Abstract
The role of Forkhead Box F1 (FoxF1) transcription factor in carcinogenesis is not well characterized. Depending on tissue and histological type of cancer, FoxF1 was shown to be either oncogene or tumor suppressor. Alveolar rhabdomyosarcoma (RMS) is the most aggressive pediatric soft tissue sarcoma. While FoxF1 is highly expressed in alveolar RMS, the functional role of FoxF1 in RMS is unknown. The present study demonstrates that expression of FoxF1 and its closely related transcription factor FoxF2 are essential for rhabdomyosarcoma tumor growth. Depletion of FoxF1 or FoxF2 in rhabdomyosarcoma cells decreased tumor growth in orthotopic mouse models of RMS. The decreased tumorigenesis was associated with the reduced tumor cell proliferation. Cell cycle regulatory proteins Cdk2, Cdk4/6, Cyclin D1 and Cyclin E2 were decreased in FoxF1- and FoxF2-deficient RMS tumors. Depletion of either FoxF1 or FoxF2 delayed G1-S cell cycle progression, decreased levels of phosphorylated Rb and increased protein levels of the CDK inhibitors, p21Cip1 and p27Kip1. Depletion of both FoxF1 and FoxF2 in tumor cells completely abrogated RMS tumor growth in mice. Overexpression of either FoxF1 or FoxF2 in tumor cells was sufficient to increase carcinogenesis in orthotopic RMS mouse model. FoxF1 and FoxF2 directly bound to and repressed transcriptional activity of p21Cip1 promoter through −556/−545 bp region, but did not affect p27Kip1 transcription. Knockdown of p21Cip1 restored cell cycle progression in the FoxF1- or FoxF2-deficient tumor cells. Altogether, FoxF1 and FoxF2 promoted RMS tumorigenesis by inducing tumor cell proliferation via transcriptional repression of p21Cip1 gene promoter. Due to robust oncogenic activity in RMS tumors, FoxF1 and FoxF2 may represent promising targets for anti-tumor therapy.
Keywords: Forkhead transcription factor FoxF1 and FoxF2, Rhabdomyosarcoma, muscle tumors, orthotopic mouse model, p21Cip1, p27Kip1 CDK inhibitor, cell cycle
INTRODUCTION
Rhabdomyosarcoma (RMS) is the most common pediatric soft tissue sarcoma. About two-thirds of all sarcomas and 7-8% of all pediatric solid tumors are RMS (1). Although the cell-of-origin of rhabdomyosarcoma remains speculative, RMS is thought to arise from primitive mesenchymal progenitors that have undergone a limited program of myogenic differentiation (2). The two main histological subtypes of RMS are embryonal RMS and alveolar RMS (3). Compared to embryonal RMS, which usually has good prognosis, alveolar RMS is an aggressive, highly metastatic disease that is associated with high mortality rates (4, 5). Alveolar RMS is often associated with recurrent translocations always involving genes from the PAX family of transcription factors fused to FKHR. Approximately 60–70% of histologically diagnosed alveolar RMS involve a t(2;13)(q35;q14) leading to PAX3-FKHR gene fusion (6) and 10–20% have a t(1;13)(q36;q14) leading to PAX7-FKHR gene fusion (7, 8). Based on gene expression arrays, there is growing evidence that RMS is a heterogeneous disease with multiple molecular subtypes (8-10). Interestingly, analysis of clinical RMS samples demonstrated that Forkhead Box F1 (FoxF1) transcription factor is one of the consistently upregulated genes in translocation-positive alveolar RMS, indicating its potential role in the aggressiveness of this disease (8-11).
The Forkhead box family of transcription factors mediates a wide variety of cellular functions including cell growth, tissue repair, cell migration and tumor formation (12). The FoxF subgroup contains two members, FoxF1 and FoxF2 (13, 14). The FoxF1 expression has been found in fetal and adult lungs, placenta, intestine, liver and prostate tissue (15, 16). FoxF1 is a mesenchyme-specific transcription factor and is normally expressed in mesenchyme-derived cells, including pulmonary capillary endothelial cells, fibroblasts, stellate cells of the liver, and visceral smooth muscle cells surrounding trachea, bronchi, stomach, small intestine, colon, and gallbladder (17-22). FoxF1 is not expressed in cardiac or skeletal muscles. FoxF1 has been recently implicated in epithelial carcinogenesis. However, its functional role remains controversial. In breast cancer cell lines, FoxF1 has been shown to function as a tumor suppressor and is inactivated via hypermethylation of its promoter (23). Hypermethylation of the FoxF1 promoter was shown in a subpopulation of invasive ductal carcinomas (23). In colon and breast cancer cell lines, FoxF1 protects tumor cells from DNA re-replication (24). Genomic analysis of human prostate adenocarcinomas showed that a subset of tumors had a loss of the 16q24 chromosome region, which contains several genes including FoxF1 (25). Based on the tumor suppressor properties of FoxF1 in breast and colon cancers, it was proposed that FoxF1 is the most probable candidate for a tumor suppressor in prostate carcinomas containing genomic deletions of 16q24, but this hypothesis had never been confirmed experimentally. In contrast, several published studies have established oncogenic roles of FoxF1. High expression of FoxF1 was found in 78% of Hedgehog (HH)-positive non-small-cell lung cancers, and was positively correlated with metastasis (26). Increased expression of FoxF1 was also found in basal cell carcinoma and medulloblastoma (8, 27). FoxF1 has been shown to be a positive regulator of stemness in lung cancer (28) and an activator of epithelial-to-mesenchymal transition in breast cancer cells (29). All these studies suggest that FoxF1 may function as an oncogene or tumor suppressor depending on the tissue and specific type of cancer.
The FoxF2 expression has been found in the mesenchyme of the oral cavity, limb buds, genitalia, central nervous system, eyes, lung, prostate, ear and placenta as well as the lamina propria and smooth muscle of the GI tract (30, 31). FoxF2 was shown to be a target of miR-200 family in lung cancer and its expression in lung tumor cells increased invasion and metastasis, indicating an oncogenic role of FoxF2 in lung cancer (32). In contrast, decreased FOXF2 expression was associated with the early-onset metastasis and poor prognosis for patients with histological grade II and triple-negative breast cancer (33), and reduced FoxF2 in intestinal fibroblasts increased colon adenoma formation (34), indicating tumor suppressive roles of FoxF2. These conflicting findings suggest that the role of FoxF transcription factors in carcinogenesis is complex and tissue specific. Given the known association of FoxF1 with alveolar RMS (8, 11) and robust expression of FoxF1 and FoxF2 in mesenchymal cells, the FoxF genes may play a role in RMS pathogenesis. However, whether FoxF1 and FoxF2 regulate RMS tumorigenesis remains unknown. The present study was designed to determine the role of FoxF1 and FoxF2 in RMS carcinogenesis. Using in vivo and in vitro models of rhabdomyosarcoma, we demonstrated that both FoxF1 and FoxF2 synergize to induce RMS tumorigenesis and stimulate proliferation of tumor cells through transcriptional repression p21Cip1 promoter.
RESULTS
FoxF1 and FoxF2 are essential for proliferation of rhabdomyosarcoma tumor cells in vitro
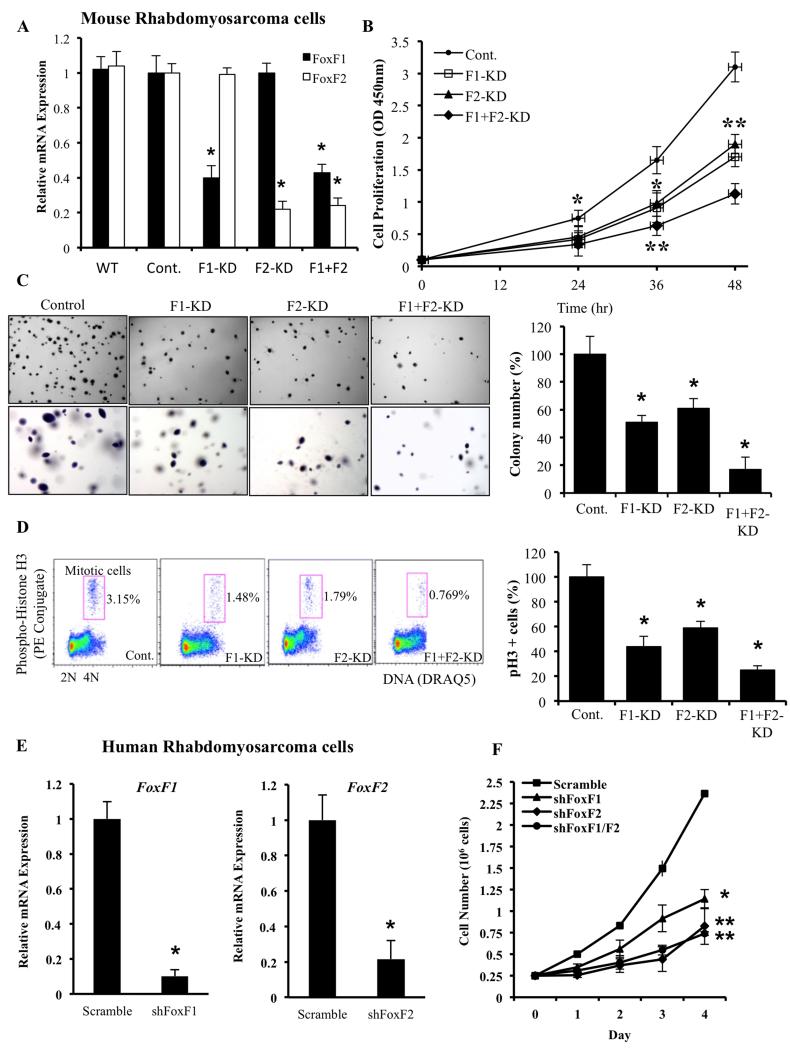
Global gene expression profiling demonstrated that FoxF1 gene is differentially expressed in human alveolar rhabdomyosarcoma (RMS) tumors and its increased expression is associated with poor prognosis in RMS patients (8-11). To examine FoxF1 requirements in RMS tumors, we used lentiviral transduction of FoxF1 shRNA to deplete the FoxF1 in mouse 76-9 RMS cells. Since 76-9 cells also express FoxF2, a protein with almost identical DNA binding domain (35), FoxF2 was also depleted either alone or in combination with FoxF1. Transduced cells were selected using lentivirus-associated GFP and subcloned, resulting in generation of 76-9 RMS tumor cell lines with stable knockdown of FoxF1 and/or FoxF2. qRT-PCR showed approximately 60% reduction in FoxF1 mRNA and 75% reduction in FoxF2 mRNA (Figure 1A). 76-9 clones with higher than 60% and 75% decrease in FoxF1 and FoxF2 levels, respectively, were unable to grow in cell culture. Control 76-9 cells were generated using a lentivirus containing shRNA against a non-targeting sequence. While depletion of either FoxF1 or FoxF2 significantly decreased tumor cell expansion in culture in a time-dependent manner (Figure 1B), the depletion of both FoxF proteins had the strongest inhibition of tumor cell growth (Figure 1B). Depletion of either FoxF1 or FoxF2 was sufficient to significantly reduce the ability of rhabdomyosarcoma cells to form colonies on soft agar, and depletion of both FoxF proteins had the strongest effect on anchorage-independent tumor cells growth (Figure 1C). To determine whether growth inhibition after FoxF1/F2 knockdown was due to proliferation defects, we performed flow cytometery analysis with PH3 antibodies, labeling cells undergoing mitosis (36). Knockdown of FoxF1, FoxF2 or double-knockdown decreased the number of tumor cells undergoing mitosis (Figure 1D), indicating that FoxF1 and FoxF2 are essential for proliferation of mouse 76-9 RMS tumor cells. Furthermore, reduced cellular proliferation was observed in human alveolar rhabdomyosarcoma Rh30 cells after depletion of FoxF1 or FoxF2 (Figure 1E-F). Thus, knockdown of FoxF1 and FoxF2 decreased proliferation of mouse and human rhabdomyosarcoma cells in vitro and the effect of FoxF1/FoxF2 depletion is cumulative.
Figure 1. Lentiviral knockdown of FoxF1, FoxF2 or both in mouse and human RMS cells decreased cellular growth in vitro.
(A) Efficiency of FoxF1 and FoxF2 lentiviral knockdown in mouse 76-9 rhabdomyosarcoma cells was shown by qRT-PCR. β-actin mRNA was used for normalization. (B) Depletion of FoxF1, FoxF2 or both decreased proliferation of 76-9 cells in vitro. Control, FoxF1-KD, FoxF2-KD and F1+F2-KD 76-9 cells were seeded in triplicates and counted at different time points using WST1 Cell Proliferation Assay. (C) Depletion of FoxF1, FoxF2 or both decreased colony formation in soft agar compared to control RMS cells. Cells were seeded in triplicates. Values are the means ± SD of three independent experiments. A p value <0.05 is shown with (*). (D) Depletion of FoxF1, FoxF2 or both decreased the number of cells in mitosis shown by flow cytometery with phospho-Histone H3 antibodies. (E) Efficiency of FoxF1 and FoxF2 lentiviral knockdown in human Rh30 rhabdomyosarcoma cells was shown by qRT-PCR. β-actin mRNA was used for normalization. (F) Depletion of FoxF1, FoxF2 or both decreased proliferation of Rh30 cells in vitro. Control, FoxF1-, FoxF2- or F1+F2-deficient Rh30 cells were seeded in triplicates and counted at different time points using hemocytometer. Data represent mean±s.d. of three independent experiments. A p value < 0.05 is shown with (*); p value < 0.01 is shown with (**).
Depletion of FoxF1 and FoxF2 decreased rhabdomyosarcoma tumor growth in mice
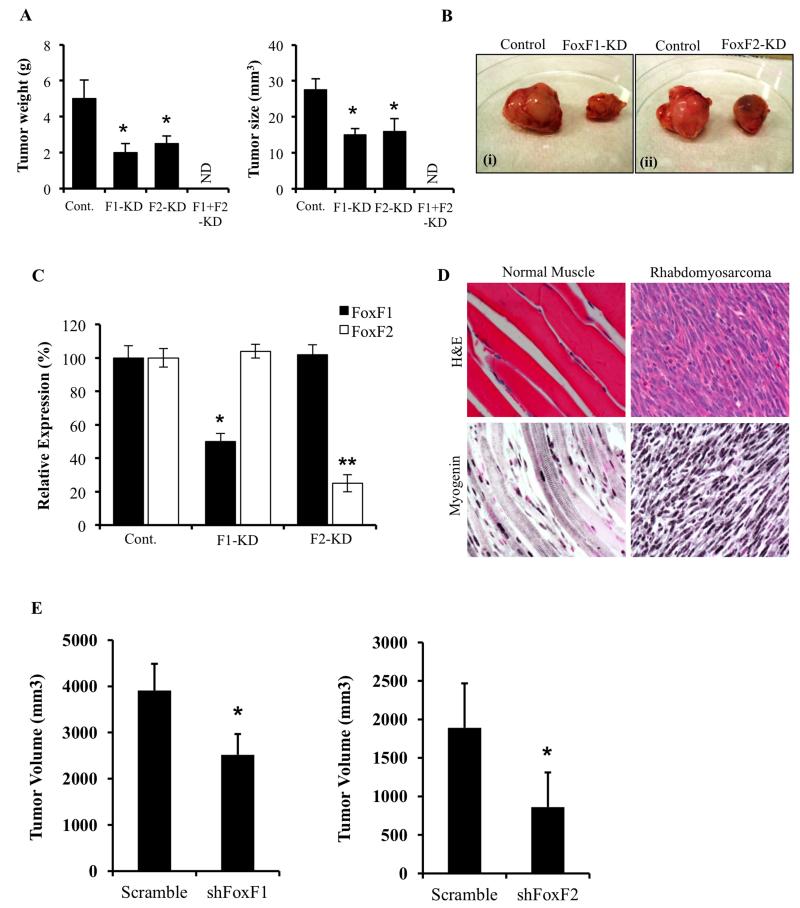
Parental, FoxF1- and FoxF2-deficient mouse 76-9 rhabdomyosarcoma cells were injected intramuscular in the left hind limb muscle of syngeneic C57Bl/6 mice. Three weeks later, tumors were harvested and measured. Knockdown of either FoxF1 (FoxF1-KD) or FoxF2 (FoxF2-KD) was sufficient to decrease RMS tumor diameter and tumor weight by approximately 50% (Figure 2A-B). Knockdown of FoxF proteins was stable during the period of tumor growth as shown by the decreased FoxF1 or FoxF2 mRNAs in harvested tumors (Figure 2C). Interestingly, knockdown of both FoxF1 and FoxF2 completely abrogated tumor growth, resulting in the absence of RMS tumors in 100% of injected mice (n=10). These results indicate that FoxF1 and FoxF2 function in synergetic manner to induce RMS tumorigenesis in vivo. Identity of rhabdomyosarcoma tumors were established by histological examination of H&E-stained paraffin sections and immunostaining of tumor tissue for myogenin, (Figure 2D), a marker of skeletal muscle cells (37). In addition, to demonstrate that the proliferation-specific role of FoxF1 and FoxF2 is not limited to mouse rhabdomyosarcoma tumors, we depleted FoxF1 and FoxF2 from human Rh30 RMS cells. FoxF1- and FoxF2-deficient Rh30 tumors were significantly smaller compared to control tumors (Figure 2E), which is consistent with the results obtained from mouse 76-9 RMS tumors. Thus, depletion of FoxF1 and FoxF2 decreased the growth of mouse and human RMS tumors in orthotopic mouse models.
Figure 2. Knockdown of FoxF1 or FoxF2 decreased RMS tumor growth in mice.
RMS tumors were harvested 3 weeks after inoculation of control 76-9 rhabdomyosarcoma cells or 76-9 cells with stable knockdown of FoxF1, FoxF2 or both (FoxF1-KD, FoxF2-KD or F1-F2 KD). (A) FoxF1 or FoxF2 deficiency decreased the growth of RMS tumors in orthotopic model. Mean weight and size of tumors (±SD) are shown (n=8 for control 76-9 cells, n=6 for each group of KD cells). ND – not determined: No tumors were found in F1-F2 KD group. (B) Representative images of excised tumors from FoxF1 and FoxF2 KD mice compared to control are shown. (C) Efficiency of FoxF1 and FoxF2 depletion in tumors are shown by qRT-PCR. Total RNA was prepared from tumors isolated from control, FoxF1-KD and FoxF2-KD mice. β-actin mRNA was used for normalization. Data represent means ± SD of three independent determinations using RMS tumor tissue from n=6-10 mice in each group. (D) Histological analysis of the excised RMS tumors was performed using H&E staining and immunostaining with antibodies to Myogenin. Magnification: 400x. (E) Human rhabdomyosarcoma Rh30 cells with stable knockdown of FoxF1 or FoxF2 were inoculated into immunocompromised NSG mice. Depletion of FoxF1 or FoxF2 decreased tumor growth compared to control tumors. N=8 per group for shFoxF1 tumors and N=6 per group for shFoxF2 tumors. A p value <0.05 is shown with (*).
Depletion of FoxF1 and FoxF2 increased expression of p21Cip1 and p27Kip1 CDK inhibitors
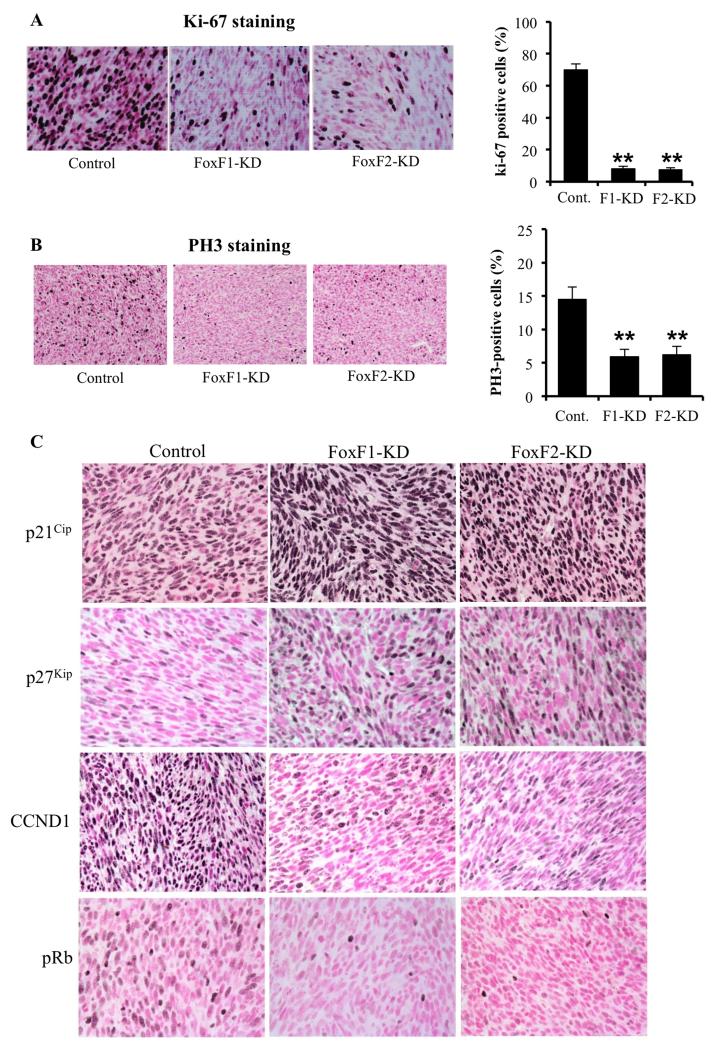
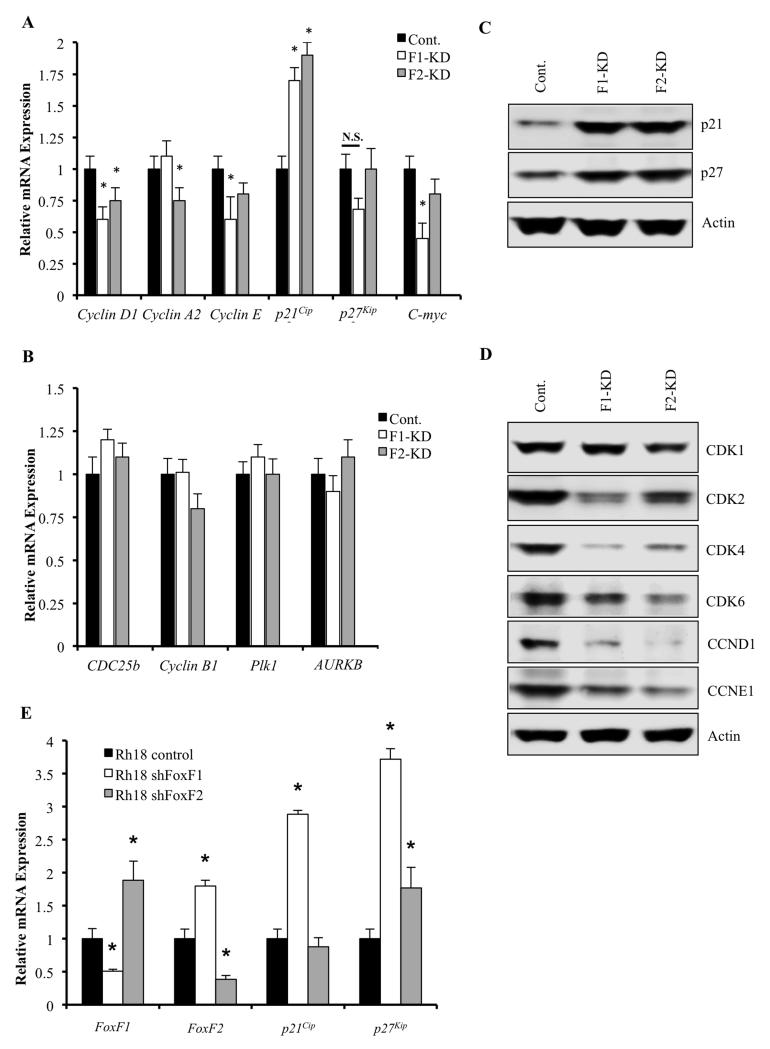
Decreased sizes of FoxF1-KD and FoxF2-KD rhabdomyosarcoma tumors were associated with reduced cellular proliferation as shown by a significant reduction in the number of Ki-67 and PH-3 positive cells (Figure 3A-B), findings consistent with the in vitro data (Figure 1A-D). Cyclin D staining was also decreased (Figure 3C). Reduced cellular proliferation in FoxF1-KD and FoxF2-KD tumors was associated with increased nuclear staining for p21Cip1 and p271Kip1 proteins, potent inhibitors of Cdk2 in G1 phase of cell cycle (38). Since reduced Cdk2 activity is associated with decreased phosphorylation of retinoblastoma protein (Rb) (39), we stained tumor sections with antibodies specific to phospho-Rb (pRb). The number of RMS tumor cells positive for pRb was reduced in FoxF1-KD and FoxF2-KD tumors (Figure 3C). Knockdown of either FoxF1 or FoxF2 in RMS tumor cells significantly decreased mRNA levels of Cyclin D1, Cyclin A2, Cyclin E and c-Myc, all of which are critical regulators of G1/S cell cycle transition and are known targets of Cdk2/pRb/E2F pathway (40) (Figure 4A). Moreover, depletion of either FoxF1 or FoxF2 in RMS cells dramatically increased the mRNA and protein levels of p21Cip1, whereas p27Kip1 was increased only at the protein level (Figure 4A and 4C). Finally, protein levels of G1/S-promoting Cdk2, Cdk4 and Cdk6 were decreased in FoxF1 KD and FoxF2 KD RMS cells, as were Cyclin D1 and Cyclin E2 proteins (Figure 4D). These results indicate that knockdown of either FoxF1 or FoxF2 reduce cellular proliferation, increase p21Cip1 and p27Kip1 protein levels and reduce expression of genes critical for G1/S transition of the cell cycle. Interestingly, neither FoxF1- nor FoxF2-depletion changed Cdk1 (Figure 4D) or influenced expression of Cdc25b, Cyclin B1, Plk1 and Aurora B (Figure 4B), that are important regulators of G2/M cell cycle transition and cytokinesis (40). In addition, knockdown of FoxF1 in human Rh18 RMS cells increased p21 mRNA (Fig. 4E). Knockdown of FoxF2 did not affect p21 mRNA, indicating that FoxF1, but not FoxF2 regulates p21 expression in human Rh18 rhabdomyosarcoma cells. Interestingly, there was a significant increase in FoxF1 after FoxF2 knockdown in Rh18 cells (Fig. 4E), suggesting a presence of compensatory mechanism and further supporting our findings in which FoxF1 and FoxF2 work synergistically to regulate cellular proliferation.
Figure 3. Depletion of FoxF1 and FoxF2 increased expression of p21Cip1 and p27Kip1 in RMS tumors.
RMS tumors were harvested 3 weeks after inoculation of control 76-9 rhabdomyosarcoma cells or 76-9 cells with stable knockdown of FoxF1 or FoxF2 (FoxF1-KD, FoxF2-KD). (A-B) Decreased cellular proliferation was demonstrated by reduced numbers of Ki-67-positive (A) and pH3-positive cells (B). Percentage of Ki-67-positive and PH3-positive cells were counted in five random microscope fields (n=3 mice per group, right panels). A p value < 0.01 is shown with (**). Magnification is 400x (upper panels), 200x (bottom panels). (C) Depletion of FoxF1 or FoxF2 increased expression of p21Cip1 and p27Kip1 CDK inhibitors and decreased protein levels of CCND1 and pRb in RMS tumors. Magnification is 200x.
Figure 4. FoxF1 or FoxF2 knockdown alters expression of p21Cip1 /p27Kip1 pathway-related genes.
Total RNA and protein from orthotopicaly-grown RMS tumors were harvested 3 weeks after 76-9 tumor cells inoculation. (A) Depletion of FoxF1 or FoxF2 (FoxF1-KD, FoxF2-KD) in RMS tumor cells decreased mRNAs of cyclin D1, cyclin A2, cyclin E and C-myc, all of which are G1/S-promoting cell cycle genes. p21 mRNA was increased in the FoxF1- and FoxF2-depleted RMS tumors. qRT-PCR was used to assess expression levels. β-actin mRNA was used for normalization. A p value <0.05 is shown with (*). (B) No changes in mRNAs of G2/M cell cycle transition and cytokinesis genes, Cdc25b, cyclin B1, Plk1 and Aurora B, were found using qRT-PCR. β-actin mRNA was used for normalization. (C) Western blot demonstrated increased expression of p21Cip1, p27Kip1 and pRb (Ser807/811) proteins in FoxF1-KD or FoxF2-KD RMS tumors compared to control. (D) Western blot showed decreased protein levels of Cdk1, Cdk2, Cdk4, Cdk6, cyclin D1, and cyclin E1 in FoxF1-KD or FoxF2-KD RMS tumors compared to control. β-Actin was used as loading control. (E) qRT-PCR was performed using human rhabdomyosarcoma Rh30 cells with stable knockdown of FoxF1 or FoxF2. β-actin mRNA was used for normalization.
Depletion of FoxF1 and FoxF2 delayed G1-S progression of cell cycle
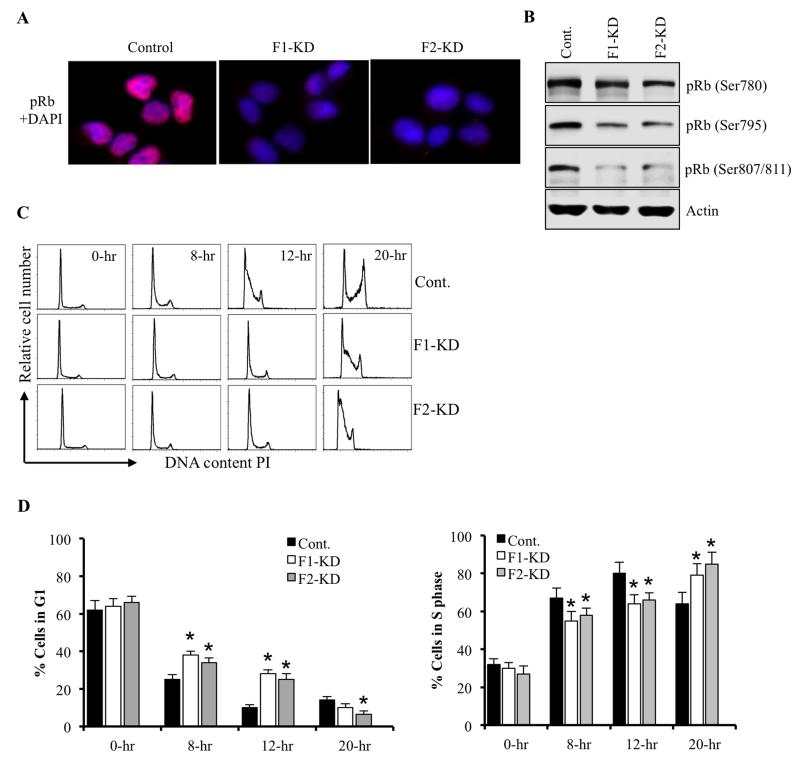
Since p21Cip1 and p27Kip1 inhibit phosphorylation of Rb (39) and we detected diminished pRb staining in FoxF1-KD and FoxF2-KD rhabdomyosarcoma tumors (Figure 3C), western blot was used to examine pRb levels in FoxF1-KD and FoxF2-KD rhabdomyosarcoma cells. Phosphorylation of Rb on Ser 780, Ser 795 and Ser 807/811 were decreased in both FoxF1-KD and in FoxF2-KD tumor cells (Figure 5B). Nuclear staining for phospho-Rb (pRb) protein was also reduced (Figure 5A). These findings are consistent with decreased proliferation of FoxF1-KD and FoxF2-KD cells in RMS tumors (Figure 3A-B). To determine what stage of cell cycle is affected by the depletion of FoxF1 and FoxF2, we serum-starved 76-9 RMS cells in vitro. After serum stimulation, cell cycle analysis was performed using flow cytometry. Depletion of FoxF1 or FoxF2 delayed entry of 76-9 RMS cells into S-phase as demonstrated by decreased number of cells in S-phase and an increase in G1-phase at 8 and 12 hours after serum stimulation (Figure 5C-E). These results are consistent with the increased levels of p21Cip1 and p27Kip1 Cdk inhibitors and reduced pRB in FoxF1- or FoxF2-depleted RMS cells (Figure 4C) and RMS tumors (Figure 3C). Altogether, these data indicate that FoxF1 and FoxF2 promote G1/S entry during cell cycle progression and inhibit p21Cip1 and p27Kip1 proteins via direct or indirect mechanisms.
Figure 5. Depletion of FoxF1 or FoxF2 delays cell cycle progression at the G1/S transition.
(A) Decreased levels of phosphorylated Rb (pRb) protein in FoxF1-KD or FoxF2-KD RMS cells were shown using immunofluorescent staining with antibodies against pRb. Nuclei were counterstained with DAPI. (B) Western blot on FoxF1- or FoxF2-deficient tumor cells showed decreased expression of pRb (Ser780), pRb (Ser795) and pRb (Ser807/811. (C) FoxF1- or FoxF2-depletion caused delay in cell cycle progression at G1/S transition. Control, FoxF1-KD or FoxF2-KD 76-9 RMS cells were serum starved overnight. After serum addition, cells were harvested at 8 hrs, 12 hrs, and 20 hrs. Total DNA content was measured by propidium iodide staining and quantification were done by flow cytometry. (D) Quantification of accumulated FoxF1- or FoxF2-deficient RMS cells in G0/G1 and S phase were performed using flow cytometry analysis. Depletion of FoxF1 or FoxF2 resulted increase accumulation of tumor cells in G0/G1 and decrease number of cells in S phase. Percent of total cells in G1 phase and S phase was presented as mean ± SD. Data represent mean±s.d. of three independent experiments. A p value <0.05 is shown with (*).
Overexpression of FoxF1 or FoxF2 in RMS tumor cells increased carcinogenesis
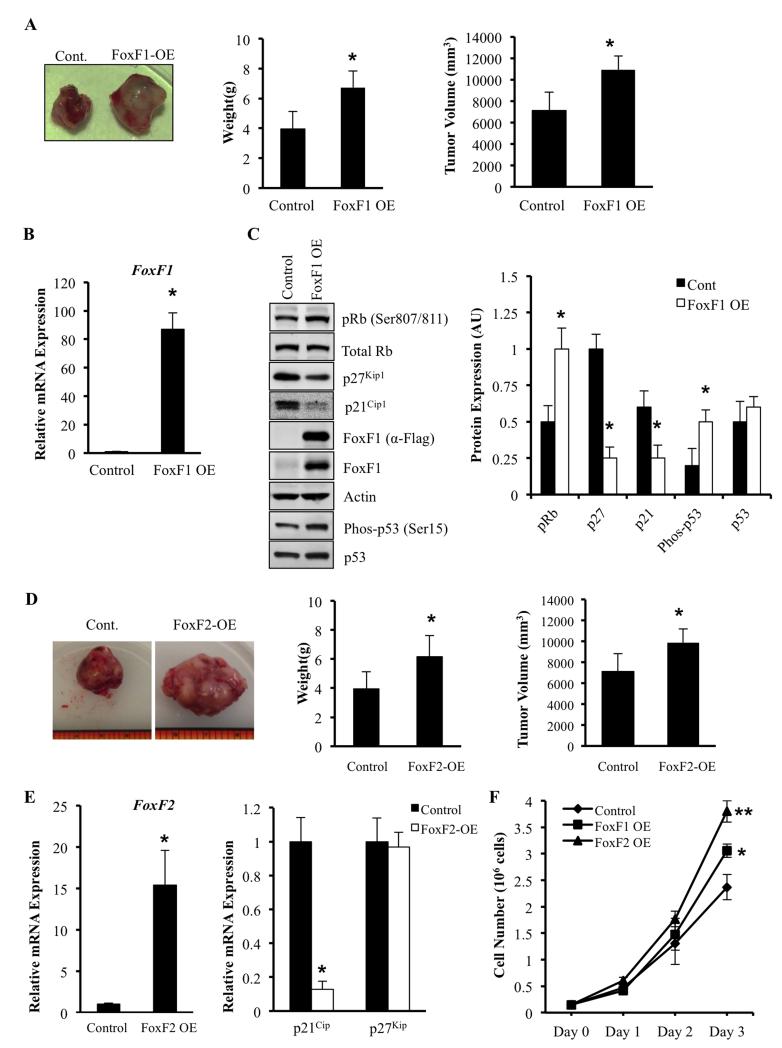
Since depletion of either FoxF1 or FoxF2 reduced formation of RMS tumors (Figure 2A-B), we tested whether overexpression of FoxF1 or FoxF2 is sufficient to increase RMS tumor growth in vivo. Mouse 76-9 cells stably expressing Flag-tagged FoxF1, FoxF2 or control vector were generated and subsequently injected into the muscle tissue of syngeneic mice. Overexpression of FoxF1 was confirmed by qRT-PCR and by Western blot using FoxF1 antibodies and Flag antibodies (Figure 6B-C). Overexpression of FoxF1 increased the tumor burden, demonstrated by the increased weight and size of RMS tumors (Figure 6A). An increase in tumor growth was associated with elevated pRb and decreased protein levels of p21Cip1 and p27Kip1 (Figure 6B-C). Overexpression of FoxF1 had no effect on total protein levels of p53, but increased the amounts of active (phospho-Ser15) p53 (Figure 6C). Overexpression of FoxF2 increased tumor growth, reduced p21Cip1, but did not affect and p27Kip1 mRNA (Figure 6D-E). In vitro studies showed that overexpression of either FoxF1 or FoxF2 significantly increased tumor cell proliferation in a time-dependent manner (Figure 6F). Altogether, our results indicate that overexpression of either FoxF1 or FoxF2 in rhabdomyosarcoma cells is sufficient to increase RMS tumor growth, increase tumor cell proliferation and reduce expression of CDK inhibitor p21Cip1.
Figure 6. Overexpression of FoxF1 or FoxF2 promotes tumor growth by decreasing expression of p21Cip1 and p27Kip1.
(A) Representative tumors formed 3 weeks after receiving intra-muscular injection of 2 ×105 76-9 cells transduced with an empty-vector control or FoxF1 overexpression retrovirus (FoxF1-OE). Tumor weights and volumes for mice (n=5) are shown as mean ± SD. (B) Efficiency of FoxF1 overexpression was confirmed by qRT-PCR using total RNA isolated from orthotopic RMS tumors. FoxF1 expression levels were normalized to β-actin mRNA. (C) Overexpression of FoxF1 decreased protein levels of pRb, p21Cip1 and p27Kip1 shown by Western blot using of total protein from RMS tumors. Blots were probed with antibodies against phosphorylated Rb (pRb), total Rb, p21CIP1, p27Waf1/Cip1, Flag, FoxF1, p53, phospho-p53, or β-actin (left panel). Relative expression levels of these proteins were determined with densitometry and normalized to β-actin (right panel). AU=arbitrary units. (D) Representative tumors formed 3 weeks after receiving intra-muscular injection of 2 ×105 76-9 cells transduced with an empty-vector control or FoxF2 overexpression retrovirus (FoxF1-OE). Tumor weights and volumes for mice (n=7) are shown as mean ± SD. (E) Efficiency of FoxF1 overexpression was confirmed by qRT-PCR using total RNA isolated from orthotopic RMS tumors (left panel). Overexpression of FoxF2 increased p21Cip1 mRNA as shown by qRT-PCR using total RNA isolated from rhabdomyosarcoma tumors (right panel). β-actin mRNA was used for normalization. All data represent mean ± SD of three independent determinations using tumor tissue from n=5–7 mice in each group. (F) Overexpression of FoxF1 or FoxF2 increased proliferation of RMS tumor cells in vitro. 76-9 control, FoxF1 OE, or FoxF2 OE cells were plated in triplicates. Cell numbers were measured every 24 hours for 3 days. Each point represents mean±SEM. Data represent mean±s.d. of three independent experiments. A p value <0.05 is shown with (*), a p value <0.01 is shown with (**).
FoxF1 and FoxF2 directly bind to and activate the p21Cip1 promoter
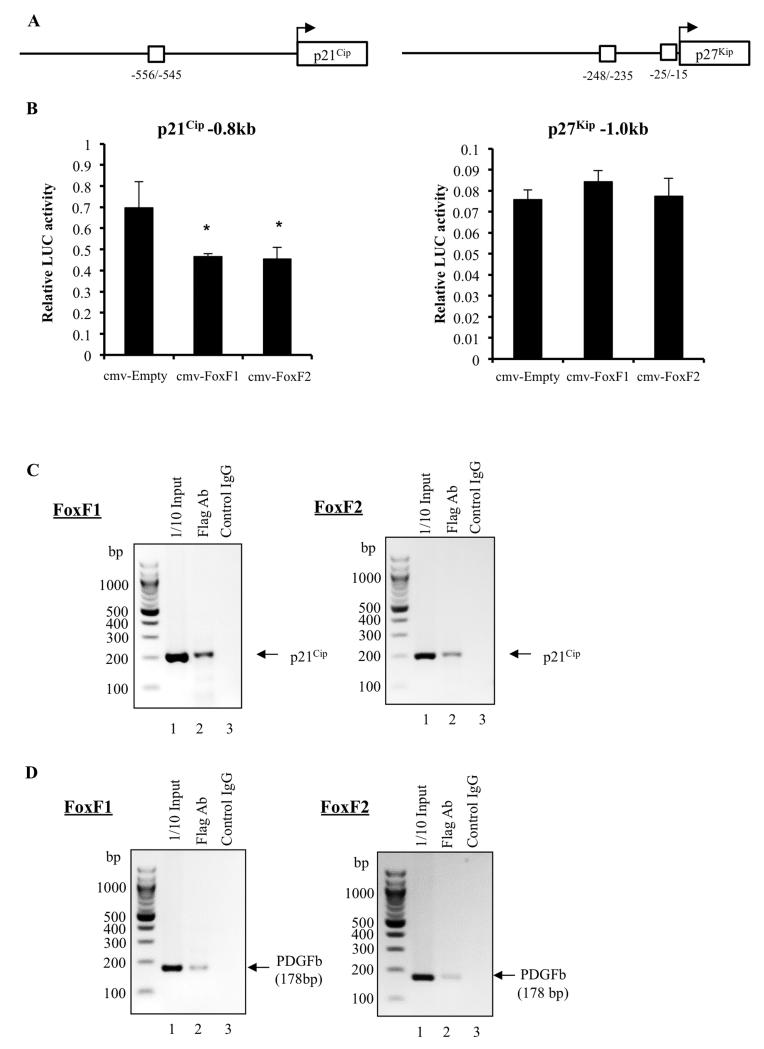
Knockdown of FoxF1 or FoxF2 increased protein levels of p21Cip1 and p27Kip1, but only p21Cip1 was increased at mRNA levels (Figure 3C, 4A and 4C). These results suggest that FoxF1 and FoxF2 inhibit transcription of p21Cip1 gene. Consistent with this hypothesis, high-affinity FoxF1/F2 binding sites were identified in −556/−545 bp region of the mouse and in the −680bp region of the human p21Cip1 gene. Next, the −0.8 kb mouse p21Cip1 promoter region containing FoxF1/F2 binding site was cloned into luciferase (LUC) reporter vector using mouse genomic DNA. Co-transfections of the −0.8 kb p21Cip1-Luc reporter with either CMV-FoxF1 or CMV-FoxF2 expression vectors demonstrated that FoxF1 and FoxF2 inhibited transcriptional activity of the p21Cip1 promoter in 76-9 rhabdomyosarcoma cells (Figure 7A). Interestingly, neither FoxF1 nor FoxF2 regulated p27Kip1 promoter activity in LUC assay, in spite of the presence of the potential FoxF1/F2 binding sites in −248/−235 and −25/−15 bp regions of the mouse p27Kip1 promoter (Figure 7B). Since FoxF1 and FoxF2 repressed transcription of the p21Cip1 promoter, we next determined whether FoxF1 and FoxF2 physically bound to the −556/−545 bp p21Cip1 promoter region. Chromatin immunoprecipitation (ChIP) assay was performed using mouse parental 76-9 RMS cells and 76-9 RMS cells stably expressing Flag-tagged FoxF1 or FoxF2. Chromatin immunoprecipitation was done with anti-Flag antibodies and compared to control IgG. Flag-tagged FoxF1 and FoxF2 specifically bound to the p21Cip1 promoter DNA as shown by ChIP (Figure 7C). Both FoxF1 and FoxF2 also bound to the PDGFb promoter (Figure 7D), a known transcriptional target of the FoxF genes (21, 41). Altogether, these data indicate that FoxF1 and FoxF2 directly inhibit p21Cip1 transcription in rhabdomyosarcoma cells.
Figure 7. p21Cip1 is a direct transcriptional target of FoxF1 and FoxF2.
(A) Schematic diagram shows the potential FoxF1/FoxF2 binding sites in the murine p21Cip1 and p27Kip1 promoters. (B) FoxF1 and FoxF2 repressed transcriptional activity of p21Cip1 promoter (left panel), but did not change transcriptional activity of p27Kip1 promoter (right panel). Cells were transfected with CMV-Empty, CMV-FoxF1 or CMV-FoxF2 expression vectors and a −0.8kb p21Cip1-pGL2 promoter plasmid or −1.0kb p27Kip1-pGL2 promoter plasmid. Dual LUC assays were used to determine LUC activity. Transcriptional activity is shown as a fold change relative to CMV-empty vector (±SD). A p value <0.05 is shown with (*). Data represent mean±s.d. of three independent experiments. (C) Direct binding of FoxF1 and FoxF2 proteins to the p21Cip1 promoter region is shown by ChIP assay. Protein/ DNA complexes were immunoprecipitated using Flag-specific antibody and 76-9 RMS cells that overexpressed His-Flag (HF)-tagged FoxF1 or FoxF2. Immunoprecipitation with isotype control antibodies was used for normalization. Data represent one of three independent experiments. (D) Binding of FoxF1 and FoxF2 to the PDGFb promoter region is presented as a positive control for the ChIP assay using the same cells.
Knockdown of p21Cip1 in the FoxF1- or FoxF2-deficient cells restored cellular proliferation
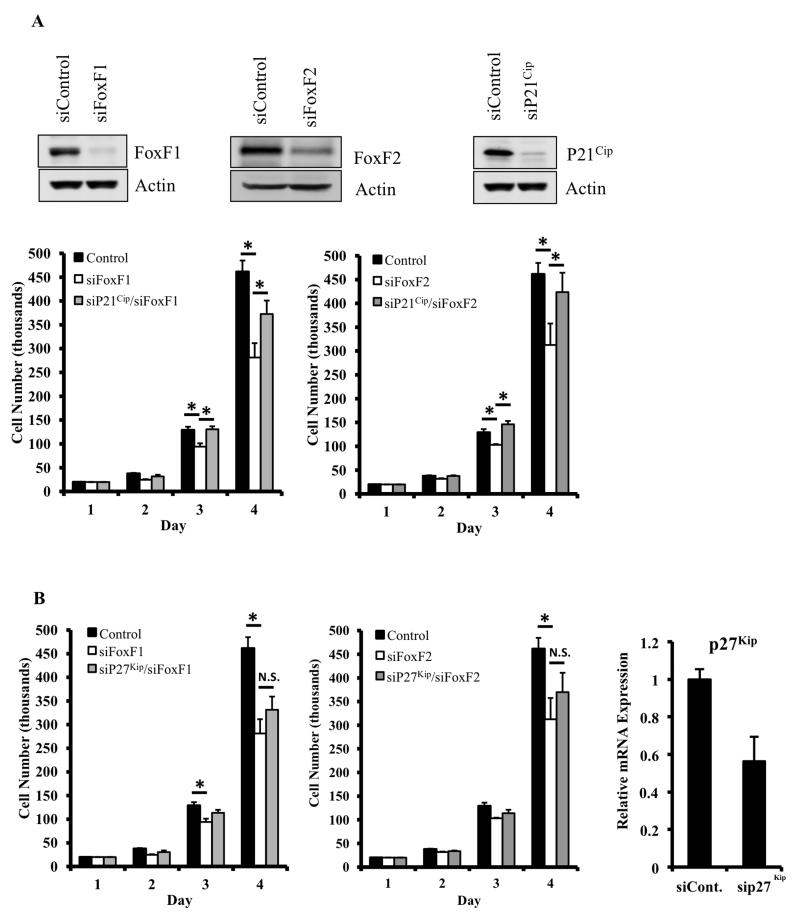
Since FoxF1 and FoxF2 repress p21Cip1 transcription (Figure 7D) and increased p21Cip1 levels were observed in FoxF1- and FoxF2-deficient RMS tumors and tumor cells (Figure 3C and 4C), we determined whether reduced proliferation in FoxF1- and FoxF2-deficient cells is due to increased p21Cip1 levels. p21Cip1–specific siRNA was used to knockdown p21Cip1 in the FoxF1- or FoxF2-deficient tumor cells (Figure 8A). Decreasing the levels of p21Cip1 in cultured FoxF1-KD or FoxF2-KD cells restored their proliferation to the level found in control cells (Figure 8A-B). In contrast, depletion of p27Kip1 was insufficient to restore cellular proliferation in FoxF1-KD and FoxF2-KD cells (Figure 8C). Thus, p21Cip1, but not p27Kip1, is a key target of FoxF proteins in rhabdomyosarcoma cells. Altogether, our results indicate that FoxF1 and FoxF2 increase proliferation of rhabdomyosarcoma cells through transcriptional inhibition of p21Cip1.
Figure 8. Knockdown of p21Cip1, but not p27Kip1, restores cell proliferation in FoxF1- and FoxF2-deficient cells in vitro.
(A) Western blots showing protein expression FoxF1 (top left), FoxF2 (top middle panel) or p21Cip1 (top right panel) of 91U cells after transfection with control siRNA or siRNA against FoxF1, FoxF2, or p21Cip1. Cell proliferation in the transfected cells was determined by daily cell counts using a hemacytometer. Knockdown of FoxF1 (bottom left) or FoxF2 (bottom right) decreased cell proliferation. Knockdown of p21Cip1 rescued cellular proliferation in both FoxF1- and FoxF2-deficient cells. Data represents the mean ± SD of triplicate wells. (B) Knockdown of p27Kip1 did not restore decreased cellular proliferation in FoxF1- or FoxF2-deficient cells (left and middle panels). Efficiency of p27Kip1 knockdown is shown by qRT-PCR (right panel). Expression levels were normalized to β-actin mRNA. A p value <0.05 is shown with (*).
DISCUSSION
Rhabdomyosarcoma was described as an aggressive pediatric disease nearly 60 years ago in which the 5-year incident free rate was below 25% (42). Since then, significant progress has been made in treatment and management of the disease with the current 5 year incident-free rate hovering around 70% for all RMS cases (42). Over the past two decades, however, attempts to advance our treatment of RMS have failed with no improvement in patient survival despite better risk stratification, escalation of treatment, more aggressive surgical resection of the tumors and improved monitoring. Rhabdomyosarcoma is a heterogeneous disease with multiple molecular subtypes. There is a compelling need to identify the key molecular drivers of the disease which will provide the new clinical targets to improve RMS treatment.
Previous studies demonstrated that the most aggressive alveolar rhabdomyosarcomas differentially express high levels of FoxF1 transcription factor (8, 11). FoxF1 and its closely related transcription factor FoxF2 have been recently implicated in epithelial carcinogenesis. However, their functional roles remain controversial. In fact, several studies demonstrated that FoxF1 functions as an oncogene in breast cancer (29), while others have reported that FoxF1 is a tumor suppressor in breast and colon cancer cells (23, 43). The same controversy exists for FoxF2, with FoxF2 functioning as an oncogene in lung tumor cells (32), but acting as a tumor suppressor in triple-negative breast cancer (33) and colon adenoma formation (34). While these studies appear to be conflicting, it’s possible that FoxF1 and FoxF2 have different functions depending on cell/ tissue specificity or mutation status of the tumor cells. It is also important to note that all of these published studies investigated epithelial-derived tumors, whereas the role of FoxF1 and FoxF2 in sarcomas remains unknown. To understand the effects of these Fox proteins in the pathogenesis of rhabdomyosarcomas, we have utilized mouse orthotopic transplantation models for RMS. By inducing or inhibiting FoxF1 and FoxF2 in rhabdomyosarcoma cells and subsequently transplanting these cells into mice, we have uncovered an oncogenic role of FoxF1 and FoxF2 in RMS growth and progression via direct transcriptional repression of p21Cip1 and indirect inhibition of p27Kip1 tumor suppressors.
P21Cip1 and p27Kip1 are critical regulators of both normal and pathological myogenesis. Activated myoblasts commit to terminal differentiation by activating the differentiation-promoting transcription factor myogenin and simultaneously withdrawing from the cell cycle via activation of both p21Cip1 and p27Kip1 Cdk inhibitors (44). Paradoxically, rhabdomyosarcoma cells express high levels of myogenin and other differentiation markers (MyoD, desmin, MHC) but fail to withdraw from the cell cycle. These studies suggest that p21Cip1 and p27Kip1 and/or their upstream regulators can affect proliferation of tumor cells, influencing RMS tumorigenesis. An important contribution of the present study is that FoxF1 and FoxF2 synergize to inhibit p21Cip1 and p27Kip1 in RMS cells. Since p21Cip1 and p27Kip1 inhibit Cdk2/cyclin complexes and prevent phosphorylation of Rb protein (40), increased p21Cip1 and p27Kip1 levels can contribute to decreased proliferation of FoxF1/F2-deficent RMS cells. Regulation of p21Cip1 by FoxF1/F2 appears to be on transcriptional level as demonstrated by physical binding of both FoxF1 and FoxF2 proteins to the p21Cip1 promoter region. The binding of FoxF1/F2 to the p21Cip1 promoter DNA resulted in transcriptional inhibition of p21Cip1 gene expression as supported by luciferase assay. Thus, FoxF1 and FoxF2 are transcriptional repressors of p21Cip1 gene. Consistent with this data, increased p21Cip1 levels are found in FoxF1/F2-deficient RMS cells and RMS tumors. Interestingly, FoxF1 and FoxF2 appear to regulate p27Kip1 indirectly. These Fox proteins did not bind the p27Kip1 promoter DNA and did not change p27Kip1 mRNA levels in spite of increased protein levels of p27Kip1in FoxF1/F2-deficient RMS cells.
Published studies implicated p53 in the regulation of FoxF1 gene expression. It was shown that p53 induced FoxF1 mRNA and protein levels in head and neck cancer cell lines (43). In breast and colorectal cancer cells, inactivation of the p53 resulted in silencing of FoxF1 (24). P53 stimulated FoxF1 gene expression via direct activation of the FoxF1 promoter (43). All these published studies suggest that FoxF1 can function downstream of p53. Interestingly, p53 activates p21Cip1 and inhibits cell cycle progression, at least in part, through increased p21Cip1 levels (40). Our studies demonstrated that FoxF1 inhibits p21Cip1 through direct transcriptional repression of the p21Cip1 promoter. In addition, we found that FoxF1 has no effect on total protein levels of p53, indicating that FoxF1 does not regulate p53 expression. Interestingly, FoxF1 overexpression increased the levels of active (phospho-Ser15) p53, indicating that FoxF1 may induce replication stress followed by the DNA damage response that enhances p53 activity. Alternatively, it is possible that increased activation of p53 in FoxF1-overexpressing cells is a result of compensatory mechanism which is directed to restore p21 levels in FoxF1-overexpressing cells. Our data suggest that expression of p21Cip1 in rhabdomyosarcoma is controlled by the complex network of transcriptional regulators. The balance between p53-mediated activation and FoxF1/F2-mediated transcriptional repression may regulate the rate of tumor cell proliferation, ultimately affecting tumor progression and outcome in RMS patients.
In summary, we identified FoxF1 and FoxF2 transcription factors as important regulators of tumor cell proliferation in rhabdomyosarcomas through transcriptional inactivation of the p21Cip1 tumor suppressor. Based on these findings, FoxF1 and FoxF2 may represent promising targets for anti-tumor therapy in rhabdomyosarcoma patients.
MATERIALS AND METHODS
Generation of RMS cell lines with stable knockdown of FoxF1 and FoxF2
Plasmids expressing shRNA of either mouse FoxF1 (5′-CAGGTCACCTACCAAGACA-3′) (GIPZ Mouse FoxF1 shRNA, Clone ID: V2LMM_74793; GE healthcare) mouse FoxF2 (5′-GTGACTACATGTAAGACAT-3′) (GIPZ Mouse FoxF2 shRNA, Clone ID: V2LMM_61137; GE healthcare), human FoxF1 (5′-GCAGCATTTGTGACACGTATT-3′) (plKO-shFoxF1), human FoxF2 (5′-AGAGCGTCTGTCAGGATATTA-3′) (plKO-shFoxF2), or shRNA control (5′-CAACAAGATGAAGAGCACCAA-3′) were used to generate lentiviruses. The stable knockdown of FoxF1 and FoxF2 was achieved by transducing murine 76-9 rhabdomyosarcoma cells (45) and human rhabdomyosarcoma cells Rh30, Rh18 ((46); a kind gift from Dr. T. Cripe) with lentiviruses followed by puromycin selection.
Generation of RMS cell lines with stable overexpression of FoxF1 and FoxF2
To generate double-tagged FoxF1 and FoxF2 constructs, murine FoxF1 and FoxF2 cDNAs were PCR-amplified and N-terminal Flag (5′-GGC-GGA-TCC-GCC-ACC-ATG-GAC-TAC-AAA-GAC-GAT-GAC-GAC-AAG-GAC-CCC-GCG-GCG-GCG-GGC-3′) and a C-terminal (His)6 –tag (5′-GGG-CCC-TCG-AGT-CAG-TGA-TGG-TGA-TGG-TGG-TGC-ATC-ACA-CAC-GGC-TTG-ATG-TCT-TGG-3′) were introduced by using PCR and cloned into BamHI and XhoI sites of bicistronic retroviral pMIEG3 vector (47) to generate pMIEG3-HF-FoxF1 and pMIEG3-HF-FoxF2 as previously described (48). PCR was performed using AccuPrime Pfx DNA Polymerase according to manufacturers’ protocol (Invitrogen, Carlsbad, CA, USA). The retroviral and lentiviral particles were made in Cincinnati Children's Viral Vector Core facility and the generation of stable cell lines was done as described previously (48). The murine 76-9 rhabdomyosarcoma cell line (45) was transduced with lentiviruses. After two days, GFP expressing cells were sorted using flow cytometry.
Orthotopic mouse model of RMS
Power analysis (DSTPLAN 4.2 software) with the power value of 0.9 predicted that the sample size for the orthotopic tumor studies should be approximately 6 mice per group. As in our previous studies (48), we used 7 mice per group. 2×105 76-9 RMS tumor cells in 50μL sterile PBS were injected in the left hind limb muscle (i.m.) of 6-8 weeks old C57BL/6 (Jackson Lab, Bar Harbor, ME) anesthetized male mice. Tumors were harvested 3 weeks later. The human Rh30 and Rh18 RMS tumor cells (5×106 cells/mL in 30% matrigel) in 100μl were injected in the left hind limb muscle of the 6-8 weeks old Nod-Scid-Gamma (NSG, Jackson Lab) anesthetized male mice. Tumors were harvested at 8 weeks (shFoxF1 experiment) or at 6 weeks (shFoxF2 experiment) and tumor volume was measured using a digital caliper. The mice were randomly selected to inoculate either control or experimental rhabdomyosarcoma cells. All animals were included in the analysis. No blinding was done. All animal studies were approved by the Animal Care and Use Committee of Cincinnati Children’s Research Foundation.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA was prepared from dissected rhabdomyosarcoma tumors or from cultured RMS cells using RNeasy mini kit (Qiagen, Valencia, CA, USA) and analyzed by qRT-PCR using the StepOnePlus Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA) as described (49). RNA was amplified with Taqman Gene Expression Master Mix (Applied Biosystems) combined with inventoried Taqman mouse gene expression assays: FoxF1, Mm00487497_m1; β-Actin, Mn00607939_s1; FoxF2, Mn00515793_m1; Cyclin D1, Mm00432359_m1; Cyclin A2, Mm00438063_m1; Cyclin E, Mm00432367_m1; p21Cip1, Mm01303209_m1; C-myc, Mm00487804_m1; Cdc25b, Mm00499136_m1; Cdc25c, Mm00486872_m1; Cyclin B1, Mm00838401_g1; Aurk b, Mm01718146_g1. Following human Taqman primers (Applied Biosystems) were used: FoxF1, Hs00230962_m1; FoxF2, Hs00230963_m1. Reactions were analyzed in triplicates and expression levels were normalized to β-actin.
Cell Growth Assay
Cells were plated in triplicate. Viable cells were counted at designated time points using hemocytometer as previously described (48). Experiments were performed in triplicates and presented as average numbers of cells ± S.D.
Soft agar assay
Anchorage-independent growth of control, FoxF1 KD and/or FoxF2 KD cells was determined in 0.33% agarose (Invitrogen, Carlsbad, CA, USA), as described previously (50, 51). The culture medium containing 10% fetal calf serum was replaced every 3 days. Colonies containing more than 50 cells were counted after 7 days. Triplicate plates were used to count colonies and determine the mean number of colonies ± SD.
Cell cycle analysis
Cell cycle analysis was done as previously described (48). 76-9 cells with stable depletion of FoxF1 and FoxF2 were serum starved for 36hr by using DMEM media supplemented with 0.1% FBS. A complete media with 10% FBS was added and cells were collected at the indicated time points, fixed in 70% ethanol, and stained with propidium iodide/RNase solution (Cell Signaling). Flow cytometry was used to count cells in G0/G1, S and G2/M phases of cell cycle. To measure the mitotic index, ethanol fixed cells were permeablized with 0.25% Triton X-100 and stained with Histone H3-pS10 primary Ab followed by secondary Ab conjugated with Alexa-488 (Molecular Probes, Carlsbad, CA). Cells were counterstained with propidium iodide and RNaseA and subjected to FACS analysis (BD FACSCanto II, BD Bioscience, San Jose, CA).
Western blot
Protein extracts were prepared from RMS cells using RIPA buffer. Western blot analysis was done as previously described (36, 52). The following antibodies were used: p21Cip1, p27Kip1, total Rb, pRb (S795), pRb(S780), pRb(S807/811), total p53(#2527), phospho-p53(S15, #9284) (Cell Signaling), Cyclin D1(BD biosciences), FoxF1 (R&D systems, Minneapolis, MN) and Flag (Sigma, St. Louis, MO). Antibodies against Cdk1, Cdk2, Cdk4, Cdk6, Cyclin E1, β-Actin were from Santa Cruz (Paso Robles, CA). The signals from the primary antibody were amplified by HRP-conjugated secondary Abs (Calbiochem) and detected with Enhanced Chemiluminescence Plus reagent (Amersham Pharmacia Biotech, Piscataway, NJ) followed by autoradiography. β-Actin was used as loading control.
Immunohistochemistry
Paraffin sections (5μm) were cut and stained with hematoxylin and eosin (H&E) for morphological examination. Immunostaining was performed using following antibodies: anti-myogenin, anti-Ki67, anti-phospho-Histone H3 (Santa Cruz), anti-pRb, anti-p27Kip1, anti-p21Cip1 (Cell Signaling Technology, Inc.), anti-Cyclin D1 (BD biosciences) were done as previously described (52, 53).
Immunofluorescence
Cells were grown on poly-d-lysine-coated coverslips, fixed with 4% paraformaldehyde and permeablized with 0.3% Triton X-100. Permeabilized cells were stained with primary antibodies followed by secondary Abs conjugated with Alexa-488. DAPI (Vector Laboratories, Burlingame, CA) was used as a counterstain to visualize nuclei. Images were taken using a Zeiss Axiovert 200M microscope (Carl Zeiss, Oberkochen, Germany).
ChIP assay
FoxF1 and FoxF2-overexpressing RMS cells were cross-linked by addition of formaldehyde, sonicated and used for immunoprecipitation with mouse anti-Flag antibody (Sigma, St. Louis, MO) as described previously (21, 54). Mouse IgG were used as a control. DNA fragments were between 500 bp and 1000 bp in size. Reversed cross-linked ChIP DNA samples were subjected to PCR amplification with oligonucleotides specific to p21Cip1 promoter region. Following PCR primers were used to amplify 207bp DNA fragment specific to mouse p21Cip1 promoter: 5′- TCACCCAGCAAAGCCTTG -3′ and 5′- GGACTTTGGGATACTACACATAAAC -3′. Primers specific to mouse Pdgfb promoter (5′-TAGATGAGTTCTGGGACTGGACT-3′ and 5′-AGACATAACCGGAGGAGAAGAAG-3′) were used as positive control.
Luciferase assay
Mouse genomic DNA was used to amplify the −800bp to +1bp region of the mouse p21Cip1 promoter (Gene Bank Number NC_000083.6) using the primers (Forward: 5′-GCGGTACCTCTCCAGACATAGTGGGA-3′ and Reverse: 5′-CGCTCGAGCTAGACTCTGACACCGC-3′). PCR products were cloned into a pGL2 firefly luciferase (LUC) reporter plasmid (Promega, Madison, WI) and verified by DNA sequencing. p21Cip1 promoter-LUC plasmid was co-transfected with CMV-FoxF1 or CMV-FoxF2 expression vectors in HEK293T cells using Lipofectamine (Invitrogen). CMV-empty vector was used as a control. In addition, CMV- Renilla was used as an internal control to normalize the transfection efficiency. A dual luciferase assay (Promega, Madison, WI) was performed 48 hours after transfection as described previously (48).
Statistical analysis
ANOVA and Student’s T-test were used to determine statistical significance. P values less than 0.05 were considered significant. Values for all measurements were expressed as the mean ± standard deviation (SD). Data were graphically displayed using GraphPad Prism v.5.0 for Windows (GraphPad Software, Inc., La Jolla, CA, USA).
ACKNOWLEDGMENTS
This work was supported by the Research Grant from the American Cancer Society 125011-RSG-13-325-01-CSM (TVK), NIH grants R01 CA142724 (TVK) and R01 HL84151 (VVK).
Abbreviations
- RMS
Rhabdomyosarcoma
- Cre
Cre recombinase
- Fox
Forkhead Box transcription factor
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.McDowell HP. Update on childhood rhabdomyosarcoma. Arch Dis Child. 2003;88(4):354–7. doi: 10.1136/adc.88.4.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merlino G, Helman LJ. Rhabdomyosarcoma--working out the pathways. Oncogene. 1999;18(38):5340–8. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- 3.Hayes-Jordan A, Andrassy R. Rhabdomyosarcoma in children. Curr Opin Pediatr. 2009;21(3):373–8. doi: 10.1097/MOP.0b013e32832b4171. [DOI] [PubMed] [Google Scholar]
- 4.Breneman JC, Lyden E, Pappo AS, Link MP, Anderson JR, Parham DM, et al. Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma--a report from the Intergroup Rhabdomyosarcoma Study IV. J Clin Oncol. 2003;21(1):78–84. doi: 10.1200/JCO.2003.06.129. [DOI] [PubMed] [Google Scholar]
- 5.Qualman SJ, Morotti RA. Risk assignment in pediatric soft-tissue sarcomas: an evolving molecular classification. Curr Oncol Rep. 2002;4(2):123–30. doi: 10.1007/s11912-002-0073-4. [DOI] [PubMed] [Google Scholar]
- 6.Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, 3rd, Emanuel BS, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5(3):230–5. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 7.Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the variant t(1;13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54(11):2869–72. [PubMed] [Google Scholar]
- 8.Lae M, Ahn EH, Mercado GE, Chuai S, Edgar M, Pawel BR, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212(2):143–51. doi: 10.1002/path.2170. [DOI] [PubMed] [Google Scholar]
- 9.Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64(16):5539–45. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 10.Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, et al. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174(2):550–64. doi: 10.2353/ajpath.2009.080631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armeanu-Ebinger S, Bonin M, Habig K, Poremba C, Koscielniak E, Godzinski J, et al. Differential expression of invasion promoting genes in childhood rhabdomyosarcoma. Int J Oncol. 2011;38(4):993–1000. doi: 10.3892/ijo.2011.921. [DOI] [PubMed] [Google Scholar]
- 12.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: Lessons from transgenic mouse models. Cell Cycle. 2011;10(3):396–405. doi: 10.4161/cc.10.3.14709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLin VA, Henning SJ, Jamrich M. The role of the visceral mesoderm in the development of the gastrointestinal tract. Gastroenterology. 2009;136(7):2074–91. doi: 10.1053/j.gastro.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Ormestad M, Astorga J, Carlsson P. Differences in the embryonic expression patterns of mouse Foxf1 and −2 match their distinct mutant phenotypes. Dev Dyn. 2004;229(2):328–33. doi: 10.1002/dvdy.10426. [DOI] [PubMed] [Google Scholar]
- 15.Hellqvist M, Mahlapuu M, Samuelsson L, Enerback S, Carlsson P. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J Biol Chem. 1996;271(8):4482–90. doi: 10.1074/jbc.271.8.4482. [DOI] [PubMed] [Google Scholar]
- 16.Peterson RS, Lim L, Ye H, Zhou H, Overdier DG, Costa RH. The winged helix transcriptional activator HFH-8 is expressed in the mesoderm of the primitive streak stage of mouse embryos and its cellular derivatives. Mech Dev. 1997;69:53–69. doi: 10.1016/s0925-4773(97)00153-6. [DOI] [PubMed] [Google Scholar]
- 17.Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128(2):155–66. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- 18.Kalinichenko VV, Lim L, Beer-Stoltz D, Shin B, Rausa FM, Clark J, et al. Defects in Pulmonary Vasculature and Perinatal Lung Hemorrhage in Mice Heterozygous Null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 19.Kalinichenko VV, Zhou Y, Bhattacharyya D, Kim W, Shin B, Bambal K, et al. Haploinsufficiency of the Mouse Forkhead Box f1 Gene Causes Defects in Gall Bladder Development. J Biol Chem. 2002;277(14):12369–74. doi: 10.1074/jbc.M112162200. [DOI] [PubMed] [Google Scholar]
- 20.Kalinichenko VV, Gusarova GA, Shin B, Costa R. The Forkhead Box F1 Transcription Factor is Expressed in Brain and Head Mesenchyme during Mouse Embryonic Development. Gene Expression Patterns. 2003;3:153–8. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 21.Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res. 2014;115(8):709–20. doi: 10.1161/CIRCRESAHA.115.304382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinichenko VV, Bhattacharyya D, Zhou Y, Gusarova GA, Kim W, Shin B, et al. Foxf1 +/− Mice Exhibit Defective Stellate Cell Activation and Abnormal Liver Regeneration Following CCl4 Injury. Hepatology. 2003;37(1):107–17. doi: 10.1053/jhep.2003.50005. [DOI] [PubMed] [Google Scholar]
- 23.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70(14):6047–58. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cell Signal. 2012;24(1):316–24. doi: 10.1016/j.cellsig.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H, et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23(19):3487–94. doi: 10.1038/sj.onc.1207474. [DOI] [PubMed] [Google Scholar]
- 26.Gialmanidis IP, Bravou V, Petrou I, Kourea H, Mathioudakis A, Lilis I, et al. Expression of Bmi1, FoxF1, Nanog, and gamma-catenin in relation to hedgehog signaling pathway in human non-small-cell lung cancer. Lung. 2013;191(5):511–21. doi: 10.1007/s00408-013-9490-4. [DOI] [PubMed] [Google Scholar]
- 27.Wendling DS, Luck C, von Schweinitz D, Kappler R. Characteristic overexpression of the forkhead box transcription factor Foxf1 in Patched-associated tumors. Int J Mol Med. 2008;22(6):787–92. [PubMed] [Google Scholar]
- 28.Wei HJ, Nickoloff JA, Chen WH, Liu HY, Lo WC, Chang YT, et al. FOXF1 mediates mesenchymal stem cell fusion-induced reprogramming of lung cancer cells. Oncotarget. 2014;5(19):9514–29. doi: 10.18632/oncotarget.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nilsson J, Helou K, Kovacs A, Bendahl PO, Bjursell G, Ferno M, et al. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Res. 2010;70(5):2020–9. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- 30.Aitola M, Carlsson P, Mahlapuu M, Enerback S, Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn. 2000;218(1):136–49. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 31.McLin VA, Shah R, Desai NP, Jamrich M. Identification and gastrointestinal expression of Xenopus laevis FoxF2. Int J Dev Biol. 2010;54(5):919–24. doi: 10.1387/ijdb.092916vm. [DOI] [PubMed] [Google Scholar]
- 32.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, et al. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2015 doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8(4):e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nik AM, Reyahi A, Ponten F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144(5):1001–11. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 35.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13(20):5002–12. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balli D, Ren X, Chou FS, Cross E, Zhang Y, Kalinichenko VV, et al. Foxm1 transcription factor is required for macrophage migration during lung inflammation and tumor formation. Oncogene. 2011 doi: 10.1038/onc.2011.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosoi H, Sugimoto T, Hayashi Y, Inaba T, Horii Y, Morioka H, et al. Differential expression of myogenic regulatory genes, MyoD1 and myogenin, in human rhabdomyosarcoma sublines. Int J Cancer. 1992;50(6):977–83. doi: 10.1002/ijc.2910500626. [DOI] [PubMed] [Google Scholar]
- 38.Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim Biophys Acta. 2001;1471(3):M123–33. doi: 10.1016/s0304-419x(01)00019-1. [DOI] [PubMed] [Google Scholar]
- 40.Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–93. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 41.Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem. 2015;290(12):7563–75. doi: 10.1074/jbc.M114.609487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pappo I, Meirshon I, Karni T, Siegelmann-Danielli N, Stahl-Kent V, Sandbank J, et al. The characteristics of malignant breast tumors in hormone replacement therapy users versus nonusers. Ann Surg Oncol. 2004;11(1):52–8. doi: 10.1007/BF02524346. [DOI] [PubMed] [Google Scholar]
- 43.Tamura M, Sasaki Y, Koyama R, Takeda K, Idogawa M, Tokino T. Forkhead transcription factor FOXF1 is a novel target gene of the p53 family and regulates cancer cell migration and invasiveness. Oncogene. 2014;33(40):4837–46. doi: 10.1038/onc.2013.427. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13(2):213–24. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigel BJ, Rodeberg DA, Krieg AM, Blazar BR. CpG oligodeoxynucleotides potentiate the antitumor effects of chemotherapy or tumor resection in an orthotopic murine model of rhabdomyosarcoma. Clin Cancer Res. 2003;9(8):3105–14. [PubMed] [Google Scholar]
- 46.Douglass EC, Valentine M, Etcubanas E, Parham D, Webber BL, Houghton PJ, et al. A specific chromosomal abnormality in rhabdomyosarcoma. Cytogenet Cell Genet. 1987;45(3-4):148–55. doi: 10.1159/000132446. [DOI] [PubMed] [Google Scholar]
- 47.Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37(6):879–86. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng XH, Black M, Ustiyan V, Le T, Fulford L, Sridharan A, et al. SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene. PLoS Genet. 2014;10(9):e1004656. doi: 10.1371/journal.pgen.1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, Kalin TV. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27(30):4137–49. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 50.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66(3):1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang IC, Meliton L, Ren X, Zhang Y, Balli D, Snyder J, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4(8):e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balli D, Ustiyan V, Zhang Y, Wang IC, Masino AJ, Ren X, et al. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32(2):231–44. doi: 10.1038/emboj.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balli D, Zhang Y, Snyder J, Kalinichenko VV, Kalin TV. Endothelial cell-specific deletion of transcription factor FoxM1 increases urethane-induced lung carcinogenesis. Cancer Res. 2011;71(1):40–50. doi: 10.1158/0008-5472.CAN-10-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol. 2008;39(4):390–9. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]