Abstract
It is generally believed that maize (Zea mays L. ssp. mays) arose as a tetraploid; however, the two progenitor genomes cannot be unequivocally traced within the genome of modern maize. We have taken a new approach to investigate the origin of the maize genome. We isolated and sequenced large genomic fragments from the regions surrounding five duplicated loci from the maize genome and their orthologous loci in sorghum, and then we compared these sequences with the orthologous regions in the rice genome. Within the studied segments, we identified 11 genes that were conserved in location, order, and orientation. We performed phylogenetic and distance analyses and examined the patterns of estimated times of divergence for sorghum and maize gene orthologs and also the time of divergence for maize orthologs. Our results support a tetraploid origin of maize. This analysis also indicates contemporaneous divergence of the ancestral sorghum genome and the two maize progenitor genomes about 11.9 million years ago (Mya). On the basis of a putative conversion event detected for one of the genes, tetraploidization must have occurred before 4.8 Mya, and therefore, preceded the major maize genome expansion by gene amplification and retrotransposition.
Maize (Zea mays L. ssp. mays), from the grass tribe Andropogoneae, is an agronomically important crop and also a traditional genetic model. The theory that maize is a tetraploid first arose from the fact that maize has a haploid chromosome number of 10 (2n = 20), whereas many closely related grasses, such as Coix aquatica, Saccharum sp., and Erianthus sp., have only five chromosomes in the haploid nucleus (Celarier 1956; Mehra and Sharma 1975; Mason-Gamer et al. 1998). Rhoades (1951) demonstrated by genetic linkage mapping that nontandem gene duplicates are common in the maize genome. Mapping results of isozymic variants were also consistent with the hypothesis that the maize genome contains large duplicated regions (Goodman et al. 1980; McMillin and Scandalios 1980; Wendel et al. 1986). More recently, molecular mapping studies using RFLP markers have shown that most of the 10 maize chromosomes contain duplicated segments (Helentjaris et al. 1988; Ahn and Tanksley 1993). Comparative genomic studies based on the collinearity of grass genomes (Moore et al. 1995; Gale and Devos 1998) indicated that the maize genome aligns with the diploid rice and sorghum genomes in two chromosome sets, implying whole-genome duplication.
Three models can explain the large-scale duplications in the maize genome, that is, segmental duplication (multiple independent duplications within a genome), autotetraploidy (intraspecific genomic duplication), and allotetraploidy (interspecific genome hybridization). Gaut and Doebley (1997) proposed a segmental allotetraploid model and suggested that one of the maize subgenomes is more closely related to sorghum than to the other maize subgenome. However, they deemed their conclusions tentative (Gaut and Doebley 1997; Gaut et al. 2000) in the absence of sorghum sequences. Although many ESTs from sorghum and maize are now available, we cannot easily distinguish orthologous and paralogous gene copies by computational methods. Furthermore, it has been shown that many genes that are amplified in the maize genome move to nonorthologous positions, and some are completely lost in various strains of maize relative to sorghum and rice (Song et al. 2002; Song and Messing 2003). Therefore, we sequenced large genomic fragments from five different loci of known duplicated regions in the maize genome (Fig. 1). We also sequenced the orthologous regions from sorghum and aligned them with the orthologous regions from rice. Genomic comparison and sequence alignment resulted in identification of orthologous and full-length protein-coding genes. Using genes orthologous across the four genomes, we investigate in this study the evolutionary relationship of sorghum and maize, examine patterns of sequence divergence among gene orthologs, explore the rate of synonymous substitution among them, study the pattern of divergence time for pairs of orthologous sequences, and evaluate the different evolutionary models for the origin of maize.
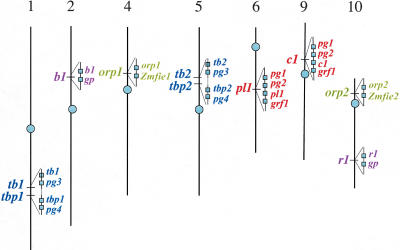
Figure 1.
Chromosomal location and physical arrangement of 11 genes' orthologs. Five duplication regions are shown in their related mapping position in the seven maize chromosomes, with tb1/2 and tbp2 closely linked. Genes (boxes) are arranged according to their gene order on an assembled BAC sequence contig (solid line within each triangle). The dashed line symbolizes about 7 cM of genetic distance between the tb1/2 and tbp1/2. For each of the BAC contigs, their actual orientations in the chromosomes are not known. The Zmfie1 and Zmfie2 are not a strict orthologous pair (see explanation in text), thus, are not included in our sequence analysis.
RESULTS
Isolation of BAC Clones and Their Sequencing
Probes for six loci (c1/pl1, tbp1/2, r1/b1, orp1/2, tb1/2, and Zmfie1/2) were used to screen maize and sorghum BAC libraries. These loci are located on seven of the 10 chromosomes (Fig. 1). These locations do not have a bias for centromeric or telomeric regions, but do reflect the segmental nature of the syntenous regions of the maize genome. For sorghum, each of the six probes identified only one overlapping series of BAC contigs, indicating that those six genes are not duplicated in the sorghum genome. In maize, the clones hybridizing with each probe belonged to two separate BAC contigs, demonstrating that those six loci are duplicated in maize. A total of 18 BAC clones, six from sorghum, and 12 from maize, were sequenced (Table 1). Two of the six loci, the genes encoding the FIE-like and ORP proteins, are physically linked. Nevertheless, because of the low gene density in maize, optimal alignment between sorghum and maize required us to select an additional six maize BAC clones to extend the size of the contigs. Table 1 shows the 24 BACs (providing more than 3 Mb of DNA) that were sequenced.
Table 1.
BAC Clones Used in This Study
| Loci | Names | BAC ID | Map position | Length (bp) | Accession no. |
|---|---|---|---|---|---|
| c1/pl1 | c1 | Z438D03 | ch9S | 184890 | AY530950 |
| c1 extension | Z214A02 | ch9S | 159000 | AY530951 | |
| pl1 | Z576C20 | ch6L | 155173 | AY530952 | |
| pl1 extension | Z264N17 | ch6L | 161000 | AY560577 | |
| Sorghum ortholog | SB35P03 | Unknown | 144120 | AF466199 | |
| Rice ortholog | OSJNBb0015B15 | ch6 | 123160 | AP005652 | |
| r1/b1 | r1 | Z138B04 | ch10L | 115734 | AF466202 |
| r1 extension | Z333J11 | ch10L | 207475 | AF466202 | |
| b1 | Z092E12 | ch2S | 147198 | AF466203 | |
| b1 extension | Z556K20 | ch2S | 90000 | AY542310 | |
| Sorghum ortholog | SB20O07 | Unknown | 157237 | AY542311 | |
| Rice ortholog | OSJNBa0065O17 | ch4 | 167446 | AL606682 | |
| Rice ortholog extension | OSJNBb0012E24 | ch4 | 127506 | AL606647 | |
| tb1/tb2 | tb1 | Z178A11 | ch1L | 130843 | AF464738 |
| tb1 extension | Z013105 | ch1L | 152337 | AY325816 | |
| tb2 | Z195D10 | ch5S | 141937 | AF466646 | |
| Sorghum ortholog | SB45l19 | Unknown | 77947 | AF466204 | |
| Rice ortholog | OSJNBa0004G17 | ch3 | 139071 | AC091775 | |
| tbp1/tbp2 | tbp1 | Z477F24 | ch1L | 212000 | AY542798 |
| tbp2 | Z474J15 | ch5S | 194000 | AY542797 | |
| Sorghum ortholog | SB32H17 | Unknown | 100707 | AF466201 | |
| Rice ortholog | OSJNBa0075A22 | ch3 | 153828 | AC133859 | |
| orp1/orp2 | orp1 | Z573F08 | ch4S | 181627 | AY555142 |
| Zmfie1 | Z078P04 | ch4S | 189000 | AY560576 | |
| orp2 | Z573L14 | ch10S | 144792 | AY555143 | |
| Zmfie2 | Z273B07/Z409L08 | ch10S | 138000 | AY560578 | |
| Sorghum ortholog | SB18C08 | Unknown | 159669 | AF466200 | |
| Sorghum extension | SB250O22 | Unknown | 84604 | AF466200 | |
| Rice ortholog | OJ1613_G04 | ch8 | 136186 | AP003896 | |
| Rice ortholog extension | P0680F05 | ch8 | 17000 | AP005620 | |
| Total length | 4293487 |
Sorghum and maize BACs were sequenced. Rice BACs were downloaded from the GenBank database.
Annotation of the BAC Clones
We predicted 62 protein-coding genes in the six sorghum BACs (total length of ∼681 kb) with an average gene density (avg) of 11 kb/gene. Within the 18 maize BACs (∼2554 kb), we identified 74 putative genes, avg = 34.5 kb/gene. Genes were embedded between large blocks of retrotransposons, including nested retrotransposons, as reported for other loci in maize (SanMiguel et al. 1996; Ramakrishna et al. 2002; Song et al. 2002; Song and Messing 2003). We found 70 candidate protein-coding genes in rice (∼798 kb), avg = 11.4 kb/gene, much lower than the predicted avg of 6.5 kb/gene (Rice Chromosome 10 Sequencing Consortium 2003). The major reason for this discrepancy could be the greater precision in our manual annotation compared with a primarily automated pipeline annotation.
Among the genes identified in the five chromosomal regions, only 11 genes, present as orthologous gene pairs in maize (Fig. 1), are also collinear in sorghum and rice. Four of the 11 gene duplicates (homoeologs), r1/b1, c1/pl1, tb1/tb2, and orp1/orp2, can (except for tb2) be distinguished by maize mutant phenotypes that segregate as single Mendelian traits, typical for the disomic nature of maize. No mutant phenotypes are known for the other seven gene duplicates. Although we can find homologs for all seven genes, only three genes have homologs that encode known protein. The tbp1/2 duplicate is a homolog of the TATA box-binding protein; grf1 is homologous with a putative growth-regulating factor 1 gene (grf1, rice homolog AAF17567, e < 1E-74); and gp apparently encodes a homolog of a putative glutathione peroxidase (gp, barley homolog CAB59895, e > 2.8E-82). The other four genes (pg1, pg2, pg3, pg4) show considerable sequence similarity to unknown proteins in Arabidopsis (pg1: At3g07660, e < 1E-30; pg2: At5g12080, e < 1E-100; pg3: At5g22090, e < 2.9E-11; pg4: At4g39410, e < 6E-08).
In the orp1/2 region (Fig. 1), we identified the Zmfie1/Zmfie2 genes, encoding a protein that is homologous to the Arabidopsis FIE protein (Ohad et al. 1999). Phylogenetic analysis of the Zmfie1/Zmfie2 sequences revealed that the ancestral gene duplicated in parallel on the lineage leading to rice and on the lineage leading to sorghum and maize. Furthermore, the analysis showed that the two maize copies represent descendants of different ancestral paralogs, indicating that each of the two maize genomic regions experienced reciprocal deletion of one of the ancestral paralogs. Because we wanted to use only orthologous genes, we did not include the Zmfie1/Zmfie2 genes in this study. Two copies of the r1/b1 genes were identified in sorghum, but because their duplication seems to postdate the divergence of maize and sorghum (data not shown), we used both copies in our analyses. Three copies of the r1/b1 genes were identified in rice. Two copies, the two closer to the 5′ end of the sequence (rice 1 and rice 2 in Fig. 4, below), appear to be non-functional due to the presence of premature stop codons, and were not included in our analyses because they may have evolved under different conditions than a functional gene. In addition to the r1/b1 gene copy identified in the rice region that is lacking a premature stop codon (rice 3 in Fig. 4, below), we also performed independent analysis using the functional and well-described r1/b1 gene homolog from the purple522 strain of Oryza sativa (GenBank accession no. U39860). Results of the two analyses did not differ. The tb2 coding sequence in our BAC clone is incomplete. We identified a maize GSS (genome survey sequences) sequence (BZ617075, 530 bp) that overlapped with the tb2 genomic sequence with 100% identity over 134 bp. The fused sequence yielded the complete tb2 gene that was used in our analysis.
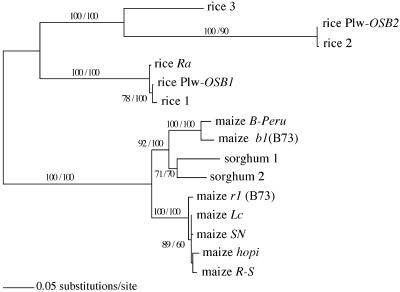
Figure 4.
Phylogenetic relationships among r1/b1 gene homologs resulting from ML (GTR+I+G) analysis. The numbers at the internodes represent bootstrap proportions from MP/ML analyses.
Phylogenetic Analyses
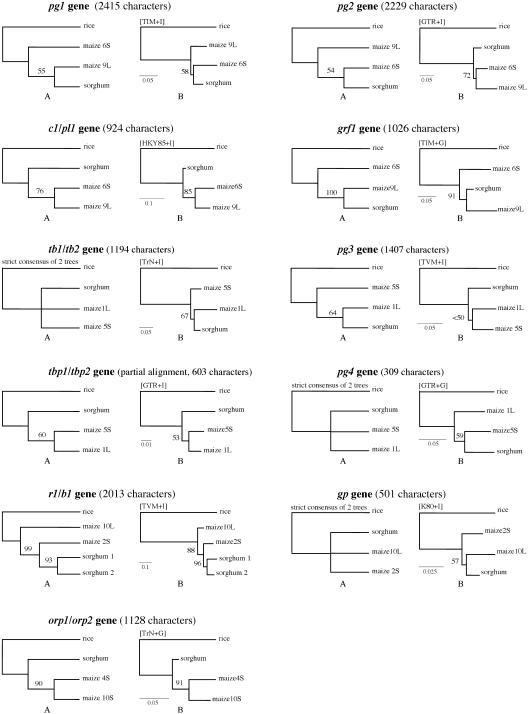
Phylogenetic analyses were carried out to resolve the relationship of the sorghum genome and the two subgenomes of maize. However, analysis of the relationship was confounded by a short internode. Only three of the 11 genes (grf1, orp1/2, and r1/b1) recover a relationship supported by >85 bootstrap values (see Fig. 2). To determine whether the topology of the ML gene tree differs significantly from a trichotomous tree (in which the three genomes diverge from the same node), we applied the likelihood ratio test (LRT). Using adjusted α according to the Bonferroni correction, we found that the orp1/2 and r1/b1 gene trees differ significantly from the trichotomous tree. However, those two gene trees show different topologies (Fig. 2).
Figure 2.
Gene trees resulting from (A) maximum-parsimony (MP) and (B) maximum-likelihood (ML) analyses. Models used in ML analyses are shown in parentheses on the branch leading to rice. MP tree was found via branch-and-bound search option. Heuristic search with 100 replicates was used in ML analyses. Numbers at the internodes represent the bootstrap proportions resulting from 1000 pseudoreplicates. Bars refer to number of substitutions per site.
Sequence Divergence and Nucleotide Substitution
Synonymous and nonsynonymous distances between gene orthologs are shown in Table 2. Synonymous distance varies 2.8-fold between the two maize orthologs and 3.2-fold between gene orthologs from maize and sorghum. Unusually low divergence at nonsynonymous sites was found for the two maize tbp1/2 orthologs. Nonsynonymous distance between maize and sorghum orthologs varies 15.5-fold. Graphs of estimated synonymous distances (Fig. 3A) show that the standard deviations overlap for the three pairs of orthologs, except in the cases of pg2 and r1/b1 genes. Therefore, we performed Z-tests for each gene for a null hypothesis (H0), which proposes that the three gene orthologs (one from sorghum and two from maize) diverged within a short time period and therefore, that the pairwise synonymous distances are equal. Using α value adjusted by the Bonferroni method, the H0 was found to be true for all but the r1/b1 gene.
Table 2.
Distances Between Gene Orthologs and Rates of Synonymous Substitution
| Sorghum-maize (6/1/10/4)
|
Sorghum-maize (9/5/2/10)
|
Maize-maize
|
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Chars. | Ks | Ka | Ks | Ka | Ks | Ka | Rate × 10−9 (95% CI) |
| pg1 (6-9)a | 2415 | 0.147 (0.022) | 0.050 (0.007) | 0.134 (0.020) | 0.043 (0.006) | 0.164 (0.023) | 0.064 (0.008) | 6.38 (±0.37) |
| pg2 (6-9) | 2229 | 0.157 (0.021) | 0.024 (0.004) | 0.207 (0.024) | 0.033 (0.005) | 0.210 (0.023) | 0.036 (0.005) | 8.29 (±0.41) |
| c1/p/1 (6-9) | 924 | 0.322 (0.063) | 0.080 (0.012) | 0.233 (0.050) | 0.072 (0.012) | 0.301 (0.055) | 0.074 (0.011) | 12.61 (±1.41) |
| grf1 (6-9) | 1026 | 0.296 (0.058) | 0.080 (0.012) | 0.244 (0.050) | 0.044 (0.008) | 0.330 (0.065) | 0.111 (0.014) | 8.39 (±0.86) |
| tb1/2 (1-5) | 1194 | 0.166 (0.032) | 0.037 (0.007) | 0.219 (0.024) | 0.070 (0.008) | 0.257 (0.031) | 0.090 (0.010) | 16.67 (±1.69) |
| pg3 (1-5) | 1407 | 0.348 (0.062) | 0.063 (0.008) | 0.278 (0.051) | 0.060 (0.008) | 0.299 (0.050) | 0.057 (0.007) | 9.49 (±0.87) |
| tbp1/2 (1-5) | 603 | 0.183 (0.053) | 0.028 (0.009) | 0.136 (0.043) | 0.029 (0.010) | 0.118 (0.006) | 0.0001 (0.00) | 11.61 (±1.49) |
| pg4 (1-5) | 309 | 0.320 (0.093) | 0.068 (0.016) | 0.216 (0.087) | 0.093 (0.020) | 0.324 (0.146) | 0.085 (0.020) | 13.03 (±2.68) |
| r1/b1 (10-2) | 2019 | 0.309 (0.042) | 0.088 (0.010) | 15.70 (±1.15) | ||||
| sorghum1 | 0.431 (0.052) | 0.088 (0.010) | 0.255 (0.035) | 0.092 (0.010) | ||||
| sorghum2 | 0.420 (0.052) | 0.073 (0.009) | 0.288 (0.039) | 0.075 (0.009) | ||||
| gp (10-2) | 501 | 0.133 (0.041) | 0.023 (0.008) | 0.137 (0.043) | 0.010 (0.005) | 0.128 (0.038) | 0.035 (0.010) | 6.75 (±0.76) |
| orp1/2 (4-10) | 1128 | 0.311 (0.042) | 0.007 (0.003) | 0.267 (0.037) | 0.006 (0.002) | 0.307 (0.043) | 0.008 (0.003) | 7.50 (±0.52) |
Ks/Ka refer to estimates of synonymous/nonsynonymous distance between two sequences. Standard deviations are shown in parentheses. Rates of synonymous substitution with 95% confidence interval are presented in the rightmost column.
Numbers in parentheses refer to chromosomal location.
Figure 3.
Estimated synonymous distances and divergence times for sorghum (s) and maize (m1, m2) gene orthologs. (A) Graph of pairwise synonymous distances for a particular gene (specified at top). (B) Divergence times between sorghum and maize gene orthologs. (C) Divergence times estimated for maize gene duplicates.
We also compared estimated distances between sorghum and maize and also between maize gene orthologs across genes. Both homogeneity tests (Gaut and Doebley 1997) were highly significant (χ2 = 100.49, P < 0.001; χ2 = 97.97, P < 0.001, respectively). The recovered heterogeneity may be caused by variable divergence times of the compared sequences, or by unequal rates of substitution across genes, or by both. Rate heterogeneity at synonymous sites was shown for the pg2, orp1/2, and tb1/tb2 genes, and therefore, these three gene pairs were not used for estimating divergence times. Nonsynonymous substitution rates varied among lineages more dramatically; only three gene pairs (pg2, pg3, and pg4) showed rate homogeneity across lineages. All other genes exhibited rate heterogeneity for at least one pair of sequences.
Assuming that rice diverged from an ancestor of sorghum and maize at ∼50 millon years ago (Mya; Wolfe et al. 1989), we estimated the rate of synonymous substitution and found that the rates vary nearly 2.6-fold among the 11 genes (Table 2). To account for the rate heterogeneity among the genes, we investigated the pattern of estimated divergence times separately for gene pairs of sorghum and of maize orthologs, and for pairs of maize homoeologs. The χ2 homogeneity test applied to the estimates of divergence time for sorghum and maize orthologs (Fig. 3B) was nonsignificant (χ2 = 26.25, P = 0.07). On the other hand, the test performed on the divergence times of maize homoeologs (Fig. 3C) was highly significant (χ2 = 37.62, P = 0). When the maize tbp1/tbp2 genes were excluded because of their unusual sequence conservation, this test resulted in a set of homogeneous time estimates (χ2 = 6.31, P = 0.39), implying that all but the tbp1/tbp2 genes diverged within the same time interval.
Estimation of Divergence Time
Phylogenetic analyses and distance analyses showed a trichotomous split of the two maize progenitor genomes and the sorghum genome. Assuming that rice diverged from the ancestor of sorghum and maize at ∼50 Mya (Wolfe et al. 1989) and excluding genes that showed rate heterogenity across lineages, we calculated the average divergence time for speciation of the three genomes to occur ∼11.9 Mya.
DISCUSSION
From cytology to genetic and molecular mapping, there is considerable evidence that the maize genome contains extensive chromosomal duplication. The large-scale genome duplication in maize could have resulted from segmental duplication or whole-genome duplication by autotetraploidy or allotetraploidy. On the basis of recovered bimodal distribution of synonymous distances between duplicated genes, Gaut and Doebley (1997) suggested that maize is a segmental allotetraploid and estimated that the two maize progenitors diverged at ∼20.5 Mya. Using sequence divergence between sorghum and maize from two genes (mdh and waxy), they further suggested that the sorghum genome and one of the duplicated maize regions might be closer relatives than are the two duplicated maize regions themselves. At that time, sorghum sequences orthologous to maize gene duplicates were unavailable. It was also unknown whether the gene duplicates in maize were true orthologs.
To establish the orthology of gene duplicates across taxa, it became necessary to isolate gene duplicates in maize along with other physically closely linked genes (microcollinear regions). To isolate such genes, we sequenced large duplicated chromosomal fragments in five different loci that are located on seven different maize chromosomes (Fig. 1; Table 1). We found that gene islands and blocks of retrotransposons are quite variable in length in the homoeologous regions of maize. We also note that tb1 and tbp1 on maize chromosome 1L are about 7 cM apart (www.agron.missouri.edu) and that both of their orthologs in the rice genome are present on chromosome 3 at a distance of about 2 Mb, indicating that microcollinearity between maize and rice is probably preserved over this interval. However, the analysis of >4,000,000 bp of DNA across taxa yielded only 11 genes that were conserved between the two duplicated regions of maize and the sorghum and rice genomes. Larger conservation was found between the sorghum and rice segments, whereas both maize regions experienced extensive genomic rearrangements. A more detailed investigation of gene mobility and gene/transposon organization can be found in the article by Lai et al. (2004). To investigate the origin of the maize genome, we particularly focused on strictly orthologous genes, as only those can be used to trace the evolution of species. On the example of the Zmfie1/2 gene, we show that not all maize gene duplicates, although present in orthologous regions, are necessarily orthologs. The two Zmfie genes in maize are derived from two closely linked paralogous sequences, and both chromosomal regions experienced reciprocal deletion of one of the ancestral gene paralog. This conclusion could not have been possible without a phylogenetic/genomic approach, and inclusion of such a gene pair in the analyses would produce incorrect results.
Sequences of the 11 orthologous genes, shared by the maize duplicated regions and the rice and sorghum genomes, were subjected to several analyses. First, phylogenetic analyses demonstrated a close relationship of the two maize progenitor genomes and the sorghum genome, indicating “a near-instantaneous” speciation of the three genomes (trichotomy). Multiple tests (the LTR test and the Z-test) showed that the r1/b1-gene tree provides a significantly nontrichotomic tree. Because the r1/b1 genes were extensively studied (Purugganan and Wessler 1994; Hu et al. 1996) partial sequence data (434 nucleotides in length) from additional grass taxa are available. Analysis of partial sequences of r1/b1-homologs from five grass species (data not shown), among which sequences of closer outgroups such as Pennisetum and Phyllostachys already exist, shows topological agreement with a gene tree on the basis of full-length coding sequences of r1/b1 orthologs from maize, sorghum, and rice (Fig. 4). Although the high support and consistent recovery of sister taxon relationship of the b1 gene of maize with the two paralogous copies from sorghum imply an allotetraploid origin of maize, the internode recovered is very short, raising a question of whether other factors might have influenced the nucleotide substitution rates in the r1/b1 genes. Furthermore, the short internode is consistent with our conclusion of a near-instantaneous speciation of all three progenitor-species.
In our second analysis, we analyzed patterns of estimated distances for each pair of sequences from maize and sorghum. In contrast to Gaut and Doebley (1997), who used the method of Nei and Gojobori (1986), we used a codon-based likelihood model for estimation of substitution rates. This model accounts for biases in transition/transversion rates and in codon frequencies. We also concentrated on synonymous substitutions, because rates of nonsynonymous substitution vary extensively among genes (Li 1997; Graur and Li 2000) and often show rate heterogeneity among lineages (Kimura and Ohta 1974), as our own results illustrate. Comparison of synonymous distances between sorghum and each of the two maize gene duplicates with the distance between the two maize gene duplicates, conducted for each gene using the Z-test, proved largely to be nonsignificant, indicating that sorghum and both maize progenitors diverged within a short period. The assumption behind the test is that the rate of nucleotide substitution of a given gene is constant over time. Our results indicate that, except for three genes (pg2, orp1/2, and tb1/tb2), the rates do not vary significantly across lineages.
Furthermore, we compared estimated distances between sorghum and maize and also between maize homoeologs across genes. Both χ2 homogeneity tests were highly significant. The heterogeneity recovered among estimated distances may be caused by variable divergence times of the compared sequences, or by unequal rates of substitution across genes, or by both. We found that the rates of substitution among the 11 genes vary 2.6-fold. This rate heterogeneity was taken into account when we performed analyses on estimated divergence times. The χ2 homogeneity test performed on data containing divergence times from all pairs of sequences was significant, but it did not reveal any distinct groups. In contrast, the test conducted on the divergence time of sorghum and maize was not significant, indicating their divergence during a common time interval. Homogeneity of the times of divergence between the maize orthologs was recovered when the tbp1/2 gene pair was excluded from the data, indicating that the tbp1/2 orthologs diverged at a different time than the other maize orthologs. Because the five chromosomal segments represent different parts of the maize genome (Fig. 1), the recovery of a common time interval for divergence of all but the tbp1/tbp2 genes in maize demonstrate that all five chromosomal regions duplicated at the same time, supporting the theory that maize arose from a doubling of the entire genome (tetraploidization) and not by a segmental duplication of genomic regions. In addition, because the variance of synonymous distance and the number of nonsynonymous substitutions of the tbp1/2 gene pair is very small, despite rather elevated rate of synonymous substitutions (Table 2), the tbp1/2 gene copies on maize chromosome 1 and 5 appear to be the product of an ectopic homoeologous conversion event that occurred ∼4.8 Mya. Finally, a Z-test comparing the mean divergence time of sorghum and of maize and the mean divergence time of maize homoeologs indicated that the time differences were not significant. This confirms the close relationship of sorghum and the two maize progenitors, in agreement with the phylogenetic analyses. Thus, our data demonstrate that maize is a product of a tetraploid event between two progenitors that are diverged to a similar extent from each other as each of them is diverged from sorghum.
If maize is indeed a tetraploid with 10 haploid chromosomes, then how did sorghum arrive at the same number of chromosomes as maize? One possibility would be that sorghum is a tetraploid as well. There is evidence from QTL and RFLP studies that the genome of sorghum (Sorghum bicolor) contains large duplicated regions (Pereira et al. 1994; Lin et al. 1995; Paterson et al. 1995, 1996). However, screening of BAC libraries with the five probes used in this study resulted in one single contig for each of the regions in the sorghum genome, disproving that any of these regions are duplicated in sorghum. Furthermore, on a genome-wide basis, the tetraploid origin cannot be supported by the comparison of the genetic maps (Gaut et al. 2000). The simplest explanation for these opposing results is that sorghum, like many plant genomes, may contain large-scale duplications that might have originated from intragenomic duplication events, similarly as reported for the sequenced genome of Arabidopsis (Vision et al. 2000; Simillion et al. 2002) and rice (Kishimoto et al. 1994; Nagamura et al. 1995; Vandepoele et al. 2003).
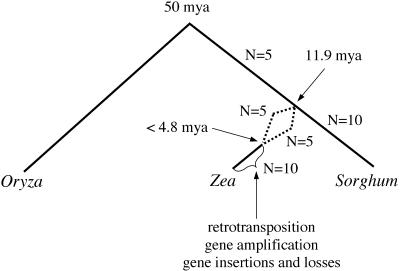
Here, we demonstrate that the two progenitor genomes of maize and the sorghum genome diverged from each other ∼11.9 Mya. The most parsimonious assumption about the ancestor of the three genomes is that the ancestor had a haploid genome of 5 (n = 5). This is consistent with the previously suggested ancestral chromosome number, n = 5, for the entire tribe Andropogoneae (Celarier 1956; Mehra and Sharma 1975) that was further supported from studies using isozyme data (Wendel et al. 1985) and DNA markers (Helentjaris et al. 1988). In the absence of any evidence of whole-genome duplication of the sorghum genome, one of the many possible scenarios could be that sorghum arrived to 10 chromosomes by chromosomal split of the five ancestral chromosomes (Fig. 5). Several reports favor such an explanation. It appears that the 10 sorghum chromosomes can be divided into two groups on the basis of centromeric sequences. Gómez et al. (1998), using a method for fluorescent in-situ hybridization (FISH), found that five of the 10 sorghum centromeres contain different repeat sequences than the other five. Then, let us hypothesize that the five progenitor chromosomes, because of their large size, contained already secondary regions with centromeric repeats, but those did not function as centromeres. This would be consistent with the report that Sorghum versicolor (n = 5) has an average chromosome length twice the size of Sorghum bicolor (n = 10) (Karper and Chisholm 1936). On the basis of such an assumption, one could propose neocentromeres to have arisen by epigenetic modification of nonactive centromere-like regions, consistent with McClintock's hypothesis about genome responses to “shocks” and exemplified by the activation of silent transposable elements through tissue culture (McClintock 1984; Peschke et al. 1987). Although such a scenario is still speculative at this time, it becomes clear that comparative genomics, in combination with other methods, may provide the means to test such hypotheses by, for instance, including genomes of n = 5 in such analyses.
Figure 5.
Hypothesized origin of maize and sorghum.
METHODS
BAC Clone Isolation and Sequencing
High-density filters for maize (Zea mays L. cv B73) BAC libraries (Yim et al. 2002) and sorghum (Sorghum bicolor cv BTx623) BAC libraries were screened with probes derived from PCR products of the maize r1, c1, orp1, tb1, Zmfie1, and tbp1 genes, based on their sequences in GenBank. All positive clones were analyzed by DNA fingerprinting using NotI- and HindIII digests, followed by Southern blot analysis. Overlapping BAC clones selected using the WebFPC program were sequenced at their ends with universal primers to identify a clone with minimal overlap to the one already sequenced.
BAC DNA was isolated using the Large Construct Kit (Qiagen, Inc.). The purified BAC DNA was physically sheared and then ligated into a pUC vector for shotgun libraries (Song et al. 2001). Sequencing was performed on an ABI 3700 DNA sequencer using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied BioSystems, Inc.). Base calling and assembly were based on the phred/phrap programs (Ewing et al. 1998). About 10× coverage was generated for all of the BACs, and sequence gaps were finished by primer walking or by transposon-based sequencing of subclones that covered the gaps, as previously described (Gordon et al. 1998). Transposon minilibraries were made according to the manufacturer's instructions (Finnzyme).
Sequence Analysis
Genes predicted by FGENESH (www.softberry.com) and GENSCAN (http://genes.mit.edu/GENSCAN.html) programs were further analyzed by homology searches using BLAST (Altschul et al. 1997). Alignment of coding sequences performed by ClustalX 1.81 (Thompson et al. 1997) was manually adjusted with reference to amino acid alignment. Maximum parsimony (MP) and maximum likelihood (ML) analyses were conducted using PAUP* 4.0b10 (Swofford 1998). Models used in ML analyses were predicted via Modeltest 3.06 (Posada and Crandall 1998). MP analysis was performed with branch-and-bound search, and ML with 100 heuristic random-addition searches. Support for individual nodes was examined by nonparametric bootstrap analysis (Felsenstein 1985) performed with 1000 pseudoreplicates. Rates of synonymous and nonsynonymous substitutions were estimated with the codon model of PAML (Yang 1997). Relative rate tests were performed with HYPHY (http://www.hyphy.org/) using the codon substitution model of Muse and Gaut (1994). To test the homogeneity of distance estimates, we used a χ2 test that incorporates variance of distance estimates (Gaut and Doebley 1997). We used the Z-test to compare substitution rates between gene orthologs separately for each gene. For calculation of rates of synonymous substitution and divergence times, see below.
To decrease the error in our rate and distance estimates, we used several approaches. First, we selected strictly orthologous genes to investigate the evolutionary history of genomes. Second, we used a codon-based likelihood model for estimation of substitution rates that account for biases in transition/transversion rates and in codon frequencies. Third, unlike Gaut and Doebley (1997) and others, we do not use a universal gene rate of synonymous substitution, represented by the rate of the Adh gene, but we use gene-specific rates of genes used in our study for estimating the divergence times. Fourth, using weighted means in our calculation, we also incorporate variance information in our estimates.
Estimation of the Rate of Synonymous Substitution
For each gene, the average rate of synonymous substitution r was calculated using the divergence time of rice and the ancestor of sorghum and maize. In our study, each gene tree has four taxa (one from sorghum, two from maize, and one from rice), that can be denoted by A, B, C, and D, where D represents the outgroup. To decrease the variance of the estimated gene rate, we calculate the rate rS,ABC from the weighted mean of the distances dAD, dBD, dCD between ingroup sequences and the outgroup as follows:
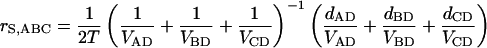 |
where VAD = Var(dAD), etc., is the estimated variance of the distance dAD, etc. The variance Var(rS,ABC) of rS,ABC is given by
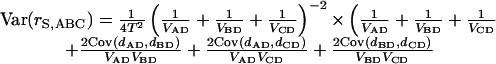 |
where Cov(dAD,dBD) can be approximated by Var(dOD), where O is the common ancestor of A and B (Nei et al. 1985), and the same holds for Cov(dAD,dCD) and Cov(dBD,dCD).
Calculation of Divergence Times
For each three-taxa gene tree (A, B, and the outgroup D), we estimated the divergence time of the ingroup sequences as follows: Let rS,AB be the rate of synonymous substitution of a gene (calculated here from the weighted mean of the distances dAD, dBD between ingroup sequences and the outgroup), that is,
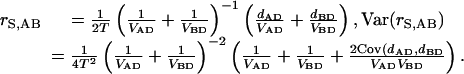 |
For the divergence time t of two ingroup sequences, we have
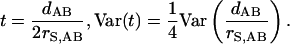 |
When, as is the present case,
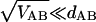 |
and
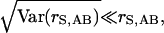 |
one can approximate the term Var(dAB/rS,AB) by the following formula:
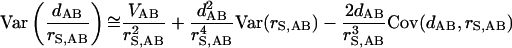 |
with
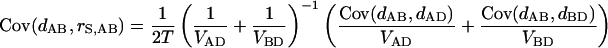 |
where the covariances Cov(dAB,dAD) and Cov(dAB,dBD) can be approximated by Var(dOA) and Var(dOB) with O the common ancestor of A and B (Nei et al. 1985).
Acknowledgments
We thank Dr. H.K. Dooner for discussions of emerging results and B.S. Gaut and J.F. Doebley for critical comments on the manuscript, and G. Fuks, Dr. Arvind Bharti, A. Bronzino Nelson, S. Kavchok, G. Keizer, and S. Young for technical assistance. This work was supported by NSF grant 9975618.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2332504.
References
- Ahn, S. and Tanksley, S.D. 1993. Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. 90: 7980-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celarier, R.P. 1956. Cytotaxonomy of the Andropogoneae. 1. Subtribes Dimeriinae and Saccharinae. Cytologia 21: 272-291. [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783-791. [DOI] [PubMed] [Google Scholar]
- Gale, M.D. and Devos, K. 1998. Plant comparative genetics after 10 years. Science 282: 656-659. [DOI] [PubMed] [Google Scholar]
- Gaut, B.S. and Doebley, J.F. 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. 94: 6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Le Thierry d'Ennequin M., Peek, A.S., and Sawkins, M.C. 2000. Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. 97: 7008-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, M.I., Islam-Faridi, M.N., Zwick, M.S., Czeschin Jr., D.G., Hart, G.E., Wing, R.A., Stelly, D.M., and Price, H.J. 1998. Tetraploid nature of Sorghum bicolor (L.) Moench. J. Hered. 89: 188-190. [Google Scholar]
- Goodman, M.M., Stuber, C.W., Newton, K., and Weissinger, H.H. 1980. Linkage relationships of 19 enzyme loci in maize. Genetics 96: 697-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Graur, D. and Li, W.H. 2000. Rates and patterns of nucleotide substitution. In Fundamentals of molecular evolution (2nd ed.), pp. 99-164. Sinauer Associates, Sunderland, MA.
- Helentjaris, T., Weber, D., and Wright, S. 1988. Identification of the genomic locations of duplicate nucleotide sequences in maize by analysis of restriction fragment length polymorphism. Genetics 118: 353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Anderson, B., and Wessler, S.R. 1996. Isolation and characterisation of rice R genes: Evidence for distinct evolutionary path in rice and maize. Genetics 142: 1021-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karper, R.E. and Chisholm, A.T. 1936. Chromosome numbers in Sorghum. Am. J. Bot. 23: 369-374. [Google Scholar]
- Kimura, M. and Ohta, T. 1974. On some principles governing molecular evolution. Proc. Natl. Acad. Sci. 71: 2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto, N., Higo, H., Abe, K., Arai, S., Saito, A., and Higo, K. 1994. Identification of the duplicated segments in rice chromosomes 1 and 5 by linkage analysis of cDNA markers of known functions. Theor. Appl. Genet. 88: 722-726. [DOI] [PubMed] [Google Scholar]
- Lai, J., Ma, J., Swigoňová, Z., Ramakrishna, W., Linton, E., Llaca, V., Tanyolac, B., Park, Y.-J., Jeong, O.Y., Bennetzen, J.L., et al. 2004. Gene loss and movement in the maize genome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Li, W.H. 1997. Rates and patterns of nucleotide substitution. In Molecular evolution, pp. 177-213. Sinauer Associates, Sunderland, MA.
- Lin, Y.-R., Schertz, K.F., and Paterson, A.H. 1995. Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141: 391-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason-Gamer, R.J., Weil, C.F., and Kellogg, E.A. 1998. Granule-bound starch synthase: Structure, function, and phylogenetic utility. Mol. Biol. Evol. 15: 1658-1673. [DOI] [PubMed] [Google Scholar]
- McClintock, B. 1984. The significance of responses of the genome to challenge. Science 226: 792-801. [DOI] [PubMed] [Google Scholar]
- McMillin, D.E. and Scandalios, J.G. 1980. Duplicated cytosolic malate dehydrogenase genes in Zea mays. Proc. Natl. Acad. Sci. 77: 4866-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra, P.N. and Sharma, M.L. 1975. Cytological studies in some central and eastern Himalayan grasses I. The Andropogoneae. Cytologia 40: 61-74. [Google Scholar]
- Moore, G., Devos, K., Wang, Z., and Gale, M.D. 1995. Grasses, line up and form a circle. Curr. Biol. 5: 737-739. [DOI] [PubMed] [Google Scholar]
- Muse, S.V. and Gaut, B.S. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates with application to the chloroplast genome. Mol. Biol. Evol. 11: 715-724. [DOI] [PubMed] [Google Scholar]
- Nagamura, Y., Inoue, T., Antonio, B.A., Shimano, T., Kajiya, H., Shomura, A., Lin, S.Y., Kuboki, Y., Harushima, Y., Kurata, N., et al. 1995. Conservation of duplicated segments between rice chromosome-11 and chromosome-12. Breed. Sci. 45: 373-376. [Google Scholar]
- Nei, M. and Gojobori, T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418-426. [DOI] [PubMed] [Google Scholar]
- Nei, M., Stephens, J.C., and Saitou, N. 1985. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol. Biol. Evol. 2: 66-85. [DOI] [PubMed] [Google Scholar]
- Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J.J., Goldberg, R.B., and Fischer, R.L. 1999. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A., Lin, Y., Li, Z., Schertz, K., Doebley, J., Pinson, S., Liu, S., Stansel, J., and Irvine, J. 1995. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714-1718. [DOI] [PubMed] [Google Scholar]
- Paterson, A., Lan, T., Reischmann, K., Chang, C., Lin, Y., Liu, S., Burow, M., Kowalski, S., Katsar, C., DelMonte, T., et al. 1996. Toward a unified genetic map of higher plants, transcending the monocot-dicot divergence. Nat. Genet. 14: 380-382. [DOI] [PubMed] [Google Scholar]
- Pereira, M.G., Lee, M., Bramel-Cox, P., Woodman, W., Doebley, J., and Whitkus, R. 1994. Construction of an RFLP map in sorghum and comparative mapping in maize. Genome 37: 236-243. [DOI] [PubMed] [Google Scholar]
- Peschke, V.M., Phillips, R.L., and Gengenbach, B.G. 1987. Discovery of a transposable element activity among progeny of tissue culture-derived maize plants. Science 238: 804--807. [DOI] [PubMed] [Google Scholar]
- Posada, D. and Crandall, K.A. 1998. MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817-818. [DOI] [PubMed] [Google Scholar]
- Purugganan, M.D. and Wessler, S.R. 1994. Molecular evolution of the plant R regulatory gene family. Genetics 138: 849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna, W., Dubcovsky, J., Park, Y.J., Busso, C., Emberton, J., SanMiguel, P., and Bennetzen, J.L. 2002. Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162: 1389-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.M. 1951. Duplicated genes in maize. Am. Nat. 85: 105-110. [Google Scholar]
- Rice Chromosome 10 Sequencing Consortium. 2003. In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300: 1566-1569. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., and Avramova, Z. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765-768. [DOI] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C., Zabeau, M., and Van De Peer, Y. 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 99: 13627-13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R. and Messing, J. 2003. Gene expression of a gene family in maize based on non-collinear haplotypes. Proc. Natl. Acad. Sci. 100: 9055-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, V., Linton, E., and Messing, J. 2001. Sequence, regulation, and evolution of the maize 22-kD zein gene family. Genome Res. 11: 1817-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca V., and Messing, J. 2002. Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12: 1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. 1998. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Simillion, C., and Van de Peer, Y. 2003. Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15: 2192-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114-2117. [DOI] [PubMed] [Google Scholar]
- Wendel, J.F., Goodman, M.M., and Stuber, C.W. 1985. Mapping data for 34 isozyme loci currently being studied. Maize Genet. Coop. News Lett. 59: 90. [Google Scholar]
- Wendel, J.F., Stuber, C.W., Edwards, M.D., and Goodman, M.M. 1986. Duplicated chromosomal segments in Zea mays L.: Further evidence from Hexokinase isozymes. Theor. Appl. Genet. 72: 178-185. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M., Yang, Y.W., Sharp, P.M., and Li, W.-H. 1989. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. 86: 6201-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood Comput. Appl. Biosci. 13: 555-556. [DOI] [PubMed] [Google Scholar]
- Yim, Y.S., Davis, G.L., Duru, N.A., Musket, T.A., Linton, E.W., Messing, J., McMullen, M.D., Soderlund, C.A., Polacco, M.L., Gardiner, J.M., et al. 2002. Characterization of three maize bacterial artificial chromosome libraries toward anchoring of the physical map to the genetic map using high-density bacterial artificial chromosome filter hybridization. Plant Physiol. 130: 1686-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genes.mit.edu/GENSCAN.html; gene prediction.
- www.agron.missouri.edu; maize map.
- www.softberry.com; gene prediction.
- http://www.hyphy.org/; software tool.