs2008134, rs2072374, and rs7311174; adjusted P value range, 0.007–0.029). None of the fine-mapping markers, including rs3741916, remained significant (P < 0.05) in the UCSD or UK sample after adjustment for multiple testing. The association with rs3741916 in the WashU sample mirrored the linkage results by showing strong allelic association in the APOE4-negative subgroup (Fig. 1 B and C), as did 21 fine-mapping markers (range, P = 0.00032–0.045; Fig. 2). Only four markers were significant in the APOE4-positive substratum (range, P = 0.01–0.05). Six of these 22 markers, located in GAPD, chromosome condensation-related SMC-associated protein 1 (CNAP1), and plakophilin 2 pseudogene 1 (PKP2P1) showed significant allelic association (P < 0.05) in the WashU series and replicated at P < 0.05 in either the UCSD or the UK series (Table 1 and Table 5, which is published as supporting information on the PNAS web site). In the combined sample, ORs for four of these six markers (rs2008134, rs7311174, rs2072374, and rs3741916) remained significant after correcting for gender, AOO, and sample set. We further tested four of the six markers (rs2008134, rs7311174, rs3741916, and rs1060621) in one affected individual per family from the linkage sample and compared them to all control samples collected in the U.S. (WashU and UCSD), because the linkage sample was also collected in the U.S. One GAPD marker (rs3741916) was significant in this comparison (P = 0.006, unadjusted). Analyses with independent controls were not done, because these were not available; analyses that determine whether these markers account for the linkage signal also were not done, because they lack power, due to the small number of sibling pairs contributing to the linkage signal on chromosome 12 (11).
Fig. 1.
Allelic P values of 223 markers are plotted for a 30-Mbp region of chromosome 12p. P values were for genotyping results of all exploratory markers tested in the WashU case-control set. (A) ALL stratum: analysis of all case and control samples in the set. (B) The APOE4 absent (APOE4-) stratum includes all samples without an APOE4 allele. (C) The APOE4 present (APOE4+) stratum includes all samples that have one or two APOE4 alleles. rs3741916 in GAPD is highlighted with a circle.
Table 1. Replicated marker statistics.
| Strata/gene/SNP ID/type/position | Sample | Cs allele frequency | Co allele frequency | P value | OR (95% CI) |
|---|---|---|---|---|---|
| All | WashU | 29.4 | 23.1 | 0.0041 | 1.38 (1.11:1.74) |
| GAPD | UCSD | 27.5 | 22.9 | 0.027 | 1.27 (1.00:1.64) |
| rs3741916 | UK | 28.1 | 30.9 | 0.89 | 0.87 (0.70:1.09) |
| UTR5/89.3 | W/UC/UK | 28.5 | 25.6 | 0.047 | 1.14 (1.00:1.30) |
| Linkage | 28.0 | 23.0 | 0.006 | 1.31 (1.06:1.61) | |
| rs1060621 | WashU | 20.6 | 16.3 | 0.033 | 1.33 (1.02:1.74) |
| Intron/90.0 | UCSD | 21.6 | 17.6 | 0.036 | 1.28 (0.98:1.69) |
| UK | 19.5 | 19.7 | 0.53 | 0.99 (0.77:1.27) | |
| W/UC/UK | 20.5 | 17.9 | 0.030 | 1.18 (1.02:1.37) | |
| Linkage | 19.0 | 16.6 | 0.027 | 1.24 (1.00:1.55) | |
| APOE4- | WashU | 27.7 | 37.5 | 0.0030 | 0.63 (0.47:0.86) |
| PKP2P1 | UCSD | 33.5 | 33.5 | 0.51 | 1.00 (0.70:1.43) |
| rs2008134 | UK | 27.0 | 33.5 | 0.024 | 0.73 (0.54:1.00) |
| MS/33.8 | W/UC/UK | 28.8 | 34.8 | 0.0025 | 0.75 (0.63:0.91) |
| Linkage | 31.0 | 35.4 | 0.092 | 1.22 (0.91:1.65) | |
| CNAP1 | WashU | 19.6 | 14.2 | 0.036 | 1.47 (1.02:2.12) |
| rs2072373 | UCSD | 21.5 | 15.7 | 0.029 | 1.47 (0.99:2.20) |
| Intron/77.2 | UK | 18.1 | 18.9 | 0.61 | 0.95 (0.66:1.36) |
| W/UC/UK | 19.5 | 16.3 | 0.040 | 1.25 (1.01:1.55) | |
| rs7311174 | WashU | 24.5 | 34.6 | 0.002 | 0.61 (0.45:0.84) |
| Intron/80.9 | UCSD | 29.0 | 31.7 | 0.24 | 0.87 (0.62:1.25) |
| UK | 23.3 | 29.0 | 0.034 | 0.74 (0.54:1.02) | |
| W/UC/UK | 25.2 | 31.7 | 0.0008 | 0.72 (0.60:0.88) | |
| Linkage | 28.4 | 33.1 | 0.074 | 1.24 (0.92:1.69) | |
| rs2072374 | WashU | 24.7 | 34.6 | 0.0023 | 0.62 (0.46:0.84) |
| Silent/83.2 | UCSD | 28.8 | 31.8 | 0.21 | 0.86 (0.61:1.23) |
| UK | 23.5 | 29.0 | 0.039 | 0.75 (0.54:1.03) | |
| W/UC/UK | 25.2 | 31.7 | 0.0009 | 0.72 (0.60:0.88) | |
| GAPD | WashU | 31.9 | 21.4 | 0.0003 | 1.72 (1.28:2.33) |
| rs3741916 | UCSD | 28.4 | 22.0 | 0.030 | 1.41 (0.98:2.02) |
| UTR5/89.3 | UK | 28.0 | 30.6 | 0.79 | 0.88 (0.65:1.20) |
| W/UC/UK | 29.7 | 24.7 | 0.0084 | 1.27 (1.06:1.53) | |
| Linkage | 27.7 | 21.7 | 0.042 | 1.38 (0.96:1.99) | |
| rs1060621 | WashU | 20.6 | 14.8 | 0.028 | 1.49 (1.04:2.14) |
| Intron/90.0 | UCSD | 22.5 | 16.8 | 0.036 | 1.43 (0.97:2.13) |
| UK | 17.3 | 19.1 | 0.75 | 0.88 (0.62:1.27) | |
| W/UC/UK | 19.9 | 17.0 | 0.065 | 1.22 (0.99:1.51) | |
| Linkage | 15.9 | 15.6 | 0.19 | 1.18 (0.81:1.72) |
Allelic P values and ORs are presented. Linkage, controls are the combined U.S.-sample set controls (WashU + UCSD). The effect of rs374916 and rs2008134 differed significantly by APOE4 (all three samples combined: P < 0.05, Breslow Day test). Position, marker positions are relative to rs758738 in kbp, located at 8,171,519 bp in the Celera genome assembly. MS, missense mutation; W/UC/UK, WashU, UCSD, and UK combined; CI, confidence interval, Cs, case; Co, control.
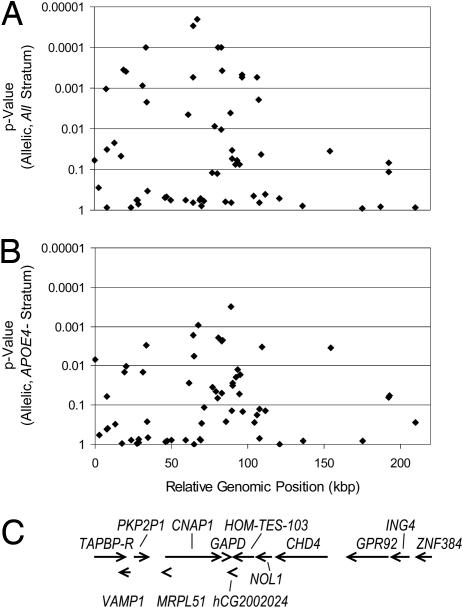
Fig. 2.
Allelic P values from 62 fine-mapping markers and rs3741916 in GAPD. P values are presented for the ALL stratum (A) and the APOE4 absent (APOE4-) stratum (B) of the WashU sample set, along with a gene map of the region based on Celera's R27 human genome assembly (C). rs758738 is located at 8,171,519 bp in the Celera genome assembly and is shown at relative genomic position 0.
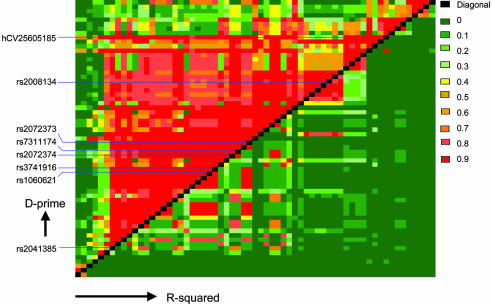
We assessed marker–marker linkage disequilibrium in this region in the APOE4-negative samples from the WashU series by calculating D′ and r2 values (Fig. 3; Fig. 5 and Table 6, which are published as supporting information on the PNAS web site). One main block of linkage disequilibrium extended from rs2041385 to hCV25605185 over 128 kbp. This block can be tagged with 10 independent markers, when markers of high-pairwise LD are defined as having D′ ≥ 0.8 and r2 ≥ 0.7 (markers below 4% allele frequency were not considered). Six of the 10 tags contain polymorphisms with allelic P < 0.05 in the WashU series. Haplotypes for several tagging marker combinations were calculated, but none gave more significant or consistent results across the three case-control series when compared with the individual markers, a result that can be expected for markers in tightly linked loci (27). The LD structure for the WashU cases and the other two series did not differ from the LD structure in the WashU controls, nor did the whole-sample results from the APOE4 negative subset (data not shown). Many of the significant markers were in LD with SNPs either in CNAP1 and PKP2P1 or in GAPD (Fig. 3 and Table 7, which is published as supporting information on the PNAS web site). After genotyping seven markers in PKP2P1 and two in the plakophilin 2 gene without finding any other replicated associations, we decided not to further characterize the PKP2P1 association.
Fig. 3.
LD metrics. The LD map was constructed from APOE4-negative subjects (cases and controls) in the WashU sample set with 63 markers, covering a 418-kbp region. D′ values are shown in the top left triangle, and r2 values are shown in the bottom right.
Population Stratification. We tested the case-control sets for population stratification as described in Materials and Methods. The 10 sets of at least 183 markers from the WashU, UK, and UCSD samples showed no evidence of stratification; i.e., the best solution for each sample was a single population. Our failure to detect evidence of stratification suggests that population stratification does not introduce significant bias into our analyses, because studies have shown that only 100 SNPs are sufficient to accurately resolve population structure (26).
SNPs in GAPD Paralogs Are Associated with LOAD. Based on the role of GAPD in neuronal apoptosis (28, 29) and the previously reported changes of GAPD expression/activity in AD patients (30, 31), we characterized the GAPD association further. We hypothesized that variation in other GAPD family members may influence risk for AD and that multilocus interactions may be important in determining an individual's risk for disease. Two functional GAPD genes, GAPD and GAPDS (or GAPD2), are known to be present in mammalian cells. Genomic studies have shown that a large number of GAPD splice variants exist, and that some of the GAPD pseudogenes are transcribed (32). To examine GAPD paralogs as well as some of the pseudogenes, we genotyped nine SNPs in GAPDS (chromosome 19, 9.5 Mbp proximal to APOE), three SNPs in hCG40445 (pseudogene on chromosome 12), and one SNP each in hCG1641716 (chromosome 10), hCG16322 (pseudogene on chromosomes 4), and hCG29012 (pseudogene on chromosomes 9), respectively. We chose these GAPD paralogs because they are on chromosomes with multiple reported linkage findings for LOAD (9, 11). The UK sample was used as the exploratory set for GAPDS, and the UCSD sample set was used for hCG16322. Polymorphisms for all other genes were tested in the WashU sample first. Two of the 15 markers, both in GAPDS, were significant in the exploratory sample and replicated in one other series. The effect of GAPDS differed significantly by age (interaction with age in all three series combined, P = 0.01), with the strongest association in patients with disease onset before 75 years of age (Table 2 and Table 8, which is published as supporting information on the PNAS web site). This observation led us to evaluate the other GAPD markers for association with early and late age of onset (AOO). This led to the identification of one additional replicated SNP, located in the GAPD pseudogene (p-GAPD) on chromosome 12. In contrast to the GAPDS finding, the significant association occurs with later AOO (75 years or greater). This polymorphism is a missense mutation within p-GAPD, but it also maps to an intron of a protein phosphatase 2C homolog (hCG40446), which is transcribed in the opposite direction to p-GAPD.
Table 2. Association of GAPD paralogs.
| Strata/gene/SNP ID/type | Sample | Cs allele frequency | Co allele frequency | P value | OR (95% CI) |
|---|---|---|---|---|---|
| All | UK | 34.8 | 42.1 | 0.0031 | 0.73 (0.60:0.90) |
| GAPDS | WashU | 37.3 | 42.3 | 0.027 | 0.81 (0.66:1.00) |
| rs12984928 | UCSD | 39.4 | 40.6 | 0.34 | 0.95 (0.75:1.20) |
| Intron | W/UC/UK | 36.9 | 41.7 | 0.0016 | 0.81 (0.72:0.93) |
| rs4806173 | UK | 34.8 | 41.9 | 0.0039 | 0.74 (0.60:0.91) |
| TFBS | WashU | 36.9 | 42.4 | 0.016 | 0.79 (0.64:0.98) |
| UCSD | 39.0 | 40.3 | 0.33 | 0.95 (0.75:1.20) | |
| W/UC/UK | 36.6 | 41.5 | 0.0011 | 0.81 (0.72:0.92) | |
| LT75 | UK | 28.3 | 44.4 | 0.00002 | 0.49 (0.35:0.69) |
| GAPDS | WashU | 34.3 | 40.7 | 0.048 | 0.76 (0.55:1.05) |
| rs12984928 | UCSD | 36.7 | 39.5 | 0.25 | 0.88 (0.63:1.26) |
| Intron | W/UC/UK | 33.1 | 41.7 | 0.00012 | 0.68 (0.57:0.83) |
| rs4806173 | UK | 27.8 | 44.4 | 0.00001 | 0.48 (0.35:0.67) |
| TFBS | WashU | 33.9 | 41.3 | 0.027 | 0.72 (0.53:1.01) |
| UCSD | 36.1 | 39.8 | 0.19 | 0.85 (0.60:1.21) | |
| W/UC/UK | 32.6 | 42.0 | 0.00003 | 0.66 (0.55:0.80) | |
| GE75 | WashU | 31.4 | 38.2 | 0.039 | 0.74 (0.56:0.99) |
| p-GAPD | UCSD | 34.9 | 33.1 | 0.66 | 1.08 (0.74:1.59) |
| rs2029721 | UK | 28.8 | 34.9 | 0.024 | 0.75 (0.57:1.00) |
| Missense | W/UC/UK | 30.8 | 35.2 | 0.018 | 0.80 (0.68:0.97) |
LT75, age of onset <75 years. GE75, age of onset ≥75 years. W/UC/UK, WashU, UCSD, and UK combined; CI, confidence interval. Cs, case; Co, control.
Expression of the GAPD Genes. We next assessed expression of the GAPD genes in brain (Fig. 6 and Supporting Text, which are published as supporting infromation on the PNAS web site). Reverse transcriptase PCR detected expression of GAPD and p-GAPD at similar levels in all tissues examined. The amplification product of the intronless p-GAPD gene was not detected without addition of reverse transcriptase, excluding the possibility of contamination from genomic DNA. Sequencing the PCR product confirmed the detected amplicon as p-GAPD. GAPDS, which is predominantly expressed in testis and also whole blood and various lymphoid cell lines (http://expression.gnf.org/cgi-bin/index.cgi), was detected at much lower levels in brain tissues we examined.
Multilocus Genotype of GAPD Genes Is Associated with LOAD. Because the three GAPD genes may function in similar pathways, we examined whether there is an association of LOAD with a multilocus genotype formed by these genes. A William's-corrected G test, where no mode of inheritance was assumed, was performed separately for each pair-wise combination of genotypes against disease status, showing significant values for GAPD-GAPDS (P < 0.01) and GAPDS-pGAPD (P < 0.005) but not GAPD-pGAPD (33). Additionally, a two-locus genotypic disequilibrium was calculated separately for cases and controls in each of the above comparisons (34). Significant disequilibrium was not observed at the P < 0.01 level. However, an interesting pattern was observed when the GAPD and p-GAPD SNPs were held constant with the genotypes CG and GG, respectively, and the genotype effect size was measured for the GAPDS SNP. All three sample sets, either individually or combined, exhibited a consistent and strong trend in risk estimates across the three GAPDS genotypes, conditional on the CG-GG two-locus genotype at the other two genes (Table 3). The combined sample result was significant even after correcting for all 27 possible genotype combinations where two genotypes are held constant (P < 0.05). No significant three-way interaction was observed in a logistic model (P = 0.17), which may have been due to low power because of small sample sizes for some multilocus genotype combinations.
Table 3. Multilocus genotype analysis.
| Sample | Multilocus genotype* | Case | Control | OR | OR 95% CI | Test of proportions P value |
|---|---|---|---|---|---|---|
| W/UC/UK | CG-CC-GG | 78 | 54 | 1.40 | 1.11: 1.76 | 0.047† |
| CG-CG-GG | 93 | 90 | 1 | n/c | ||
| CG-GG-GG | 17 | 38 | 0.43 | 0.31: 0.6 | ||
| UCSD | CG-CC-GG | 17 | 16 | 1.24 | 0.77: 1.99 | 0.047 |
| CG-CG-GG | 18 | 21 | 1 | n/c | ||
| CG-GG-GG | 2 | 10 | 0.23 | 0.1: 0.54 | ||
| UK | CG-CC-GG | 33 | 25 | 1.41 | 1.01: 1.98 | 0.025 |
| CG-CG-GG | 44 | 47 | 1 | n/c | ||
| CG-GG-GG | 7 | 16 | 0.47 | 0.28: 0.77 | ||
| WashU | CG-CC-GG | 28 | 13 | 1.53 | 0.99: 2.36 | 0.045 |
| CG-CG-GG | 31 | 22 | 1 | n/c | ||
| CG-GG-GG | 8 | 12 | 0.47 | 0.28: 0.81 |
Each individual sample set P value was calculated with a two-tailed test of proportions. n/c, not calculated; CI, confidence interval.
The multilocus genotype represents rs3741916-rs4806173-rs2029721 (GAPD-GAPDS-pGAPD). The OR was calculated relative to the most common genotype
W/UC/UK sample P value was derived with Fisher's combined P value from a two-tailed test for the UCSD sample and one-tailed tests for the other two samples. Dunn-Sidak correction for 27 tests was applied (uncorrected, P 0.0018)
Discussion
Heterogeneity of LOAD Genes on Chromosome 12. Although several studies have reported evidence of linkage to chromosome 12, the location of the linkage peaks has been somewhat variable (for review of these results, see ref. 1). Two regions, one located on the short arm and one on the long arm of the chromosome, have been suggested to contain an AD susceptibility gene (11, 12, 14, 35, 36), implying heterogeneity among the linkage samples that were studied. Despite these differences in the precise location of the peak, the most significant linkage observations were consistently observed in the APOE4-negative subgroup of families. A similar result was observed for the SNPs showing evidence of association in this study. The association was strong in the WashU sample but weaker in the other case-control series. This heterogeneity among datasets may reflect a type1 error in the original dataset, insufficient power in the replication datasets, or differences in the underlying genetic risk for disease in these different populations, as suggested by the variable linkage data. structure results argue that there is no population stratification in these datasets, ruling this out as a possible reason for the heterogeneity. Consistent with the common disease–common variant hypothesis, many of the genetic variants that alter the risk for complex diseases are expected to have only a small effect on disease outcome (see, for example, ref. 37). The power to obtain typical thresholds of P value significance after applying multiple testing corrections is limited for such markers, because significance is a function of sample size, allele frequency, and OR. Therefore, replication may be a more practical measure of overall significance, especially for markers with small effect sizes and/or when multiple test adjustment for large numbers of markers limits the available power given a fixed sample size.
Role of GAPD in Neuronal Apoptosis and Neurodegeneration. Despite this heterogeneity in the association of individual markers with LOAD, we observed a consistent association with the compound genotype of the three GAPD genes. Not only is GAPD a key enzyme in cellular energy production, but it also plays an important role in several other cellular processes, including neuronal apoptosis and neurodegenerative diseases, including AD. It is known to bind Aβ precusor protein (38) as well as Aβ (39). In several models of experimentally induced neuronal apoptosis, it is consistently observed that cell death is preceded by (i) an increase in the levels of GAPD mRNA and protein and (ii) translocation of GAPD protein from the cytoplasm to the nucleus (28, 29). GAPD-mediated apoptosis can be inhibited by antisense oligodeoxyribonucleotides to GAPD and tacrine, a Food and Drug Administration-approved medication for AD patients (40). Recently, it was shown that donepezil and tacrine reduce GAPD promoter activity (41). Other drugs targeting GAPD directly (e.g., TCH346 from Novartis) are under development for AD and other neurodegenerative diseases. Furthermore, differences in GAPD activity or conformation between AD and normal subjects may exist. GAPD activity has been reported to be ≈50% higher in brain samples from AD patients than control subjects (30), and GAPD has been found to be overexpressed in Down's syndrome brain (42). Further research is required to ascertain the role of the variants showing association with AD in this study, not only at the molecular but also the cellular level. One prediction is that neuronal cells bearing the risk alleles are more susceptible to certain apoptotic stimuli.
LD of GAPD and p-GAPD with Other Reported Chromosome 12 Candidate Genes. Although it is possible that our significant findings are due to LD with other reported chromosome 12 candidate genes (e.g., A2M, LBP1, LRP, and OLR1; for review, see ref. 17), it is extremely unlikely to be the case. The region of LD for GAPD extends for ≈130 kbp, whereas the closest candidate gene (A2M) is ≈2.5 Mbp further distal to GAPD. However, to address this possibility, we genotyped five markers in A2M in the WashU case-control series without detecting significant association. None of these five markers was in LD (D′ < 0.15, r2 < 0.005) with the markers in Table 1. The distance between GAPD and the next candidate gene (OLR1) is 7 Mbp. The closest candidate gene to p-GAPD (LRP1) is 5.6 Mbp away.
Conclusion
We have used a systematic approach to analyze potentially functional SNPs in 154 genes from the AD linkage region on the short arm of chromosome 12. SNPs within the GAPD gene showed evidence of association in two case-control series. Analysis of functional SNPs in GAPD paralogs on other chromosomes further supported the role of GAPD in risk for LOAD.
Note. While this work was under review, a publication by Laschet et al. appeared in the Journal of Neuroscience (43) and described GAPD as a GABAA receptor kinase, therefore providing another molecular mechanism for the direct involvement of GAPD in neurotransmission.
Supplementary Material
Acknowledgments
We are grateful to the families for their participation in these studies. We thank Amanda Myers and Sam Broder for critical review of the manuscript and our colleagues at Celera Diagnostics for providing expert technical support. Mary Coats and Elizabeth Grant coordinated the Washington University case material, and Mary Sundsmo coordinated the University of California at San Diego case material. Funding for this work was partly provided by the National Institutes of Health [Alzheimer's Disease Research Center Grants P50 AG05681 (to J.C.M.), P50 AG05131 (to L.T.), RO1 AG16208 (to A.G.), and PO1 AG03991 (to J.C.M.)], the Medical Research Council, U.K. (J.W., M.O., S.L., and J.P.), and the Alzheimer's Research Trust (J.W., M.O., and S.L.). J.H. was supported by the National Institute on Aging intramural program and thanks the Verum Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AD, Alzheimer's disease; APOE, apolipoprotein E; GAPD, glyceraldehyde-3-phosphate dehydrogenase; LOAD, late-onset AD; SNP, single-nucleotide polymorphism; WashU, Washington University; UCSD, University of California at San Diego; UK, Wales College of Medicine and King's College London; LD, linkage disequilibrium; OR, odds ratio; Aβ, β-amyloid.
Footnotes
Roses, A., Devlin, B., Conneally, P. M., Small, G., Saunders, A. M., Pritchard, M., Locke, P. A., et al. (1995) Am. J. Hum. Genet. Suppl. 57, A202 (abstr.).
References
- 1.Myers, A. J. & Goate, A. M. (2001) Curr. Opin. Neurol. 14, 433-440. [DOI] [PubMed] [Google Scholar]
- 2.Tanzi, R. E. & Bertram, L. (2001) Neuron 32, 181-184. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy, J. L., Farrer, L. A., Andreasen, N. C., Mayeux, R. & St George-Hyslop, P. (2003) Science 302, 822-826. [DOI] [PubMed] [Google Scholar]
- 4.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe, D. J. (2002) Science 298, 789-791. [DOI] [PubMed] [Google Scholar]
- 6.Roses, A. D. (1998) Ann. N.Y. Acad. Sci. 855, 738-743. [DOI] [PubMed] [Google Scholar]
- 7.Warwick Daw, E., Payami, H., Nemens, E. J., Nochlin, D., Bird, T. D., Schellenberg, G. D. & Wijsman, E. M. (2000) Am. J. Hum. Genet. 66, 196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehoe, P., Wavrant-De Vrieze, F., Crook, R., Wu, W. S., Holmans, P., Fenton, I., Spurlock, G., Norton, N., Williams, H., Williams, N., et al. (1999) Hum. Mol. Genet. 8, 237-245. [DOI] [PubMed] [Google Scholar]
- 9.Blacker, D., Bertram, L., Saunders, A. J., Moscarillo, T. J., Albert, M. S., Wiener, H., Perry, R. T., Collins, J. S., Harrell, L. E., Go, R. C., et al. (2003) Hum. Mol. Genet. 12, 23-32. [DOI] [PubMed] [Google Scholar]
- 10.Myers, A., Holmans, P., Marshall, H., Kwon, J., Meyer, D., Ramic, D., Shears, S., Booth, J., DeVrieze, F. W., Crook, R., et al. (2000) Science 290, 2304-2305. [DOI] [PubMed] [Google Scholar]
- 11.Myers, A., Wavrant De-Vrieze, F., Holmans, P., Hamshere, M., Crook, R., Compton, D., Marshall, H., Meyer, D., Shears, S., Booth, J., et al. (2002) Am. J. Med. Genet. 114, 235-244. [DOI] [PubMed] [Google Scholar]
- 12.Pericak-Vance, M. A., Bass, M. P., Yamaoka, L. H., Gaskell, P. C., Scott, W. K., Terwedow, H. A., Menold, M. M., Conneally, P. M., Small, G. W., Vance, J. M., et al. (1997) J. Am. Med. Assoc. 278, 1237-1241. [PubMed] [Google Scholar]
- 13.Pericak-Vance, M. A., Grubber, J., Bailey, L. R., Hedges, D., West, S., Santoro, L., Kemmerer, B., Hall, J. L., Saunders, A. M., Roses, A. D., et al. (2000) Exp. Gerontol. 35, 1343-1352. [DOI] [PubMed] [Google Scholar]
- 14.Rogaeva, E., Premkumar, S., Song, Y., Sorbi, S., Brindle, N., Paterson, A., Duara, R., Levesque, G., Yu, G., Nishimura, M., et al. (1998) J. Am. Med. Assoc. 280, 614-618. [DOI] [PubMed] [Google Scholar]
- 15.Hirschhorn, J. N., Lohmueller, K., Byrne, E. & Hirschhorn, K. (2002) Genet. Med. 4, 45-61. [DOI] [PubMed] [Google Scholar]
- 16.Wu, W. S., Holmans, P., Wavrant-DeVrieze, F., Shears, S., Kehoe, P., Crook, R., Booth, J., Williams, N., Perez-Tur, J., Roehl, K., et al. (1998) J. Am. Med. Assoc. 280, 619-622. [DOI] [PubMed] [Google Scholar]
- 17.Kamboh, M. I. (2004) Ann. Hum. Genet. 68, 381-404. [DOI] [PubMed] [Google Scholar]
- 18.McKhann, G., Drachman, D., Folstein, M., Katzman, R., Price, D. & Stadlan, E. M. (1984) Neurology 34, 939-944. [DOI] [PubMed] [Google Scholar]
- 19.Germer, S., Holland, M. J. & Higuchi, R. (2000) Genome Res. 10, 258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agresti, A. (1990) Categorical Data Analysis (Wiley, New York).
- 21.Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. (2002) Am. J. Hum. Genet. 70, 425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armitage, P. (1955) Biometrics 26, 535-546. [Google Scholar]
- 23.Pritchard, J. K. & Rosenberg, N. A. (1999) Am. J. Hum. Genet. 65, 220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritchard, J. K., Stephens, M., Rosenberg, N. A. & Donnelly, P. (2000) Am. J. Hum. Genet. 67, 170-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard, J. K., Stephens, M. & Donnelly, P. (2000) Genetics 155, 945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turakulov, R. & Easteal, S. (2003) Hum. Hered. 55, 37-45. [DOI] [PubMed] [Google Scholar]
- 27.Morris, A. P., Whittaker, J. C. & Balding, D. J. (2004) Am. J. Hum. Genet. 74, 945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawa, A., Khan, A. A., Hester, L. D. & Snyder, S. H. (1997) Proc. Natl. Acad. Sci. USA 94, 11669-11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishitani, R. & Chuang, D. M. (1996) Proc. Natl. Acad. Sci. USA 93, 9937-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soucek, T., Cumming, R., Dargusch, R., Maher, P. & Schubert, D. (2003) Neuron 39, 43-56. [DOI] [PubMed] [Google Scholar]
- 31.Mazzola, J. L. & Sirover, M. A. (2001) J. Neurochem. 76, 442-449. [DOI] [PubMed] [Google Scholar]
- 32.Tso, J. Y., Sun, X. H., Kao, T. H., Reece, K. S. & Wu, R. (1985) Nucleic Acids Res. 13, 2485-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokal, R. & Rohlf, F. (1995) Biometry (Freeman, New York).
- 34.Weir, B. S. (1996) Genetic Data Analysis II (Sinauer, Sunderland, MA)..
- 35.Zubenko, G. S., Hughes, H. B., 3rd, & Stiffler, J. S. (1999) Neurology 52, 725-732. [DOI] [PubMed] [Google Scholar]
- 36.Mayeux, R., Lee, J. H., Romas, S. N., Mayo, D., Santana, V., Williamson, J., Ciappa, A., Rondon, H. Z., Estevez, P., Lantigua, R., et al. (2002) Am. J. Hum. Genet. 70, 237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefansson, H., Sarginson, J., Kong, A., Yates, P., Steinthorsdottir, V., Gudfinnsson, E., Gunnarsdottir, S., Walker, N., Petursson, H., Crombie, C., et al. (2003) Am. J. Hum. Genet. 72, 83-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulze, H., Schuler, A., Stuber, D., Dobeli, H., Langen, H. & Huber, G. (1993) J. Neurochem. 60, 1915-1922. [DOI] [PubMed] [Google Scholar]
- 39.Oyama, R., Yamamoto, H. & Titani, K. (2000) Biochim. Biophys. Acta 1479, 91-102. [DOI] [PubMed] [Google Scholar]
- 40.Katsube, N., Sunaga, K., Aishita, H., Chuang, D. M. & Ishitani, R. (1999) J. Pharmacol. Exp. Ther. 288, 6-13. [PubMed] [Google Scholar]
- 41.Tsuchiya, K., Tajima, H., Yamada, M., Takahashi, H., Kuwae, T., Sunaga, K., Katsube, N. & Ishitani, R. (2004) Life Sci. 74, 3245-3258. [DOI] [PubMed] [Google Scholar]
- 42.Lubec, G., Labudova, O., Cairns, N. & Fountoulakis, M. (1999) Neurosci. Lett. 260, 141-145. [DOI] [PubMed] [Google Scholar]
- 43.Laschet, J. J., Minier, F., Kurcewicz, I., Bureau, M. H., Trottier, S., Jeanneteau, F., Griffon, N., Samyn, B., Van Beeumen, J., Louvel, J., et al. (2004) J. Neurosci. 24, 7614-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.