Abstract
We present here an unbiased and extremely versatile insertional library of yeast genomic DNA generated by in vitro mutagenesis with a multipurpose element derived from the bacterial transposon Tn7. This mini-Tn7 element has been engineered such that a single insertion can be used to generate a lacZ fusion, gene disruption, and epitope-tagged gene product. Using this transposon, we generated a plasmid-based library of ∼300,000 mutant alleles; by high-throughput screening in yeast, we identified and sequenced 9032 insertions affecting 2613 genes (45% of the genome). From analysis of 7176 insertions, we found little bias in Tn7 target-site selection in vitro. In contrast, we also sequenced 10,174 Tn3 insertions and found a markedly stronger preference for an AT-rich 5-base pair target sequence. We further screened 1327 insertion alleles in yeast for hypersensitivity to the chemotherapeutic cisplatin. Fifty-one genes were identified, including four functionally uncharacterized genes and 25 genes involved in DNA repair, replication, transcription, and chromatin structure. In total, the collection reported here constitutes the largest plasmid-based set of sequenced yeast mutant alleles to date and, as such, should be singularly useful for gene and genome-wide functional analysis.
Since their discovery by Barbara McClintock in the early 1950's, transposable elements have come to be recognized both as key agents of genetic variation (Boeke and Pickeral 1999) and as highly useful laboratory tools (DesEtages et al. 2001). In the latter regard, endogenous and heterologous transposon systems have been engineered to facilitate a wide variety of functional studies and biological applications in organisms ranging from H. influenza (Gwinn et al. 1997) to M. musculus (Horie et al. 2003). Most commonly, transposons have been used as insertional mutagens to disrupt gene activity for phenotypic study. Relative to other mutagens, transposons provide a number of advantages: Of fore-most importance, the transposon insertion serves as an easily detected tag for the subsequent identification and/or cloning of the mutated DNA sequence. The utility of DNA transposition, however, is not limited to this application. Transposons have been modified to serve as “traps,” typically for exons and enhancer elements (Springer et al. 1995). Bacterial and yeast transposons have been employed as tools in several DNA sequencing strategies wherein the transposon insertion serves as a sequencing primer (Devine and Boeke 1994; York et al. 1998). Transposons have also been engineered to carry epitope tags or reporter genes such that a random insertion in-frame with its surrounding gene-coding sequence can be used to generate an epitope-tagged protein or reporter chimera (Hoekstra et al. 1991; Ross-Macdonald et al. 1997).
Transposon mutagenesis is scalable and, therefore, equally effective as applied to a single gene or an entire genome. Genome-wide collections of insertional mutants now exist for several organisms including A. thaliana and D. melanogaster (Alonso et al. 2003; Parks et al. 2004; Thibault et al. 2004). In the budding yeast Saccharomyces cerevisiae, we have previously employed large-scale shuttle mutagenesis to generate several genome-wide insertional libraries (Ross-Macdonald et al. 1999). Each insertional library was constructed from a plasmid-based library of yeast genomic DNA mutagenized in vivo in Escherichia coli using a modified form of the bacterial transposon Tn3. This Tn3 transposon was engineered to be multifunctional such that a single integrated insertion could be used to derive a gene disruption, reporter gene fusion, and epitope-tagged allele (Ross-Macdonald et al. 1995). By Tn3 mutagenesis, we generated and screened in excess of 150,000 Tn3 insertions in yeast genomic DNA. Sequence analysis of 10,174 insertion alleles identified insertions affecting 2553 different genes (of ∼5800 predicted yeast genes in total). This set of defined insertion alleles represents the largest such collection in S. cerevisiae; however, the collection is nonetheless incomplete, owing to apparent biases in Tn3 transposition.
Several alternative transposition systems are currently available for large-scale insertional mutagenesis. The bacterial transposon Tn7 was developed into an efficient in vitro transposition system exhibiting little target-site selectivity (Biery et al. 2000). This system employs a gain-of-function allele of the Tn7-encoded protein TnsC, a regulatory protein mediating attachment of the Tn7 transposase (TnsAB) to either TnsD, a site-specific DNA-binding protein directing insertion into the attTn7 site of the E. coli chromosome, or to TnsE, which directs transposition at lower frequency to many non-attTn7 chromosomal sites. Whereas wild-type Tn7 integrates into the E. coli chromosome through TnsD or TnsE, the isolated gain-of-function TnsC mutant can act independently of either protein, enabling transposition into a wide variety of sites (Stellwagen and Craig 1997). The Tn7 transposon has been further modified to yield several mini-Tn7 derivatives containing truncated and translationally open termini (Biery et al. 2000), although none of these derivatives had been engineered to generate epitope-tagged alleles or to be multifunctional.
We report here the construction, analysis, and large-scale application of a Tn7-derived multifunctional transposon for insertional mutagenesis of the S. cerevisiae genome. This Tn7-derived transposon contains truncated terminal repeats, a reporter/gene trap, epitope coding sequence, and lox sites for the Cre recombinase; it possesses identical functionality to our previous Tn3 transposon and can be used to monitor gene expression, generate gene disruptions, and epitope-tag genes. We used this Tn7 transposon to mutagenize a plasmid-based library of yeast genomic DNA in vitro and have individually introduced ∼75,000 resulting insertion alleles into yeast. By DNA sequencing, we determined the exact genomic site of 7176 Tn7 insertions. We screened a nonredundant subset of these defined yeast insertional mutants for hypersensitivity to the DNA-alkylating drug cisplatin, to both illustrate the utility of this library and to investigate yeast cellular response to this drug. We further present an analysis of Tn7 insertion site specificity and a comparison of this transposon system to our previous Tn3 in vivo system.
RESULTS AND DISCUSSION
Development of a Multifunctional Tn7 Mini-Transposon for Mutagenesis of the Yeast Genome
To maximize the utility of transposon mutagenesis as a tool for functional genomics, the employed transposon system should exhibit little bias in target-site selection, and the transposon tool itself should facilitate a variety of functional studies. Accordingly, we present here an unbiased and multifunctional transposon system derived from the bacterial transposon Tn7.
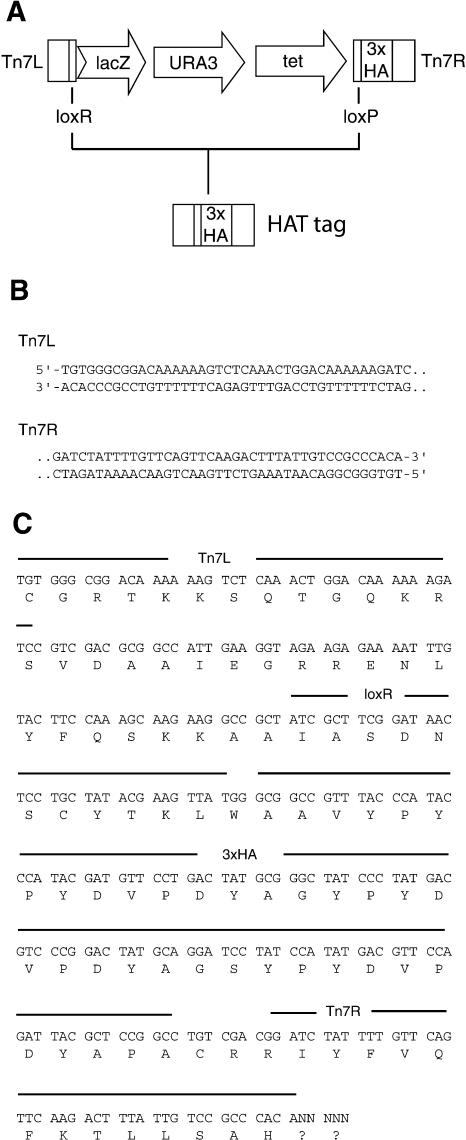
Briefly, this multipurpose Tn7 transposon contains the following functional elements (Fig. 1A). The transposon itself is bounded by Tn7 end sequences that act as substrates and binding sites for recombination proteins mediating Tn7 transposition (Fig. 1B). To enable selection in E. coli and yeast, this Tn7 mini-transposon carries the tet and URA3 genes, respectively. The transposon also carries a promoter-less and 5′-truncated lacZ gene that serves as both a reporter and a simple gene trap for protein-coding sequence. The lacZ reporter is terminated by a series of stop codons that creates a truncation of the mutagenized gene at the insertion site. This Tn7 mini-transposon has been further modified to contain two lox elements, one located near each terminal repeat. The lox sites are located internal to transposon sequence encoding three copies of the HA epitope, such that Cre-mediated recombination between the lox sites results in excision of the central body of the transposon. Post excision, the residual transposon insertion encodes 99 amino acids consisting primarily of the triplicate HA epitope and Tn7 end sequences (Fig. 1C). Thus, a single transposon insertion in-frame with its surrounding gene can be used to generate a reporter gene fusion, gene disruption, and epitope-tagged allele—the full range of functionalities associated with our previous Tn3-based multipurpose transposons.
Figure 1.
A multipurpose Tn7-based transposon for mutagenesis of yeast DNA. (A) Schematic diagram of the multipurpose mini-Tn7 transposon indicating left and right termini, bacterial and yeast selectable markers, lox sites, and triplicate HA epitope-coding sequence. The 5′-truncated lacZ reporter and selectable markers are indicated by arrows. The epitope insertion element (or HAT tag) is indicated below the brackets depicting Cre-lox recombination. (B) DNA sequence of the Tn7 truncated left (Tn7L) and right (Tn7R) termini. (C) DNA and amino acid sequence of the Tn7-encoded HAT tag. Tn7 transposition results in a 5-base pair duplication of target site sequence. As the exact sequence of this duplication depends upon the site of Tn7 insertion, the duplicated residues are indicated here as five unspecified (N) carboxy-terminal nucleotides.
In order to utilize the transposon as a tool for generating reporter gene fusions and epitope-tagged alleles, each Tn7 terminus must be translationally open in a reading frame consistent with the transposon-encoded reporter and epitope tag sequence. Furthermore, each terminus needs to be as short as possible such that the residual insertion element following transposition (and Cre-lox recombination) is not disruptive. To accommodate these needs, we significantly modified each Tn7 end sequence from its wild-type form. Specifically, each end of the mini-Tn7 transposon contains a 41-base pair sequence derived from the terminal right end of Tn7 (Fig. 1B). This terminal sequence is severely truncated; it consists of a 28-base pair terminal inverted repeat (containing a binding site for the transposase protein TnsB) and additional sequence comprising most of a second TnsB binding site. The mini-Tn7 termini were further modified such that each is translationally open in all three reading frames. Following Cre-lox recombination, the Tn7 termini contribute to a 99-codon insertion element encoding three copies of the HA epitope (Fig. 1C). This transposon-encoded HA insertion has been used extensively to tag yeast proteins for subsequent immunolocalization (Kumar et al. 2002a).
Despite the truncations and modifications applied to this transposon, the mini-Tn7 element is reasonably efficient in transposition: following a standard in vitro transposition reaction using this Tn7 derivative, ∼14% of donor substrate plasmids were converted to product bearing a simple Tn7 insertion (data not shown). Previous studies of Tn7 transposition illustrated effective recombination in vitro with 70-base pair termini (Biery et al. 2000); transposon constructs bearing the 41-base pair termini employed here exhibit a 75% reduction in transposition efficiency relative to these 70-base pair termini. However, this efficiency of reaction is still sufficient to generate in excess of 5000 recoverable transposition products in a typical in vitro reaction containing 100 ng of donor DNA—an efficiency of transposition highly suitable for genome-scale mutagenesis.
In addition to its design, this transposon is notable in that transposition can be carried out in vitro on a variety of substrates, including PCR products and extracted genomic DNA. For example, in a previous study, Bachman et al. (2002) utilized a Tn7-based transposon to mutagenize a pool of fragmented genomic DNA from yeast. Furthermore, similar Tn7 transposons carrying appropriate selectable markers may be designed for mutagenesis of many organisms, provided that the substrate DNA can be efficiently introduced into these organisms for subsequent analysis. Accordingly, Tn7 transposons have been utilized for large-scale mutagenesis of Candida glabrata (Castano et al. 2003), and other Tn7-derived systems may find widespread future applications.
Tn7-Based Shuttle Mutagenesis of the Yeast Genome
To generate a genome-wide collection of Tn7 insertional mutants in yeast, we used large-scale shuttle mutagenesis (Fig. 2). By this approach, Tn7 insertions are generated in a library of yeast genomic DNA, and insertion alleles are then “shuttled” into yeast for subsequent analysis (Seifert et al. 1986). Nonspecific Tn7 transposition is achieved in vitro using three purified proteins: TnsA, TnsB, and a TnsC gain-of-function mutant (Fig. 2A). Paired with the TnsAB transposase and appropriate cofactors (i.e., ATP and Mg2+), the TnsC mutant permits transposition into sites other than attTn7 as previously described. In addition, the Tn7 transposon is subject to transposition immunity, wherein DNA molecules containing at least one Tn7 terminus are immune from further insertions. Collectively, these properties of the in vitro Tn7 system can be exploited to generate a library of plasmids or PCR products, each bearing a single nondirected transposon insertion.
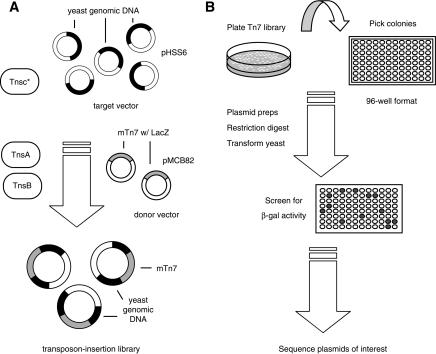
Figure 2.
Tn7 mutagenesis and high-throughput screening in yeast. (A) Simplified diagram of in vitro Tn7 mutagenesis. The TnsABCA225V complex enables transposition of mTn7 from its donor plasmid (pMCB82) to its target (pHSS6-based library of yeast genomic DNA), generating a Tn7 insertional library as shown. (B) Screening the Tn7 library in yeast. Ali-quots of the Tn7 insertional library are spread onto plates, and individual bacterial colonies are picked into 96-well plates. Plasmid DNA is prepared from each colony and is individually introduced into a diploid strain of yeast by standard methods of DNA transformation. The resulting yeast transformants are assayed for mTn-encoded β-galactosidase activity using a filter-based assay.
We used this in vitro system with our multifunctional Tn7 transposon to mutagenize 10 pools of a plasmid-based yeast genomic library derived from a strain lacking both its mitochondrial genome [ρ-] and 2-micron DNA [cir0]. Each pool is a separate library containing five genome equivalents of genomic DNA; each library consists of size-fractionated 2-3-kb genomic DNA fragments cloned into the vector pHSS6-Sal (Seifert et al. 1986). The pools were separately mutagenized in order to obtain at least 30,000 independent insertions per pool. In total, we estimate the insertion libraries encompass in excess of 300,000 independent insertions in 50 genome equivalents of DNA. As ∼30,000 Tn7-mutagenized transformants should be screened to ensure 95% coverage of the genome, we expect this library to be comprehensive.
Following Tn7 mutagenesis, this library was recovered in E. coli, and insertion alleles were screened individually in yeast by the high-throughput approach outlined in Figure 2B. This approach is described by Kumar et al. (2000). Briefly, transposon insertion alleles carried on an integrative yeast plasmid were introduced into a diploid strain of yeast; by homologous recombination, the mutagenized genomic DNA fragment replaced its native chromosomal locus, thereby generating a heterozygous diploid yeast strain carrying a chromosomal Tn7 insertion. Yeast transformants were subsequently screened for transposon-encoded β-galactosidase activity in order to identify productive Tn7 insertions within gene-coding sequence. The precise genomic site of transposon insertion within these strains was determined by sequencing each corresponding plasmid-borne insertion allele. From previous PCR-based analysis of genomic mTn insertions, we estimate that >97% of these transposon insertion alleles integrate within the yeast genome by homologous recombination (Burns et al. 1994).
Screening Summary
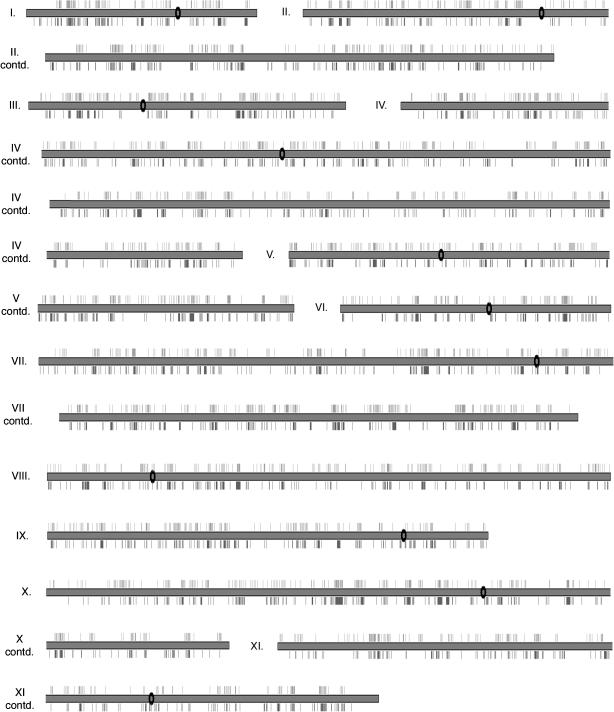
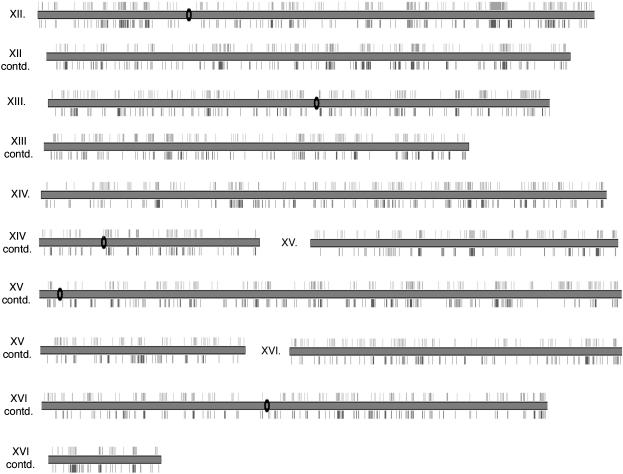
Using this high-throughput approach, we performed ∼85,000 plasmid preparations and yeast transformations, generating 10,272 yeast strains producing detectable levels of β-galactosidase under conditions of vegetative growth. We sequenced the corresponding plasmid alleles used to derive each of these yeast strains, and we successfully determined the site of transposon insertion in 9032 cases. This collection of defined mutants encompasses nearly 45% of all known yeast genes; that is, we have identified insertions within the coding sequence or neighboring regulatory regions of 2613 annotated yeast open reading frames. A summary of these sequenced insertions and affected genes is presented in Table 1. On average, we identified roughly three insertional mutant strains for each gene represented in our collection. We often found multiple yeast strains carrying insertions within the same gene; however, these insertions are typically at separate sites. As expected, the frequency of this occurrence increases in proportion to the size of the gene. Similarly, insertions were identified in all 16 yeast chromosomes, and the number of insertions observed per chromosome correlates fairly well with the size of each chromosome (Table 1). A graphic display of identified Tn7 insertions is shown in Figure 3.
Table 1.
Summary of Sequenced Tn7 Insertions
| Chromosome | Size (kb) | Sequenced insertions | Genes affected |
|---|---|---|---|
| I | 230 | 167 | 50 |
| II | 813 | 560 | 170 |
| III | 316 | 258 | 94 |
| IV | 1,554 | 961 | 308 |
| V | 577 | 472 | 149 |
| VI | 271 | 179 | 58 |
| VII | 1,091 | 843 | 240 |
| VIII | 589 | 339 | 128 |
| IX | 440 | 309 | 98 |
| X | 745 | 519 | 161 |
| XI | 666 | 350 | 120 |
| XII | 2,352 | 1,441 | 252 |
| XIII | 924 | 659 | 215 |
| XIV | 784 | 530 | 171 |
| XV | 1,091 | 764 | 207 |
| XVI | 948 | 681 | 192 |
| Total (I-XVI) | 13,392 | 9,032 | 2,613 |
Figure 3.
Distribution of Tn7 and Tn3 insertions in the S. cerevisiae genome. Chromosomal map of sequenced transposon insertions wherein each chromosome is represented by a gray rectangle; chromosomes are split over multiple lines as necessary. The black oval indicates centromeric sequence (not drawn to scale). Tn7 insertions are shown facing upwards. Previously identified Tn3 insertions are shown facing downwards.
Tn7 and Tn3-Based Insertional Mutagenesis
In previous studies, we had employed a similar approach using a multifunctional transposon derived from the bacterial element Tn3 (Ross-Macdonald et al. 1999; Kumar et al. 2000). In contrast to the Tn7 system utilized here, Tn3 mutagenesis was performed in vivo in E. coli. Resulting Tn3 insertional mutants were screened using a protocol similar to the one described above, and specific insertions were identified by DNA sequencing. The availability of these data provides an interesting opportunity to compare and contrast Tn7 in vitro transposition with Tn3 transposition in vivo. For purposes of comparison, a total of 11,329 Tn3 insertions are displayed below Tn7 insertions in Figure 3. Note that the insertion patterns are not mirror images.
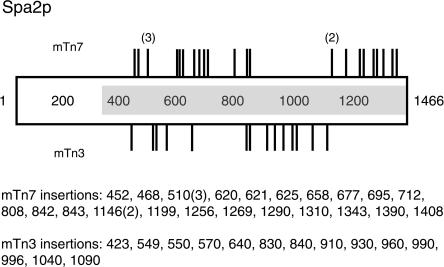
This point is more evident when considering a single gene. Figure 4 depicts both Tn7 and Tn3 insertions in the yeast polarity gene SPA2. The SPA2 gene encodes a 1466-amino acid protein important for polarized morphogenesis during processes of budding, mating, and pseudohyphal growth (Snyder 1989). To initially assess the transposition characteristics of Tn7, we mutagenized in vitro a plasmid carrying coding sequence for the C-terminal 1100 amino acids of Spa2p; previously, we had performed an identical pilot study of this same construct using Tn3 (Ross-Macdonald et al. 1997). By DNA sequencing, we defined 27 Tn7 insertions within the SPA2 fragment and 14 Tn3 insertions. As illustrated in Figure 4, Tn7 and Tn3 insertion sites are distinct: no overlap was observed. For example, in the region encoding amino acids 900 to 1100, seven Tn3 insertions were identified, whereas no Tn7 insertions were recovered in this same stretch of coding sequence. Even in regions of SPA2 for which both Tn7 and Tn3 insertions were recovered, insertion sites typically varied by at least 40 to 60 base pairs between transposons. Thus, each transposon system seems to exhibit separable characteristics defining insertion site specificity.
Figure 4.
Tn7 and Tn3-tagging of the polarity protein Spa2p. The Spa2p protein is indicated as a rectangle. The numbered residues correspond to amino acids; Spa2p is 1466 amino acids in length. The gray shaded region indicates SPA2 coding sequence subjected to transposon mutagenesis. Sequenced Tn7 insertions are shown facing upwards; Tn3 insertions are shown facing downwards. The precise amino acid position of each insertion is indicated below the figure. Bracketed numbers indicate multiple identified transposon insertions at a given site.
Sequence Analysis of Tn7 and Tn3 Insertion Sites
To consider Tn7 and Tn3 insertion site specificity more rigorously, we performed an extensive computational analysis of all insertion sites for which we have sequence data and an unambiguous indication of transposon orientation. In total, we analyzed 7176 Tn7 insertion sites and 10,174 Tn3 insertion sites for consensus sequences and/or unusual base composition (e.g., GC and AT content). For purposes of this analysis, we defined Tn7 and Tn3 directionality by the orientation of the transposon-encoded lacZ reporter; the 5′ end of the lacZ gene is nearest the left terminus of each mini-transposon. This directionality is obvious in regards to Tn3, as our mini-transposon was derived from left and right Tn3 termini; the imposed directionality is some-what more arbitrary in regards to Tn7, as this mini-transposon was derived from two variants of Tn7 right terminal sequence. A complete listing of identified Tn3 and Tn7 insertion coordinates is available online as Supplemental data.
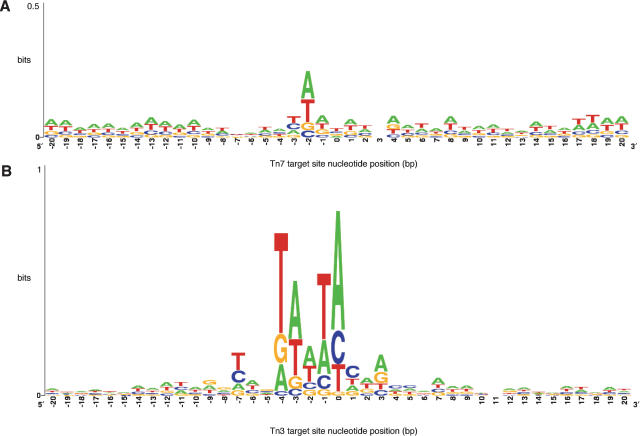
To identify a consensus sequence for Tn7 and Tn3 transposition, we tallied the frequency of adenine, thymine, guanine, and cytosine at every position in a 41-base pair window encompassing 20 base pair 5′ and 20 base pair 3′ of each transposon insertion. The results of this analysis are presented in Figure 5 as a sequence logo, in which the degree of sequence conservation at a given position is indicated by the total height of a stack of letters, and the frequency of each base at a given position is indicated by the relative height of that letter. In Figure 5, position 0 indicates an arbitrarily defined “insertion point” corresponding to the nucleotide immediately upstream of the transposon. Although Tn7 and Tn3 transpose by different mechanisms, each generates a 5-base pair duplication in target-site sequence at the point of insertion (Huang et al. 1986; Bainton et al. 1991). In Figure 5, this 5-base pair duplication corresponds to the region from position 0 to position -4; the duplication itself is not presented in the consensus.
Figure 5.
Tn7 and Tn3 target site consensus sequences. (A) Consensus sequence logo drawn from 7176 defined Tn7 insertion sites. In this logo, the nucleotide frequency at each position is indicated by the relative height of each letter; the degree of sequence conservation at each position is indicated as the total height of a stack of letters, measured in arbitrary units called “bits.” The y-axis is measured in bits, with two bits possible at each position. Note the y-axis is truncated at 0.5 bits to conserve space. The x-axis indicates nucleotide position in a 41-base pair window centered on the transposon insertion. The “0” position indicates the nucleotide immediately 5′ of the insertion. The negative positions are upstream of the insertion; the positive positions are downstream of the insertion. (B) Consensus sequence logo drawn from 10,174 defined Tn3 insertion sites. Here, the y-axis is truncated at 1 bit.
As shown in Figure 5B, Tn3 insertion sites exhibit an AT-rich 5-base pair sequence of TA(A/T)TA corresponding to the target site duplicated by Tn3 transposition. In contrast, Tn7 insertion sites (Fig. 5A) reveal relatively little sequence preference, other than a tendency towards the occurrence of an A or T at the central position in its duplicated sequence (position -2). Note that this 5-base pair region exhibits a slight tendency to be palindromic about its central position; although not strongly preferred, the sequence AT(A/T)AT can be derived by compiling frequently observed bases at each position in the duplicated region.
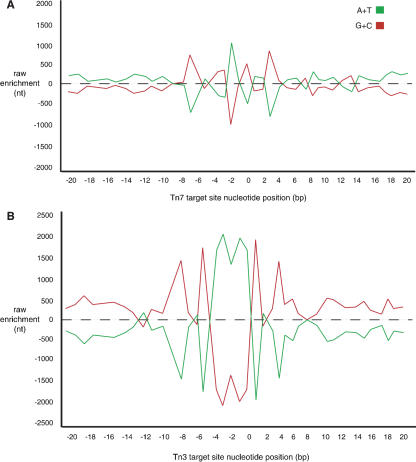
Target-site specificity may be considered further by examining AT and GC content local to each transposon insertion. Using the approach described above, we tallied the frequencies of AT and GC nucleotides at every position within the aforementioned 41-base pair window encompassing each transposon insertion. To account for a known genome-wide bias in GC content, we calculated the average A/C/G/T content of the yeast genome as a whole; we used this value as the expected nucleotide composition for each position in our window and subsequently determined the actual AT and GC enrichment relative to this expected value. The results of this analysis are graphically depicted in Figure 6. Again, a strong local AT bias is evident in Tn3 target-site sequence corresponding to the 5-base pair duplicated region (Fig. 6B). This AT-rich sequence is flanked, however, by GC-rich sequence extending seven base pairs upstream and downstream of the AT-rich target. Again, Tn7 target sequences show significantly less enrichment for either AT or GC (Fig. 6A), although some enrichment for AT is evident at a position central within the target-site duplication as discussed above. Interestingly, Tn7 insertion sites also exhibit a region of GC enrichment flanking this AT-rich sequence; however, the degree of enrichment is decreased relative to Tn3. As this mild AT enrichment is restricted to a region immediate to Tn7 insertion, we expect little difficulty in obtaining Tn7 insertions within GC-rich gene coding sequence. Note that 70% of the yeast genome is predicted to encode protein; these regions are noticeably GC-rich, yet Tn7 insertions have been identified fairly evenly throughout the genome.
Figure 6.
Nucleotide composition of Tn7 and Tn3 target sites. (A) Graph charting enrichment of A/T and G/C at Tn7 target sites within a 41-base pair window centered on the position immediately upstream of transposon insertion (position “0” on the x-axis). As in Figure 5, the x-axis indicates nucleotide position within the window. As the yeast genome tends to be GC-rich, the y-axis here indicates the enrichment of A/T or G/C at each position in raw numbers of nucleotides. Any point near 0 on the y-axis indicates little deviation from the norm in A/T or G/C content. (B) Graph of A/T and G/C enrichment at Tn3 target sites. Note the AT-rich sequence present in the 5-base pair duplicated region associated with Tn3 transposition (positions “-4” to “0”).
A Refined Tn3 Insertion Site Consensus Sequence
Our analysis of Tn3 insertion site specificity is particularly note-worthy in light of previous studies describing Tn3 target sequences. Specifically, Davies and Hutchison III (1995) previously characterized a 19-base pair asymmetric target consensus from sequence analysis of 223 Tn3 insertion sites. From this analysis, they identified a consensus sequence of “TNGG(T/C)(C/T)GTA(A/T)TAGGGGCGA”, where the central 5-base pair duplication is underlined. From our analysis of 10,174 insertion sites, we suggest a refinement of this consensus. Target site preference for TA(A/T)TA is clear; however, the flanking sequence is not strongly conserved. In fact, the most frequently observed residue at each position immediately downstream of this AT-rich target site is C,A,A, respectively, rather than G,G,G. This Tn3 consensus sequence is, therefore, most accurately limited to the 5-base pair sequence highlighted in Figure 6.
Of clear importance, the Tn3 transposon is differently, and apparently more strongly, biased than Tn7. We therefore expect the Tn7 insertional library to be an excellent complement to previous Tn3 libraries and, as such, a strong resource for functional genomics and phenotypic analysis in yeast.
Directed Phenotypic Screening for Cisplatin Sensitivity
Although phenotypic screening may be accomplished in yeast by a wide variety of approaches, the availability of genome-wide plasmid-based collections of defined yeast mutant alleles has enabled the development of a high-throughput screening strategy termed directed allele replacement technology (DART; Bidlingmaier and Snyder 2002).
For purposes of DART analysis, we generated a nonredundant and representative collection of plasmid-borne insertion alleles, each carrying a defined Tn7 insertion affecting a different annotated and nonessential yeast gene. We subsequently introduced each insertion allele individually into a haploid strain of yeast by standard methods of DNA transformation adapted for application in 96-well format (Kumar et al. 2002b). By homologous recombination, the transposon-mutagenized DNA integrated at its native chromosomal locus, thereby replacing its wild-type copy. The resulting yeast mutants were assayed in arrayed format (Methods) for hypersensitivity to the chemotherapeutic agent cis-diamminedichloroplatinum(II) (cisplatin).
At present, cisplatin is one of the most widely used anticancer drugs; it is a potent DNA-damaging agent with demonstrated chemotherapeutic activity, particularly in the treatment of testicular and ovarian cancer (Loehrer and Einhorn 1984). Cisplatin is thought to generate cytotoxicity through the induction of bi-functional platinum-DNA adducts that block DNA and RNA polymerases, preventing fundamental processes of replication and transcription (Burger et al. 2000). However, its complete mechanism of action remains to be determined. Saccharomyces cerevisiae has proven to be an effective model system for the analysis of cisplatin toxicity; as expected, cisplatin has been found to be toxic to yeast strains impaired in pathways mediating DNA damage repair (Simon et al. 2000).
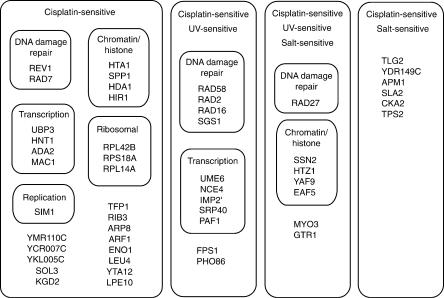
By screening a nonredundant collection of 1327 defined insertion alleles in a haploid strain of yeast, we identified 51 genes conferring cisplatin hypersensitivity upon disruption. (Fig. 7, Table 2). Seven of these strains contained disruptions in genes known to function in yeast DNA damage repair pathways; these genes include REV1 (error-prone damage tolerance), RAD58 (double-strand break formation and resection), and RAD27 (long-patch base excision repair; Simon et al. 2000). Cisplatin sensitivity was evident in strains mutated for RAD2, RAD7, and RAD16—three genes functioning in nucleotide excision repair as subunits of nuclear excision repair factor (NEF)3 (Rad2p) and NEF4 (Rad7p and Rad16p). In addition, disruption of SGS1 resulted in cisplatin hypersensitivity; SGS1 is homologous to RecQ in E. coli and is an ortholog of the human proteins defective in Bloom's syndrome and Werner's syndrome (Yamagata et al. 1998). Of the remaining 44 identified cisplatin-sensitive mutants, eight contain insertional mutations in genes encoding proteins mediating chromatin structure and remodeling. For example, disruption of SPP1 generates cisplatin sensitivity; Spp1p is likely involved in chromatin remodeling as a member of the Set1p complex (Miller et al. 2001). Several affected genes encode histone-associated proteins. HDA1 encodes a component of histone deacetylase A (Rundlett et al. 1996), and Htz1p is a histone-related protein that can suppress histone H4 point mutations (Dhillon and Kamakaka 2000). An additional nine cisplatin-sensitive mutants carry insertions within genes known to be involved in processes of transcription and DNA replication. For example, the gene MAC1 encodes a metal-binding transcriptional activator known to mediate resistance to cadmium, zinc, and lead exposure (Jungmann et al. 1993). Therefore, in total, 25 of 51 identified cisplatin-sensitive mutants carry an insertion in a gene involved in processes of DNA damage repair, structure, replication, or transcription. Of the remaining 26 identified mutants, four contain insertions in functionally uncharacterized genes (YMR110C, YCR007C, YDR149C, and YKL005C).
Figure 7.
Diagrammatic summary of phenotypic screening for cisplatin sensitivity. Fifty-one genes conferring sensitivity to cisplatin upon disruption are indicated in functional groups related to DNA damage/repair, transcription, replication, chromatin structure, and ribosomal function. Ungrouped genes function in processes that were not enriched among this set. Standard gene names have been used when available. YCR007C, YDR149C, YKL005C, and YMR110C are functionally uncharacterized.
Table 2.
Cisplatin-Sensitive Tn7 Insertional Mutants
| Strain | Mutagenized gene | Standard name | Description of gene product | UV | Salt |
|---|---|---|---|---|---|
| TN7-70C2 | YBL008W | HIR1 | Involved in cell-cycle regulation of histone transcription | WT | WT |
| TN7-93F1 | YBR114W | RAD16 | Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad7p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4); member of the SWI/SNF family | Sens | WT |
| TN7-69D7 | YBR279W | PAF1 | RNA polymerase II-associated, nuclear protein that may serve as both a positive and negative regulator of a subset of genes, perhaps operating in parallel with Gal11p | Sens | WT |
| TN7-29F8 | YCR007C | N/A | Putative integral membrane protein, member of DUP240 gene family | WT | WT |
| TN7-12A9 | YDL125C | HNT1 | Hint homolog, member of the histidine triad superfamily of nucleotide-binding proteins | WT | WT |
| TN7-28H7 | YDL185W | TFP1 | Encodes a protein with three regions (ABC) that is spliced to yield the extein AC and the intein B; AC is a 69K vacuolar (H+)-ATPase and B is 50K site-specific endonuclease named VDE (PI-Scel) that is homologous to HO | WT | WT |
| TN7-56E10 | YDL192W | ARF1 | Implicated in signal transduction and intracellular protein transport to or within the Golgi apparatus | WT | WT |
| TN7-1G1 | YDR074W | TPS2 | Trehalose-6-phosphate phosphatase | WT | Sens |
| TN7-71D4 | YDR148C | KGD2 | Dihydrolipoyl transsuccinylase component of α-ketoglutarate dehydrogenase complex in mitochondria | WT | WT |
| TN7-37F4 | YDR149C | N/A | Unknown gene product | WT | Sens |
| TN7-12E10 | YDR207C | UME6 | Regulator of both repression and induction of early meiotic genes. Ume6p requires Ume4 for mitotic repression and interacts with and requires Ime1p and Rim11p for induction of meiosis-specific transcription | Sens | WT |
| TN7-17H11 | YDR225W | HTA1 | Histone H2A | WT | WT |
| TN7-73H9 | YDR443C | SSN2 | Required for stable association of Srb10p-Srb11p kinase with RNA polymerase holoenzyme; regulates YGP1 expression; component of RNA polymerase II holoenzyme and Kornberg's mediator (SRB) subcomplex | Sens | Sens |
| TN7-44F7 | YDR448W | ADA2 | Transcription factor, member of ADA and SAGA, two transcriptional adaptor/HAT (histone acetyltransferase) complexes | WT | WT |
| TN7-70F9 | YDR450W | RPS18A | Homology to rat S18 and E. coli S13 | WT | WT |
| TN7-32H8 | YDR487C | RIB3 | Riboflavin biosynthesis | WT | WT |
| TN7-30D9 | YEL018W | EAF5 | Esa1p-Associated Factor-5 | Sens | Sens |
| TN7-9H11 | YER151C | UBP3 | Possible role for UBP3 in controlling the activity or assembly of the SIR protein complex | WT | WT |
| TN7-1F5 | YGR254W | ENO1 | Enolase I | WT | WT |
| TN7-42C4 | YGR258C | RAD2 | Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein | Sens | WT |
| TN7-48D6 | YHR141C | RPL42B | Homology to rat L36a and human L36a | WT | WT |
| TN7-29G4 | YHR163W | SOL3 | Weak multicopy suppressor of los1-1 | WT | WT |
| TN7-60C8 | YIL123W | SIM1 | Putatively involved in control of DNA replication | WT | WT |
| TN7-18E8 | YIL154C | IMP2′ | Protein involved in nucleo-mitochondrial control of maltose, galactose, and raffinose utilization | Sens | WT |
| TN7-33E10 | YJL117W | PHO86 | May collaborate with Pho87p and Pho84p in phosphate uptake | Sens | WT |
| TN7-49C9 | YJR052W | RAD7 | Protein that recognizes and binds damaged DNA in an ATP-dependent manner (with Rad16p) during nucleotide excision repair; subunit of Nucleotide Excision Repair Factor 4 (NEF4) | WT | WT |
| TN7-60F6 | YKL005C | N/A | Unknown gene product | WT | WT |
| TN7-62C5 | YKL006W | RPL14A | Homology to mammalian L14 | WT | WT |
| TN7-22H5 | YKL113C | RAD27 | 5′ to 3′ exonuclease, 5′ flap endonuclease, required for Okazaki fragment processing and maturation as well as for long-patch base-excision repair; member of the S. pombe RAD2/FEN1 family | Sens | Sens |
| TN7-30D10 | YKL129C | MYO3 | Myosin I | Sens | Sens |
| TN7-39D4 | YKR092C | SRP40 | Suppressor of mutant AC40 subunit of RNA polymerase I and III (high serine) | Sens | WT |
| TN7-9B3 | YLL043W | FPS1 | Suppressor of tps1/fdp1 and member of the MIP family of transmembrane channels; may be involved in glycerol efflux | Sens | WT |
| TN7-49F7 | YML121W | GTR1 | Involved in the function of the Pho84 phosphate transporter | Sens | Sens |
| TN7-45G8 | YMR021C | MAC1 | Metal-binding transcriptional activator | WT | WT |
| TN7-29A6 | YMR089C | YTA12 | Mitochondrial ATPase (similar to E. coli FtsH protein) that resides in the inner mitochondrial membrane | WT | WT |
| TN7-10H1 | YMR110C | N/A | Unknown gene product | WT | WT |
| TN7-66H1 | YMR190C | SGS1 | Involved in maintaining genome stability. Homologous to E. coli RecQ and human BLM and WRN protein that are defective in Bloom's syndrome and Werner's syndrome, respectively | Sens | WT |
| TN7-34F12 | YMR224C | RAD58 | Disp. for premeiotic DNA synthesis, but required for both double strand break formation and resection, synaptonemal complexes, meiotic recombination, and viable spores. Localizes to discrete sites in rad50 mutants | Sens | WT |
| TN7-69H9 | YNL021W | HDA1 | Component of histone deacetylase A | WT | WT |
| TN7-15D2 | YNL104C | LEU4 | Leucine biosynthesis | WT | WT |
| TN7-12A2 | YNL107W | YAF9 | Homolog of the human leukemogenic protein AF9; member of a yeast chromatin modifying complex | Sens | Sens |
| TN7-66D1 | YNL243W | SLA2 | Protein involved in membrane cytoskeleton assembly, required for cell polarization and endocytosis | WT | Sens |
| TN7-15A8 | YOL012C | HTZ1 | Histone-related protein that can suppress histone H4 point mutation | Sens | Sens |
| TN7-2F12 | YOL018C | TLG2 | Member of the syntaxin family of t-SNAREs | WT | Sens |
| TN7-23F2 | YOR061W | CKA2 | May have a role in regulation and/or execution of the eukaryotic cell cycle | WT | Sens |
| TN7-3A1 | YOR141C | ARP8 | Actin-related protein | WT | WT |
| TN7-2D10 | YOR346W | REV1 | Deoxycytidyl transferase involved in DNA-directed DNA polymerase activity and DNA repair mechanisms | WT | WT |
| TN7-14C3 | YPL024W | NCE4 | Negative regulator of Cts1 Expression | Sens | WT |
| TN7-54H4 | YPL060W | LPE10 | Mitochondrial protein with homology to MRS2 | WT | WT |
| TN7-61B11 | YPL138C | SPP1 | Likely involved in chromatin remodeling; member of Set1p complex | WT | WT |
| TN7-31H4 | YPL259C | APM1 | Medium subunit of the clathrin-associated protein complex | WT | Sens |
For all assays, strain sensitivity indicates an optical density reading at 600 nm <50% wild type.
Further Phenotypic Analysis of Cisplatin-Sensitive Mutants
As cisplatin is thought to block transcription and replication, mutants impaired in processes affecting DNA metabolism are expected to be cisplatin-sensitive; however, cisplatin interacts with many biological molecules, and additional pathways may contribute to the yeast cisplatin response. To consider the pathway specificity and function of genes implicated in cisplatin sensitivity, we further screened the 51 cisplatin-sensitive Tn7 insertional mutants described above for UV and salt sensitivity. UV treatment is an effective phenotypic assay for defects in DNA damage repair (Hampsey 1997). As such, it provides a complement to assays for cisplatin sensitivity and should identify a similar set of target genes. On the other hand, assays for salt sensitivity target separate pathways. Mutants hypersensitive to cisplatin and UV treatment are very likely impaired in processes affecting DNA dynamics, whereas mutants sensitive to both cisplatin and high salt (but not UV treatment) may be compromised in other pathways (Hampsey 1997).
From our analysis (Fig. 7, Table 2), 18 insertional mutants exhibited both UV and cisplatin sensitivity: 14 of these mutants contained a Tn7 insertion in a known gene involved in DNA damage repair, transcription, or chromatin structure/remodeling. Seven of these mutants were sensitive to high salt concentrations as well, and an additional six mutants were hypersensitive to both salt and cisplatin but not UV treatment. No mutant sensitive strictly to cisplatin and high salt contained a Tn7 insertion affecting a gene known to function in a DNA damage or structure-related pathway (Fig. 7, far right). The 11 disrupted genes conferring solely cisplatin and UV sensitivity very likely affect processes of DNA damage repair and transcription, with the exception of FPS1 and PHO86, neither of which is thought to function in these processes. In contrast, the six mutants hypersensitive to both cisplatin and salt treatment likely mediate cisplatin sensitivity through a separate mechanism(s), highlighting the fact that pathways apparently unrelated to DNA structure, repair, and dynamics also mediate cellular cisplatin response in yeast.
Although the Tn7 insertional library is highly useful for phenotypic analysis, one must be careful to ensure that any single insertion mutant chosen for study truly represents a null mutant. For example, phenotypic analysis of a strain carrying a Tn7 insertion within the RAD55 gene revealed a wild-type response to cisplatin; however, Rad55p is involved in the recombinational repair of double-strand DNA breaks, and, therefore, a rad55 disruption mutant would be expected to be cisplatin-sensitive (Lovett and Mortimer 1987). In the rad55 insertional mutant tested in the present study, Tn7 lies at codon position 355 of 406. This carboxy-terminal truncation is not likely to yield a null phenotype. To safeguard against this possibility, whenever possible, we selected yeast mutants carrying a Tn7 insertion at a position approximately one-third the length of the complete coding sequence. We estimate that the significant majority of these Tn7 mutants are null for the indicated gene: of nine Tn7 mutants carrying an insertion within a known gene involved in DNA repair, seven were found to be cisplatin-sensitive, as expected.
Applications of a Tn7 Insertional Library for Phenotypic Screening
From our analysis, the Tn7-based insertional library reported here is unbiased; as such, it represents an excellent tool for functional genomics and phenotypic analysis in yeast. Specifically, our transposon insertion library may be used as a tool for genetic screening by either of two approaches. First, insertion alleles may be transformed en masse into yeast for the subsequent identification of mutants exhibiting a desired phenotype. Because the Tn7-encoded sequence serves as a marker or tag, sites of transposon insertion can be rapidly identified by PCR amplification. Second, transposon insertion libraries may be used to generate genome-wide collections of defined insertion alleles. Plasmid-borne insertion alleles can be individually introduced into yeast by high-throughput DNA transformation; yeast mutants containing Tn7 insertions within protein-coding sequence can be selected by screening for transposon-encoded reporter gene activity. Assuming integration by homologous recombination, precise genomic sites of Tn7 insertion may be identified from the DNA sequence of each corresponding plasmid. Plasmids and yeast strains of interest can be permanently stored, generating a streamlined collection of yeast insertional mutants. By this approach, gene function in yeast may be investigated on a genome-wide scale through a survey of 5800 defined mutants, rather than through a screen of several hundred thousand uncharacterized mutations. Regardless of which approach is employed, the availability of a representative Tn7 insertional library promises to significantly expedite the functional analysis of yeast genes by simplifying the manner in which genetic screens are performed. Furthermore, transposon insertion libraries are uniquely useful in analyzing essential genes (by the generation of conditional mutations), unlike deletion strain collections. As a result, this Tn7 insertion collection is significant as a complement to current yeast genomic resources, and, additionally, represents a means of genome-scale mutagenesis applicable to any organism with well defined selectable markers and a high rate of DNA integration by homologous recombination.
METHODS
Strains, Reagents, and Standard Procedures
Bacterial and yeast strains were cultured according to standard protocols (Sambrook et al. 1989; Guthrie and Fink 1991). All yeast strains are derivatives of Y800 (Ross-Macdonald et al. 1999). The genomic DNA library utilized in this study was derived from diploid strain Y2278; Y2278 is isogenic to Y800 except that it has been cured of its 2-micron plasmid (cir0) and lacks mitochondrial DNA (rho0). Phenotypic screens were performed in Y2279 [MATa ura3-52 trp1Δ1 ade2-101 lys2-801 leu2Δ98]. All ade2 mutant strains were grown in medium supplemented with 0.3 mM adenine. Cisplatin was obtained from Sigma.
Tn7 Mini-Transposon Construction
Tn7 “read-in” or left terminal sequence was generated by annealing the following complementary oligonucleotides: 5′-AGAAGCTTGAATTCGGCTTATTGCTCTAAATGTGGGCGGACAAAAAAGTCTCAAACTGGACAAAAAAGATCCGTCGACGCGGCCATT-3′ and 5′-AATGGCCGCGTCGACGGATCTTTTTTGTCCAGTTTGAGACTTTTTTGTCCGCCCACATTTAGAGCAATAAGCCGAATTCAAGCTTCT-3′. After annealing, the resulting DNA product was cloned into pHSS6 containing the mini-transposon mTn-3xHA/lacZ (Ross-MacDonald et al. 1999) such that it replaced the corresponding left terminal sequence of this Tn3-based transposon. Similarly, Tn7 “read-out” or right terminal sequence was generated by annealing the following complementary oligonucleotides: 5′-AGATTACGCTCCGGCCTGTCGACGGATCTATTTTGTTCAGTTCAAGACTTTATTGTCCGCCCACATTTAGAGCAATAAGCCGAATTCCGCGG-3′ and 5′-CCGCGGAATTCGGCTTATTGCTCTAAATGTGGGCGGACAATAAAGTCTTGAACTGAACAAAATAGATCCGTCGACAGGCCGGAGCGTAATCT-3′. After annealing, this DNA product was also cloned into pHSS6 such that it replaced the right terminal sequence of mTn-3xHA/lacZ. The resulting Tn7 mini-transposon was subcloned into pBluescript SK (Stratagene). This mini-Tn7 element was subsequently transposed into pBW30 (Biery et al. 2000), a conditional replicon carrying an R6Kγ origin. This conditional replicon can only be maintained in cells carrying the pir gene; thus, unreacted donor plasmid will be unable to replicate in commonly used E. coli strains (e.g., Dh5α), reducing potential background upon in vitro transposition. This mTn7 donor plasmid is designated pMCB82 (mTn7-3xHA/lacZ::R6Kγ).
Shuttle Mutagenesis and High-Throughput Screening
Tn7 transposition was performed in vitro as described (Bainton et al. 1993; Biery et al. 2000). Briefly, ∼400 ng of a pHSS6-based library of yeast genomic DNA (Kanr) was mixed with 25 ng of a gain-of-function TnsC mutant (TnsCA225V) in a 100-μL reaction volume containing the following (at final concentration): 26 mM HEPES, 4.2 mM Tris pH 7.6, 50 μg/mL BSA, 2 mM ATP, 2.1 mM DTT, 0.05 mM EDTA, 0.2 mM MgCl2, 0.2 mM CHAPS, 28 mM NaCl, 21 mM KCl, and 1.35% glycerol. This mixture was “pre-incubated” at 30°C for 20 min. Subsequently, 40 ng TnsA, 25 ng TnsB, MgAc (15 mM), and 100 ng donor plasmid pMCB82 (Tetr) were added to this mixture and incubated at 30°C for an additional 2 h. Products were phenol-extracted and ethanol-precipitated in the presence of 5 μg yeast tRNA. Precipitated product was then collected, washed, dried, and resuspended in 20 μL water with RNaseA. This reaction was typically repeated five or six times per pool of library DNA; the resulting product was introduced by electroporation into competent DH5α. Transformants were plated on LB medium supplemented with tetracycline (3 μg/mL) and kanamycin (40 μg/mL). Transformants were scraped into a cell suspension and stored as frozen stock in 15% glycerol.
Plasmids from this Tn7 insertional library were individually introduced into diploid strain Y800 by DNA transformation as described (Kumar et al. 2002b). Yeast transformants were screened for β-galactosidase activity under conditions of vegetative growth and sporulation using a filter-based assay (Kumar et al. 2000).
DNA Sequencing
Individual plasmids from the Tn7 insertional library were prepared by standard alkaline lysis in 96-well format using a Hydra 96 liquid-handling system (Matrix Technologies; Kumar et al. 2000). Plasmids containing insertion alleles resulting in β-galac tosidase activity (after introduction into yeast) were sequenced using the following primer (5′-GGCCTTCTTGCTTTGGAAGTAC-3′) corresponding to a region ∼60 base pairs from the Tn7 left-end Read-In terminus.
Transposon Insertion Site Analysis
Using purpose-written Perl scripts, the genomic regions flanking the Tn3 and Tn7 insertion sites were examined, generating a multiple sequence alignment of genomic regions into which transposons successfully integrated. The full list of transposon insertion points (defined as the nucleotide immediately 5′ of the site of transposon insertion) is available online as Supplemental material, and these coordinates were used to extract spans of yeast genomic sequence corresponding to a region from -20 base pairs through +20 base pairs relative to the insertion point. As the point of insertion was always represented by position 21 of this extracted sequence, a separate alignment step was unnecessary. Rather, the sequences were compared directly as a multiple sequence alignment, and nucleotide frequencies at each position were calculated to determine consensus sequences and GC enrichment.
Once flanking 41-base pair regions were extracted for each of the 7176 Tn7 and 10,174 Tn3 insertions examined, we created a matrix of nucleotide frequencies at each position, building on the work of Davies and Hutchison III (1995). We then computed positional AT/GC enrichment relative to the average AT/GC content of the yeast genome at each position of the alignment. Further, a sequence logo was generated for each set of sequences using WebLogo (http://weblogo.berkeley.edu/), showing the composition of any consensus sites and facilitating comparison of Tn3 insertion sites with those of Tn7.
Phenotypic Screening
A nonredundant collection of Tn7 insertion alleles was introduced into Y2279 by lithium acetate-mediated DNA transformation (Kumar et al. 2000). Liquid cultures of yeast transformants were grown in 96-well microtiter plates to mid-log phase in YPAD supplemented with cisplatin at a concentration of 50 μg/mL. A 1 mg/mL stock solution of cisplatin in water was prepared by sonicating the cisplatin mixture for 5-10 30-sec pulses; cisplatin solutions were prepared fresh prior to each use. Cell density was quantified (OD600) using the Spectrafluor Plus 96-well plate reader (Tecan-US). Strains were characterized as being cisplatin-sensitive if cisplatin-treated samples exhibited less than 50% the optical density of mock-treated samples. Cisplatin-sensitive strains were further screened for UV sensitivity: liquid cultures of cisplatin-sensitive strains spotted onto YPAD plates were exposed to 10 mJoules irradiation in a UV Stratalinker (Stratagene). Irradiated plates were incubated in the dark at 30°C for 2 d prior to scoring. To assess salt sensitivity, cisplatin-sensitive strains were spotted onto YPAD plates supplemented with 0.9M NaCl; growth was scored after 2-d incubation at 30°C.
Figure 4.
Acknowledgments
We thank S. Dellaporta for the generous use of his Tecan SpectraFluor Plus microplate reader. The Tn7 insertional library may be obtained from Anuj Kumar; individual insertion alleles should be requested from Mike Snyder at ygac.med.yale.edu. This research was supported by NIH grants to M.S.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2875304.
References
- Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R. et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Bachman, N., Biery, M., Boeke, J., and Craig, N. 2002. Tn7-mediated mutagenesis of Saccharomyces cerevisiae genomic DNA in vitro. Methods Enzymol. 350: 230-247. [DOI] [PubMed] [Google Scholar]
- Bainton, R., Gamas, P., and Craig, N. 1991. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell 65: 805-816. [DOI] [PubMed] [Google Scholar]
- Bainton, R., Kubo, K., Feng, J.-n., and Craig, N. 1993. Tn7 Transposition: Target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell 72: 931-943. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier, S. and Snyder, M. 2002. Large-scale identification of genes important for apical growth in Saccharomyces cerevisiae by directed allele replacement technology (DART) screening. Funct. Integr. Genomics 1: 345-356. [DOI] [PubMed] [Google Scholar]
- Biery, M., Stewart, F., Stellwagen, A., Raleigh, E., and Craig, N. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28: 1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. and Pickeral, O. 1999. Retroshuffling the genomic deck. Nature 398: 108-109. [DOI] [PubMed] [Google Scholar]
- Burger, H., Capello, A., Schenk, P., Stoter, G., Brouwer, J., and Nooter, K. 2000. A genome-wide screening in Saccharomyces cerevisiae for genes that confer resistance to the anticancer agent cisplatin. Biochem. Biophys. Res. Comm. 269: 767-774. [DOI] [PubMed] [Google Scholar]
- Burns, N., Grimwade, B., Ross-Macdonald, P.B., Choi, E.-Y., Finberg, K., Roeder, G.S., and Snyder, M. 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes & Dev. 8: 1087-1105. [DOI] [PubMed] [Google Scholar]
- Castano, I., Kaur, R., Pan, S., Cregg, R., Penas, A.D., Guo, N., Biery, M., Craig, N., and Cormack, B. 2003. Tn7-based genome-wide random insertional mutagenesis of Candida glabrata. Genome Res. 13: 905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, C. and Hutchison III, C.H. 1995. Insertion site specificity of the transposon Tn3. Nucleic Acids Res. 23: 507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesEtages, S., Kumar, A., and Snyder, M. 2001. Transposons as tools. In Encyclopedia of genetics. Academic Press, San Diego, CA.
- Devine, S. and Boeke, J. 1994. Efficient integration of artifical transposons into plasmid targets in vitro: A useful tool for DNA mapping, sequencing, and genetic analysis. Nucleic Acids Res. 22: 3765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon, N. and Kamakaka, R.T. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6: 769-780. [DOI] [PubMed] [Google Scholar]
- Guthrie, C. and Fink, G. 1991. Guide to yeast genetics and molecular biology, pp. 3-20. Academic Press, San Diego, CA.
- Gwinn, M., Stellwagen, A., Craig, N., Tomb, J., and Smith, H. 1997. In vitro Tn7 mutagenesis of Haemophilus influenza Rd and characterization of the role of atpA in transformation. J. Bacteriol. 179: 7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey, M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099-1133. [DOI] [PubMed] [Google Scholar]
- Hoekstra, M.F., Burbee, D., Singer, J., Mull, E., Chiao, E., and Heffron, F. 1991. A Tn3 derivative that can be used to make short in-frame insertions within genes. Proc. Natl. Acad. Sci. 88: 5457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, K., Yusa, K., Yae, K., Odajima, J., Fischer, S.E.J., Keng, V.W., Hayakawa, T., Mizuno, S., Kondoh, G., Ijiri, T., et al. 2003. Characterization of sleeping beauty transposition and its application to genetic screening in mice. Mol. Cell. Biol. 23: 9189-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Heffron, F., Twu, J., Schloemer, R., and Lee, C. 1986. Analysis of Tn3 sequences required for transposition and immunity. Gene 41: 23-30. [DOI] [PubMed] [Google Scholar]
- Jungmann, J., Reins, H.A., Lee, J., Romeo, A., Hassett, R., Kosman, D., and Jentsch, S. 1993. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12: 5051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., DesEtages, S., Coelho, P., Roeder, G., and Snyder, M. 2000. High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol. 328: 550-574. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Agarwal, S., Heyman, J.A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., et al. 2002a. Subcellular localization of the yeast proteome. Genes & Dev. 16: 707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Vidana, S., and Snyder, M. 2002b. Insertional mutagenesis: Transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 350: 219-229. [DOI] [PubMed] [Google Scholar]
- Loehrer, P. and Einhorn, L. 1984. Drugs five years later. Cisplatin. Ann. Intern. Med. 100: 704-713. [DOI] [PubMed] [Google Scholar]
- Lovett, S. and Mortimer, R. 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: Effects of temperature, osmotic strength and mating type. Genetics 116: 547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T., Krogan, N.J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J.F., and Shilatifard, A. 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. 98: 12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, A.L., Cook, K.R., Belvin, M., Dompe, N.A., Fawcett, R., Huppert, K., Tan, L.R., Winter, C.G., Bogart, K.P., Deal, J.E. et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288-292. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Burns, N., Malcynski, M., Sheehan, A., Roeder, S., and Snyder, M. 1995. Methods for large-scale analysis of gene expression, protein localization, and disruption phenotypes in Saccharomyces cerevisiae. Meth. Mol. Cell. Biol. 5: 298-308. [Google Scholar]
- Ross-Macdonald, P., Sheehan, A., Roeder, G.S., and Snyder, M. 1997. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 94: 190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413-418. [DOI] [PubMed] [Google Scholar]
- Rundlett, S.E., Carmen, A.A., Kobayashi, R., Bavykin, S., Turner, B.M., and Grunstein, M. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. 93: 14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Seifert, H., Chen, E., So, M., and Heffron, F. 1986. Shuttle mutagenesis: A method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 83: 735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J., Szankasi, P., Nguyen, D., Ludlow, C., Dunstan, H., Roberts, C., Jensen, E., Hartwell, L., and Friend, S. 2000. Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res. 60: 328-333. [PubMed] [Google Scholar]
- Snyder, M. 1989. The SPA2 protein of yeast localizes to sites of cell growth. J. Cell Biol. 108: 1419-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, P., McCombie, W., Sundaresan, V., and Martienssen, R. 1995. Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science 268: 877-880. [DOI] [PubMed] [Google Scholar]
- Stellwagen, A. and Craig, N. 1997. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics 145: 573-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, S.T., Singer, M.A., Miyazaki, W.Y., Milash, B., Dompe, N.A., Singh, C.M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H.L. et al. 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283-287. [DOI] [PubMed] [Google Scholar]
- Yamagata, K., Kato, J., Shimamoto, A., Goto, M., Furuichi, Y., and Ikeda, H. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: Implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. 95: 8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, D., Welch, K., Goryshin, I., and Reznikoff, W. 1998. Simple and efficient generation in vitro of nested deletions and inversions: Tn5 intramolecular transposition. Nucleic Acids Res. 26: 1927-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://weblogo.berkeley.edu/; provides the source code for WebLogo, a Web-based application for the generation of sequence logos.