ABSTRACT
Listeria monocytogenes is a bacterial pathogen that is found in a wide variety of anthropogenic and natural environments. Genome sequencing technologies are rapidly becoming a powerful tool in facilitating our understanding of how genotype, classification phenotypes, and virulence phenotypes interact to predict the health risks of individual bacterial isolates. Currently, 57 closed L. monocytogenes genomes are publicly available, representing three of the four phylogenetic lineages, and they suggest that L. monocytogenes has high genomic synteny. This study contributes an additional 15 closed L. monocytogenes genomes that were used to determine the associations between the genome and methylome with host invasion magnitude. In contrast to previous findings, large chromosomal inversions and rearrangements were detected in five isolates at the chromosome terminus and within rRNA genes, including a previously undescribed inversion within rRNA-encoding regions. Each isolate's epigenome contained highly diverse methyltransferase recognition sites, even within the same serotype and methylation pattern. Eleven strains contained a single chromosomally encoded methyltransferase, one strain contained two methylation systems (one system on a plasmid), and three strains exhibited no methylation, despite the occurrence of methyltransferase genes. In three isolates a new, unknown DNA modification was observed in addition to diverse methylation patterns, accompanied by a novel methylation system. Neither chromosome rearrangement nor strain-specific patterns of epigenome modification observed within virulence genes were correlated with serotype designation, clonal complex, or in vitro infectivity. These data suggest that genome diversity is larger than previously considered in L. monocytogenes and that as more genomes are sequenced, additional structure and methylation novelty will be observed in this organism.
IMPORTANCE Listeria monocytogenes is the causative agent of listeriosis, a disease which manifests as gastroenteritis, meningoencephalitis, and abortion. Among Salmonella, Escherichia coli, Campylobacter, and Listeria—causing the most prevalent foodborne illnesses—infection by L. monocytogenes carries the highest mortality rate. The ability of L. monocytogenes to regulate its response to various harsh environments enables its persistence and transmission. Small-scale comparisons of L. monocytogenes focusing solely on genome contents reveal a highly syntenic genome yet fail to address the observed diversity in phenotypic regulation. This study provides a large-scale comparison of 302 L. monocytogenes isolates, revealing the importance of the epigenome and restriction-modification systems as major determinants of L. monocytogenes phylogenetic grouping and subsequent phenotypic expression. Further examination of virulence genes of select outbreak strains reveals an unprecedented diversity in methylation statuses despite high degrees of genome conservation.
KEYWORDS: L. monocytogenes, 100K Pathogen Genome Project, SMRT sequencing, methylation, inversion, infection, bacterial epigenetics, genetic epidemiology, DNA methylation, Listeria, genome analysis, virulence regulation
INTRODUCTION
Listeria monocytogenes is a facultative, aerobic, intracellular, foodborne pathogen found in a wide variety of anthropogenic, natural, and food sources (1–3). As causative agents of foodborne illness, Salmonella, Clostridium perfringens, and Campylobacter spp. outnumber L. monocytogenes in the yearly number of cases, yet listeriosis remains one of the leading causes of deaths in the United States resulting from foodborne infections (16%). With pathogen surveillance progressing toward the use of next-generation sequencing (NGS), improved insight into the unique genotypic traits of each pathogen will be paramount to the future of outbreak detection, trace back, and management (4).
The first L. monocytogenes genome, that of strain EGD-e, was sequenced in 2001 (5). To date, draft and closed genome sequences for 358 strains are publically available, 57 of which are closed genomes. Comparative genomic analyses from Listeria sequenced prior to 2013 suggest that gene content and genomic synteny are highly conserved (6–8) despite the diversity of phase variation and pathogenic potential (9, 10). Current categorization schemes (i.e., clonal complex and serotype) also fail to capture phenotypic variations (11), despite numerous publically available genomes. This phenotypic diversity is also observed in many food-associated bacteria, where isolates displaying little genetic variation have remarkably different phenotypes that are attributed to plasmid content (12–14) and epigenetic regulation (15–17). However, there is no large-scale study that links disease or phenotype with genotype that would adequately represent the genomic space of Listeria.
Coupled with informatics capabilities, sequencing technologies enable a deeper insight into the nuances between highly similar genomes. The ability to produce closed genomes via single-molecule real-time (SMRT) sequencing allows for the identification of genome rearrangements previously undetectable with draft genomes. While large chromosomal inversions have been documented in Lactococcus (18), Lactobacillus (19), Salmonella, and Escherichia coli, similar structural variations have yet to be described in L. monocytogenes (5, 8, 13). These genome rearrangements may result in global shifts in gene expression brought on by shuffling of methyltransferase specificities (20), and the bacteria may ultimately exhibit new phenotypes represented by variations in cell and colony morphologies and fitness (21, 22). The large number of reported phenotype variations suggests that additional genomes needed to be sequenced before the underlying variation will be observed, which is congruent with the report by Weimer et al. (23) predicting that at least 500 genomes of each bacterial species are needed to robustly encapsulate the diversity in variation within bacterial genomes. Additionally, determination of methylation on the large scale is a challenge that is only now being investigated. Chen et al. (24) demonstrated methods for use in large-scale genomic studies to discover new genes associated with epigenetic variation while gene content is conserved.
DNA methylation, a chemical modification of the canonical primary nucleotides by methyltransferases, is of critical importance in the biology of bacteria and may regulate virulence in many pathogens (21, 25). Many methyltransferases are encoded in the vicinity of restriction endonucleases, suggesting their involvement in restriction-modification systems (RMS) (26), which are the primary bacterial defense against invading foreign DNA (27). Methyltransferases, usually associated with RMS, are an abundant and diverse component of the pan-genome of L. monocytogenes (28). With the exception of a Sau3AI-like RMS, which is found in an epidemic-associated subpopulation of the species, little is known about the diversity and function of methyltransferases in L. monocytogenes (29, 30). This lack of current understanding warrants additional examination to attribute modification loci to expression changes and subsequent phenotypic differences to biological consequences (26). SMRT sequencing paired with RMS calling via REBASE allows for the detection of novel RMS enzymes and corresponding motifs (31), facilitating the understanding of the role of epigenetics in gene regulation despite genome structure conservation.
In this study, we report the genome and methylation pattern comparison of RMS gene distribution between 302 L. monocytogenes genomes. Subsequent SMRT sequencing of 15 L. monocytogenes outbreak isolates was used to identify genomic variation that may contribute to phenotypic diversity, such as infectivity and serotype designation. Epigenome comparison of these isolates was included for a comprehensive analysis of genomic variability. Despite genome conservation between isolates in this study and L. monocytogenes genomes sequenced to date, SMRT sequencing revealed previously undescribed genome features and diversity. Our structural and epigenome analyses indicated an unprecedented frequency of large chromosome rearrangements and unanticipated diversity in methylation abundance and motif modification. Serotype-determining and virulence genes displayed diverse methylation loci and patterns that were unique to strains belonging to the same outbreak. Isolate-specific methylation patterns and chromosome structures were not singly predictive of the diversity of in vitro host infection capabilities.
RESULTS
Genome comparison of 318 Listeria isolates reveals lineage and clade-specific RM motifs.
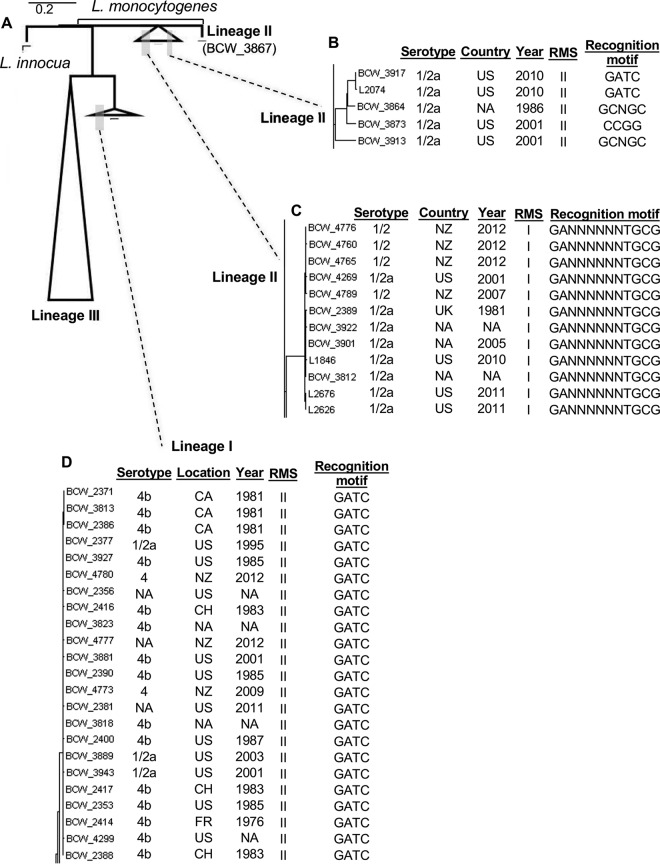
Genome distances were calculated for 302 environmental and outbreak-associated Listeria isolates acquired internationally between 1976 and 2013 to determine the classification and amount of genomic similarity. L. monocytogenes isolates clustered into three major clades distinguished by lineages I, II, and III, with Listeria innocua isolates forming a distinct branch separate from the three lineages (Fig. 1A) spread over time (Fig. 1B). Analysis of restriction-modification systems (RMS) revealed lineage- and clade-specific patterns for genes encoding the RMS. Type II RMS was the most heavily represented RMS with 256 genomes, followed by 110 genomes containing type I RMS, 73 genomes containing type IV RMS, and 25 genomes containing type III RMS (Table 1). Numerous isolates contained multiple RMS types with isolates encoding the same group of RMS genes clustering within the same clade. Of the 19 RMS motifs identified among these genomes, three motifs were present throughout all three lineages: GATC, GCWGC, and GTCGAC. Lineage-specific motifs were also identified (Table 2). Isolates with RMS enzymes recognizing the same RMS motif tend to have the least genomic variation among each other, despite being isolated decades apart from different geographical locations. This observation holds true for the lineage-specific motifs (Fig. 1C), as well as the promiscuous GATC motif (Fig. 1D). Interestingly, exceptions to this observation did exist, where a clade contained multiple RMS motifs and the same lack of temporal influence on genome distances (Fig. 1B).
FIG 1.
L. monocytogenes possesses promiscuous and clade-specific methyltransferase specificities. (A) An L. monocytogenes phylogenetic tree was constructed based on genome distances and displays overall clustering by lineage. Additional metadata for all strains are provided in Table S1 in the supplemental material. (B) L. monocytogenes clade representing multiple methyltransferase specificities. (C and D) Lineage II-specific motif (GANNNNNNTGCG) (C) and promiscuous motif (GATC) (D) that is associated with a single clade independent of geographical location and time of isolation. US, United States; NA, not available; NZ, New Zealand; UK, United Kingdom; CA, Canada; CH, Switzerland; FR, France.
TABLE 1.
RMS gene distribution among L. monocytogenes genomesa
| Gene category | No. of genomes with RMS type: |
|||
|---|---|---|---|---|
| I | II | III | IV | |
| Total genomes | 110 | 256 | 26 | 73 |
| Restriction endonuclease | 109 | 301 | 25 | 75 |
| Methyltransferase | 109 | 494 | 26 | |
| Specificity subunit | 172 | 4 | ||
| Controller protein | 26 | |||
| Helicase domain | 1 | |||
RMS genes were identified using REBASE.
TABLE 2.
Percent distribution of methyltransferase recognition motifs in L. monocytogenes lineages I, II, and IIIa
| Motif | % of isolates recognizing motif |
||
|---|---|---|---|
| Lineage 1 (n = 115) | Lineage 2 (n = 140) | Lineage 3 (n = 75) | |
| ACGT | 5.7 | ||
| CCGG | 5.0 | 8.7 | |
| CTGGAG | 1.7 | ||
| CTSAG | 15.7 | ||
| GAAGAC | 1.7 | 0.7 | |
| GACNNNNNGGT | 0.7 | ||
| GACNNNNNRTTC | 2.9 | ||
| GANNNNNNTGCG | 10.4 | ||
| GATC | 14.8 | 28.6 | 28.3 |
| GCNGC | 0.9 | ||
| GCSGC | 2.2 | ||
| GCWGC | 5.2 | 0.7 | 17.4 |
| GGCC | 2.2 | ||
| GTATCC | 17.1 | ||
| GTCGAC | 5.2 | 12.9 | 4.3 |
| GTGCAG | 1.7 | ||
| TACBNNNNNGGT | 2.1 | ||
| TACBNNNNNNGTNG | 5.0 | ||
| TTAGNNNNNNTTC | 1.4 | ||
Methyltransferase recognition motifs identified using REBASE were overlaid onto metadata provided during sample collection.
Presence of large chromosomal inversions encompassing the terminus of replication in the L. monocytogenes genome.
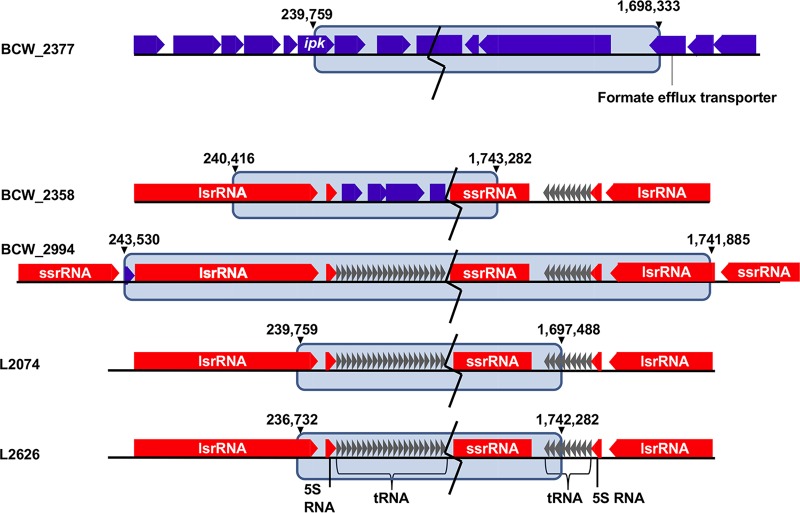
SMRT sequencing of 15 outbreak-associated L. monocytogenes isolates produced closed genomes. Analysis revealed large chromosomal rearrangements in five of the 15 strains (BCW_2358, BCW_2377, BCW_2994, L2074, and L2625) (Fig. 2; see Table S1 in the supplemental material). The inversions observed using SMRT sequencing were confirmed by PCR to be correct (see Fig. S1 and Tables S2 and S3 in the supplemental material). Inversion sites involved noncoding regions and RNA-coding genes (Fig. 3). The inversions were not a result of duplications (Fig. 2 and 3) and contained no identifiable methylation motifs at the inversion junctions. Furthermore, the presence of these inversions did not influence in vitro host association capabilities (see Fig. S2 in the supplemental material).
FIG 2.
MAUVE alignments of L. monocytogenes genomes. Whole-genome alignments of L. monocytogenes isolates were carried out using MAUVE. Isolates are arranged by serotype, and genomes are displayed as locally colinear blocks (LCBs), which are labeled 1 to 6. Additional metadata for all strains are provided in Tables S1 and S2 in the supplemental material. The isolates used for this figure are as follows, with the original identification and serotype in parentheses: BCW_2992 (J1776, serotype 4b), BCW_2993 (J1817, serotype 4b), L1846 (serotype 1/2a), L2624 (serotype 1/2b), L2625 (serotype 1/2a), L2676 (serotype 1/2a), BCW_2360 (C1-387, serotype 1/2a), BCW_2366 (J2-064, serotype 1/2b), BCW_2410 (N1-011, serotype 1/2b), BCW_2398 (J2-031, serotype 1/2a), BCW_2377 (J2-1091, serotype 4b), BCW_2358 (R2-502, serotype 1/2b), BCW_2994 (J1926, serotype 4B), L2074 (serotype 1/2a), and L2626 (serotype 1/2a).
FIG 3.
MAUVE alignments of L. monocytogenes large-scale inversion junctions. The gene contents of large-scale inversion junctions are illustrated. Due to the large inversion sizes, breaks in the inversion are displayed as black zigzag lines to highlight junction regions. rRNA genes are represented as red arrows, tRNA genes are represented as gray arrows, and all other genes are represented as blue arrows. The inversion region is highlighted by the blue blocks, with inversion genome coordinates listed at the ends of each block. Additional metadata for all strains are provided in Tables S1 and S2 in the supplemental material.
Whole-genome sequence alignments of the isolates against one another corroborated previous reports of genomic synteny within L. monocytogenes for some of the isolates (see Fig. S3 in the supplemental material). Gene order within inversion regions was also conserved. MAUVE alignments of all genomes grouped L. monocytogenes genomes into six locally colinear blocks (LCBs). Inversion-containing isolates L2626, L2074, and BCW_2994 displayed identical arrangements of all LCBs (Fig. 2). BCW_2358 (R2-502) and BCW_2377 (J2-1091) both exhibited displacement and inclusion of LCB 4 in the inverted region. LCB 5 was also displaced and inverted in BCW_2377 (J2-1091).
Methylomes are highly variable among isolates.
Using the kinetic information acquired from SMRT sequencing, it was possible to determine the specificities of active methyltransferases in bacteria (32). Epigenomes of the sequenced isolates were determined by modification type and sequence motif to observe near saturation of all motifs except one (Table 3). Six different N6-methyladenines were found, five of which were previously undescribed in L. monocytogenes, and these encompassed type I, type II, and one undetermined methylation system. Eleven isolates exhibited one active methyltransferase, one isolate showed two, and three isolates unexpectedly lacked methylation (Table 4).
TABLE 3.
Epigenomes of 15 L. monocytogenes isolates
| Methyltransferase specificitya | Modified base | Previously known | % of genomic position methylated in serogroupb: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1/2a |
1/2b |
4b |
|||||||||||||||
| 2360 (C1-387) | 2398 (J2-031) | L1846 | L2074 | L2625 | L2626 | L2676 | 2358 (R2-502) | 2366 (J2-064) | 2410 (N1-011A) | L2624 | 2377 (J2-1091) | 2992 (J1776) | 2993 (J1817) | 2994 (J1926) | |||
| 5′-GATC-3′ | m6A | Yes | 99.5 | 98.9 | |||||||||||||
| 3′-CTAG-5′ | |||||||||||||||||
| 5′-GACN5GGT-3′ | m6A | No | 98.9 | ||||||||||||||
| 3′-CTGN5CCA-5′ | 98.9 | ||||||||||||||||
| 5′-GAN6TGCG-3′ | m6A | No | 99.7 | 100 | 99.8 | ||||||||||||
| 3′-CTN6ACGC-5′ | 99.6 | 99.8 | 99.9 | ||||||||||||||
| 5′-TACBN6GTNG-3′ | m6A | No | |||||||||||||||
| 3′-ATGVN6CANC-5′ | |||||||||||||||||
| 5′-TAGRAG-3′ | m6A | No | 99.3 | ||||||||||||||
| 3′-ATCYTC-5′ | |||||||||||||||||
| 5′-GTATCC-3′ | m6A | No | 99.6 | 99.1 | 99.2 | ||||||||||||
| 3′-CATAGG-5′ | 99.7 | 98.8 | 98.0 | ||||||||||||||
| 5′-GATC-3′ | X | No | 12.2 | 15.8 | 8.4 | ||||||||||||
| 3′-CTAG-5′ | |||||||||||||||||
Methyltransferase specificities in bold were previously undescribed in L. monocytogenes.
Percentage of genomic positions detected as methylated by SMRT sequencing analysis. Blank fields indicate methylation detection signals of <1%.
TABLE 4.
DNA modifications, methyltransferase specificities, and predicted methyltransferases
| Isolate | Serotype | Clonal complex | Modified basea | Methyltransferase specificityb | Predicted RMS | REBASE |
|---|---|---|---|---|---|---|
| BCW_2398 (J2-031) | 1/2a | Type I, premature stop in specificity subunit | No | |||
| BCW_2360 (C1-387) | 1/2a | No, mrr-like type IV restriction endonuclease present | Yes | |||
| L1846 | 1/2a | 7 | m6A | 5′-GM6AN6TGCG-3′ | Type I | Yes |
| L2074 | 1/2a | X | 5′-GAXTC-3′ | Type II | Yes | |
| L2625 | 1/2a | X | 5′-GAXTC-3′ | Type II | Yes | |
| L2626 | 1/2a | 7 | m6A | 5′-GM6AN6TGCG-3′ | Type I | Yes |
| L2676 | 1/2a | 7 | m6A | 5′-GM6AN6TGCG-3′ | Type I | Yes |
| L2624 | 1/2b | 5 | m6A | 5′-Gm6ACN5GGT-3′ | Type I | Yes |
| BCW_2410 (N1-011A) | 1/2b | 3 | m6A | 5′-Gm6ATC-3′ | Type II | Yes |
| BCW_2410 (N1-011A) | 1/2b | 3 | m6A | 5′-TAGRm6AG-3′ | Type III | Yes |
| BCW_2366 (J2-064) | 1/2b | 5 | Type I | No | ||
| BCW_2358 (R2-502) | 1/2b | 3 | m6A | 5′-Gm6ATC-3′ | No RMS detected | Yes |
| BCW_2377 (J2-1091) | 4b | 1 | X | 5′-GAXTC-3′ | Type II; restriction enzyme Sau3AIc | Yes |
| BCW_2992 (J1776) | 4b | 6 | m6A | 5′-GTM6ATCC-3′ | No, DNA adenine methylase (dam) most likely candidate | Yes |
| BCW_2993 (J1817) | 4b | 6 | m6A | 5′-GTM6ATCC-3′ | No, DNA adenine methylase (dam) most likely candidate | Yes |
| BCW_2994 (J1926) | 4b | 6 | m6A | 5′-GTM6ATCC-3′ | No, DNA adenine methylase (dam) most likely candidate | Yes |
X, no previously observed modification signals for thymidine base methylation.
Methylated bases are in bold, and methylation on the opposite DNA strand is represented by underlining.
Cut site matches methyltransferase specificity: 5′…↓GATC…3′/3′…CTAG↑…5′.
Even within the same serotype, active methyltransferases varied markedly; the epigenome of every strain in serotype 1/2b was unique. Isolates from the same serotype (1/2a) and monophyletic cluster within lineage II (L1846, L2626, and L2676) exhibited the same methylation pattern and sequence motif, 5′-Gm6AN6TGCG-3′ (the methylated base is in bold, and methylation on the opposite DNA strand underlined). This observation holds true for isolates of the same 2002 U.S. multistate outbreak (BCW_2992 [J1776], BCW_2993 [J1817], and BCW_2994 [J1926]) in regard to the 5′-GTm6ATCC-3′ motif.
These closed genomes allowed a comprehensive analysis of predicted methyltransferase genes responsible for the observed methylation patterns (http://rebase.neb.com/rebase/private/pacbio_Weimer24.html). In many cases, the observed methylation could be assigned to the corresponding methyltransferase gene (Table 4). Two interesting type I systems were observed in one of the isolates (L2624), having two distinct target recognition domains present in conjunction with one methyltransferase. This likely represents an example where internal inversions permit different methylation specificities to be expressed (20). Methylation at the sequence context 5′-GTATCC-3′ had been previously observed in three strains (1816, H7550-Cds, and H7858), but the specific base methylation was previously undetermined. The SMRT sequencing data allowed confident assignment of methylation as 5′-GTm6ATCC-3′ in these isolates.
In addition to the observed methylation signals, three L. monocytogenes strains (BCW_2377, L2074, and L2625) displayed kinetic signals not explained by previously observed signals for DNA base modification. In these strains, the T residues of 8 to 16% of genomic locations with the 5′-GATC-3′ motif exhibited kinetic signals (Tables 3 and 4). In some cases, only one strand of the DNA displayed a modification signal; in other locations, both DNA strands within the 5′-GATC-3′ motif exhibited modification. While some of these strains contain a predicted 5-methylcytosine methyltransferase (m5C), the signal levels were not consistent with previously observed m5C methylation (33). We hypothesized potential phosphorothioation, but no evidence for dnd genes was found (34). These novel observations are yet to be linked to a functional role, but they were not uniquely associated with a single clonal complex, lineage, multilocus sequence type (MLST), serotype, or virulence.
O-antigen and teichoic acid biosynthesis genes and virulence genes display strain-specific methylation patterns.
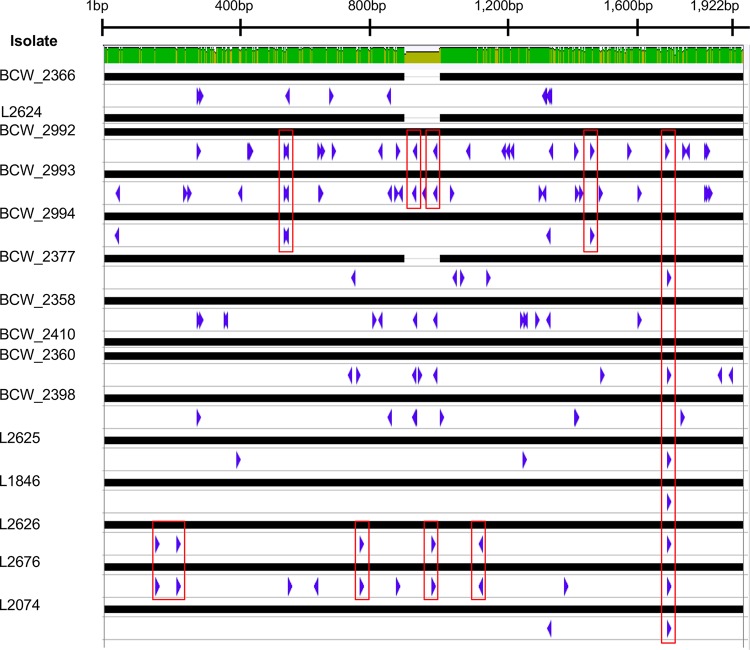
We resolved the methylation statuses of genes encoding enzymes involved in L. monocytogenes O-antigen and teichoic acid biosynthesis pathways to further examine the influence of DNA methylation on traditional L. monocytogenes categorization methods. This analysis encompassed all known methylation motifs. The methylation frequency of each gene varied from isolate to isolate independent of serotype, clonal complex, promoter region methylation (150 bp upstream of the start site), and strandedness (see Tables S4 and S5 in the supplemental material). Examination of serotype-determining genes revealed a marked difference between serotype 4b and serotypes 1/2a and 1/2b (Table S4). Methylation patterns of established virulence factors in L. monocytogenes, i.e., inlA, inlB, and virulence genes within LIPI-1 (prfA, plcA, hly, mpl, actA, plcB), were determined (Table S5). Similar to the results from methylation analysis of the O-antigen and teichoic acid synthesis genes, the virulence genes exhibited a wide range of specific methylation patterns that was not associated with clonal complex or serotype. Methylation analysis across each virulence gene did reveal patterns of methylation events between specific strains (see Fig. S4 in the supplemental material). Within actA, L2626 and L2676 displayed similar methylation patterns, while BCW_2992 (J1776), BCW_2993 (J1817), and BCW_2994 (J1926) shared a unique set of methylation patterns (Fig. 4).
FIG 4.
actA methylation patterns across all strains. Methylation patterns on actA are displayed as blue arrows. The direction of the arrow indicates the strandedness of the methylation event. Red boxes indicate methylation patterns shared by specific strains.
We also examined genes encoding established virulence factors important for host cell invasion by L. monocytogenes (inlA and inlB) and genes contained in the LIPI-1 pathogenicity island for mutations. In addition to methylation differences, this analysis revealed that actA and plcB contained indels in some isolates. In particular, previously described deletions of a proline-rich repeat (35) were detected in the actA genes of BCW_2366 (J2-064), L2624, and BCW_2377 (J2-1091); these deletions did not correlate with invasion efficiency (Fig. 4; see Fig. S2 and Table S6 in the supplemental material), Despite gene size conservation, single-nucleotide polymorphism (SNP) analysis of all virulence genes revealed serotype-specific SNP patterns (data not shown). Serotype-specific SNPs were present in actA (see Fig. S5 in the supplemental material), but differences in virulence gene methylation profiles could not be attributed to SNP location, as methylation events were detected regardless of SNP presence (Fig. 4).
L. monocytogenes host association capabilities are isolate specific and independent of serotype and clonal complex.
In vitro host adhesion and invasion assays using outbreak-associated L. monocytogenes isolates revealed extensive variation in host association phenotypes (Fig. S2). Neither serotype, clonal complex, nor genome features significantly correlated with host association (P > 0.05). While serotype was not significantly associated with invasion (P > 0.05), isolates BCW_2992 (J1776), BCW_2993 (J1817), BCW_2994 (J1926), and BCW_2377 (J2-1091), all belonging to serotype 4b and were among the least invasive isolates in this study. Conversely, isolate L2074, belonging to serotype 1/2a, displayed maximum invasiveness yet possessed the same unknown methylation pattern as the less invasive isolate BCW_2377. Initially, the clonal complex and serotype of each isolate were hypothesized to correlate with in vitro invasiveness; however, isolates of the same clonal complex were found to significantly differ in invasiveness (P ≤ 0.01). L1846 was isolated from a 2010 hog head cheese outbreak, and L2074 was isolated from a 2010 celery outbreak (Table S1). Both isolates originated from outbreaks occurring in the same year and belong to serotype 1/2a, yet they displayed significantly different invasion amounts (P ≤ 0.001) (Fig. S2).
DISCUSSION
Comparative genomics of 318 L. monocytogenes isolates reveals RMS motif-dependent clades.
L. monocytogenes is a foodborne pathogen with an increasing presence in current food recalls due to its ability to survive harsh environmental conditions. Previous whole-genome comparisons of L. monocytogenes characterized this pathogen as having a highly conserved and syntenic genome, an observation which was confirmed in our study. While genome distance-based phylogenetic comparison of 302 draft and finished L. monocytogenes genomes categorized the isolates into three lineages, the clades formed within each lineage could not be attributed to isolate source, outbreak, geographical location, or year of isolation. In spite of the discriminatory power of NGS to identify new genomic variants (36), the genotypes of these isolates were not linked to any of the specific phenotypic measures used in this study (Fig. 3 and 4; see Fig. S2 in the supplemental material). This observation is corroborated by work from Weimer et al. (23) and Weis et al. (64), highlighting the need for large numbers of genomes to link the genotype to the phenotype, especially in more slowly evolving genomes such as Listeria.
Interestingly, the majority of methyltransferase motif specificity was clade specific, with the exception of the GATC motif, which was scattered throughout isolates across all three lineages (Fig. 1). The lack of influence of isolation time and geography on clade organization was surprising, as SNPs and structural alterations to the genome are expected to accumulate as time progresses, particularly under stress-inducing conditions. RMS are traditionally considered to be the bacterial version of a defense mechanism against phage infection, with little consideration as to how they may be influencing genome evolution. Further research is needed to elucidate the role of RMS in the maintenance of genome conservation.
Large structural variations in L. monocytogenes.
In accordance with previous sequencing efforts, SMRT sequencing of outbreak-associated L. monocytogenes isolates revealed some genomic synteny among isolates. Interestingly, chromosome rearrangements were detected in the form of inversions in five isolates BCW_2358 (R2-502), BCW_2377 (J2-1091), BCW_2994 (J1926), L2074, and L2625 (Fig. 2). Except for one isolate, all inversions took place within regions of RNA clusters encompassing tRNA- and rRNA-encoding genes (Fig. 3). These inversions likely occurred as a result of homologous recombination events at the inversion junctions, since they occurred within multiple tRNA and rRNA sites. It is unlikely that these inversions occurred as a result of RMS activity due to the lack of methylation motifs detected at inversion junctions. These observations indicate that further sequencing of L. monocytogenes will reveal additional genome rearrangements that will expand the pan-genome.
Evolutionarily, genomes are organized for maximum fitness of the organism. If an inversion is tolerated, extraneous regions of the genome may reorganize to reestablish maximize overall fitness. Large-scale bacterial chromosome inversions alter cell physiologies such as growth rate and the expression of cell wall proteins (37). While rRNA-associated inversions have been documented in E. coli (38), this paper reports rRNA-associated inversions in L. monocytogenes; the effects of such rearrangements remain to be delineated. Inversions involving rRNA operons may cause a decrease in cell fitness due to a collision of the replisome with the transcription complex during growth (39). These inversions did not appear to influence the isolates' host association capacity, as no significant differences in invasiveness were observed between inversion-positive isolates and other isolates of the same outbreak.
Large chromosomal inversions, such as those detected in our isolates, often include the origin of replication or the terminus region and occur as a result of tandem repeats or duplications (40). Eisen et al. (40) described these inversions as instances of evolution when genomes were compared across different genera within the same phylogenetic class. In addition to rRNA gene involvement, inversions in BCW_2358 (R2-502), BCW_2377 (J2-1091), BCW_2994 (J1926), L2074, and L2625 encompassed the terminus region of the chromosome, likely as a result of homologous recombination due to complementary sequences flanking the inversion sites.
SMRT sequencing reveals clade-specific methylation patterns that can be attributed to specific methyltransferases/RMS.
Genome distance matrices of our isolates revealed the delineation of our isolates into the three lineages commonly associated with listeriosis outbreaks (Fig. 1). RMS were shared between isolates of the same phylogenetic clade, suggesting adaptation of the RMS to the genome in which it resides. The cooccurrence of a particular RMS with a specific genome may arise from toxin/antitoxin interactions between the RMS and phage elements present on the chromosome (41).
Three L. monocytogenes isolates displayed an unexplained kinetic signal that did not correlate with any known DNA modification-associated kinetic signals detected by SMRT sequencing (33, 42). We hypothesized that this kinetic signal represents a novel DNA modification mechanism that is unique from RMS and phosphorothioation. The only RMS identified using REBASE prediction algorithms was a type II RMS involving the Sau3AI restriction enzyme, which recognizes the same target sequence as that associated with the unexplained kinetic signal. Resistance to Sau3AI digestion has been described for many epidemic-associated L. monocytogenes isolates, although the significance of this correlation has yet to be elucidated (29, 30). While the Sau3AI restriction endonuclease and the unidentified kinetic signal are both associated with the same motif (GATC), the kinetic signal for the Sau3AI recognition site does not correspond with that of the unknown modification. Furthermore, the Sau3AI modification occurs on cytosine and the base modification of the unidentified signal on thymine. The possibility of the unidentified kinetic signal representing the cytosine methylation is slim, although studies are under way to characterize the chemical properties of this modification.
Presence of L. monocytogenes O-antigen and teichoic acid synthesis genes is serotype specific yet displays isolate-specific methylation patterns.
Analysis of SMRT sequencing results revealed extensive methylation of many O-antigen and teichoic acid biosynthesis genes with isolate-specific methylation depth and strandedness (see Table S4 in the supplemental material). Examination of the methylation patterns of genes involved in O-antigen and teichoic acid biosynthesis uncovered no correlation between gene-specific methylation patterns and the corresponding serotype. While such extensive and diverse modifications likely resulted from an active RM system, populations of these modifications were located in the promoter region (150 bp upstream of the start site) of each gene (Table S4), indicating a potential role in transcriptional regulation.
Despite the discrepancy in methylation patterns, differences in gene content between the serotype 4b isolates and serotypes 1/2a and 1/2b were apparent. Serotype 4b isolates lacked O-antigen biosynthesis enzymes, glucose-1-phosphate thymidylyltransferase (EC 2.7.7.24), dTDP–d-glucose 4,6-dehydratase (EC 4.2.1.46), and dTDP-4-dehydrorhamnose 3,5-epimerase (EC 5.1.3.13). Additionally, serotypes 1/2a and 1/2b lacked glycerophosphotransferase (EC 2.7.8.12), the enzyme involved in teichoic acid biosynthesis. These findings are in accordance with previous reports of serotype-specific representation of L. monocytogenes O-antigen-associated genes (7, 43). Although differences in sugar metabolism between isolates are not expected to affect L. monocytogenes host colonization abilities (43), the consequences of these missing genes for O-antigen structure, as well as recognition by the host, remain unknown. Glycan structures on the Listeria cell surface not only act as serotype determinants but also modulate host association (44). Although it is beyond the scope of this study to elucidate the effects of gene copy number and gene-specific methylation patterns on the expression and activity of O-antigen and teichoic acid synthesis genes, the diversity uncovered in our analysis of these genes offers potential insights into the influence of serotype-associated genes on L. monocytogenes virulence mechanisms.
L. monocytogenes invasiveness of Caco-2 cells is predicated by neither serotype-dependent SNPs nor strain-specific methylation patterns.
Methylation of the L. monocytogenes pathogenicity island LIPI-1 (prfA, plcA, hly, mpl, actA, and plcB), as well as host entry factors inlA and inlB, was analyzed (45). Similar to the case for the O-antigen and teichoic acid biosynthesis genes, methylation of neither the virulence genes nor their promoter regions was associated with L. monocytogenes invasion capabilities. All isolates contained full-length virulence genes, with the exception of truncations detected in actA and plcB. These truncations did not appear to impact in vitro invasiveness, likely because these two virulence factors are specifically involved in cell to cell spread and not initial host entry (45) (see Fig. S2 and Table S5 in the supplemental material). Additionally, invasiveness could not be attributed to the presence of serotype-dependent SNPs within L. monocytogenes virulence genes (see Fig. S5 in the supplemental material). The observed shared methylation patterns between specific strains are likely a result of shared methyltransferase specificities (Fig. S4; Table 4). Of the strains in this study, L2626 and L2676 share predicted type I RMS with identical methyltransferase specificities. This is also observed in BCW_2992 (J1776), BCW_2993 (J1817), and BCW_2994 (J1926), where these strains share a methyltransferase specificity that is unique to these three strains (Table 4).
Virulence and invasiveness are not specific to serotype or clonal complex in L. monocytogenes.
L. monocytogenes is a foodborne pathogen that is highly adaptable to diverse environments and survives many conditions for long periods in food and the environment (3). In addition to adaptability, L. monocytogenes displays diverse host association capabilities within a single-host model (Fig. S2) with invasiveness that is unrelated to serotype, clonal complex, and temporal influences, a finding supported by a study by Roberts et al. (46). Neither clonal complex, time of isolation, nor serotype can be used as a predictive determinant of in vitro invasiveness even when isolates belong to the same serotype or clonal complex, confirming that previously assigned classification phenotypes are not predictors of invasiveness. As an organism under high purifying selection in systems of food production (47), L. monocytogenes relies on the presence of phage elements to introduce phenotypic variability, allowing for rapid adaptation to different environments (48). Genome sequencing and subsequent analyses revealed unexpected differences in genome structure and diverse and unique methyltransferase motifs and methylome patterns. While this is the largest genome comparison of L. monocytogenes with draft and closed genomes, it is clear that additional sequencing will likely find more isolates with divergent genomes.
Conclusion.
SMRT sequencing of 15 L. monocytogenes isolates from outbreaks spanning 2 decades revealed conservation of the core genome but an unexpectedly diverse array of structural variations that give rise to phenotypic multiplicity. Though L. monocytogenes genomes were observed to be syntenic, advanced NGS technologies are exposing an additional diversity of methylation. Each of these genomic variations has the potential to alter the transcriptional profiles of their respective genomes, rendering the reliance of outbreak surveillance on clonal complex designations an outdated practice. Many more factors are involved in governing the expression of persistence and virulence genes than just the presence of the gene itself. A greater understanding of the implications of genome attributes for pathogen persistence will contribute to the rapidly evolving landscape of food safety surveillance, facilitating faster outbreak management and potentiating microbe- and isolate-specific personalized treatment, allowing the bypass of broad-spectrum antibiotics and thereby minimizing the spread of antibiotic-resistant pathogens throughout the food industry and the community. The increasing availability of closed genomes and the corresponding epigenomes in the public domain continues to be a tremendous contribution toward the improvement of foodborne pathogen outbreak management.
MATERIALS AND METHODS
Bacterial strains.
L. monocytogenes cultures were grown overnight in brain heart infusion (BHI) broth (BD, Franklin Lakes, NJ) for DNA extraction (see Table S1 in the supplemental material). Approximately 1010 cells in liquid culture were centrifuged at 16,000 × g for 5 min. The pellets were collected and stored in a −80°C freezer for DNA extraction.
Infectivity assay.
The in vitro infectivity of each L. monocytogenes strain was determined in cultured intestinal epithelial monolayers (Caco-2, ATCC HTB-037) using a gentamicin protection assay as previously described (49) and modified by Shah et al. (50, 51). Briefly, Caco-2 monolayers were infected with L. monocytogenes (n = 3, multiplicity of infection [MOI] = 100) for 60 min, followed by 2 h of incubation in cell culture medium supplemented with 1 mg/ml gentamicin (Sigma-Aldrich, St. Louis, MO). Samples were lysed with Warnex lysis buffer (AES Chemunex Canada, Inc.) following the manufacturer's instructions. Quantification was done using quantitative PCR (qPCR), with results reported as L. monocytogenes CFU/Caco-2 cell. All experiments were conducted in biological triplicate; results were graphed using GraphPad InStat 3 (GraphPad Software, Inc., La Jolla, CA), and statistical analysis was done using analysis of variance (ANOVA) in conjunction with Tukey's test on JMP version 10 (SAS Institute, Triangle Park, NC). Data are displayed as the mean ± standard error of the mean (SEM). A P value of <0.05 was considered to be statistically significant.
DNA extraction.
L. monocytogenes DNA was extracted as described by Kong et al. (52) Briefly, L. monocytogenes cell pellets were resuspended in 1 ml of lysis buffer (500 mM NaCl, 50 mM Tris-HCl, 50 mM EDTA, 1% SDS, pH 8.0) and 0.2 g of 0.1-mm glass beads. Bacterial cells were lysed by bead beating and then incubated at 70°C for 15 min. The lysates were centrifuged at 4°C for 5 min at 16,000 × g and the supernatant transferred to a new 2-ml tube. Ammonium acetate was added to the supernatant to achieve a final concentration of 0.1 M. Samples were incubated on ice for 5 min and centrifuged at 4°C for 10 min at 16,000 × g. The supernatant was transferred to a new tube with a final concentration of 50% isopropanol. Samples were mixed by gentle inversion and incubated on ice for 30 min, followed by centrifugation at 4°C for 15 min at 16,000 × g. The supernatant was discarded, follow by washing with 70% ethanol, and the pellet was left to air dry. Sample pellets were dissolved in 200 μl Tris-EDTA buffer. Genomic DNA (gDNA) contaminants were removed by using QIAamp DNA stool minikit 51304 (Qiagen, Valencia, CA) per the manufacturer's recommendations. Quality control was done as described by the 100K Pathogen Genome Project and included measures of protein and organic contamination (A260/A280 and A260/A230 ratios of ≥1.8 to 2.0, respectively), as well as intact gDNA (52).
Library preparation and SMRT sequencing of 15 L. monocytogenes isolates.
Genomic DNA was sheared to fragments of ∼10 to 15 kb using Covaris g-TUBEs (Woburn, MA). SMRTbell sequencing libraries were made according to the manufacturer's instructions (Pacific Biosciences, Menlo Park, CA). Briefly, fragmented DNA underwent DNA damage repair, end repair, and ligation to hairpin adapters. SMRTbell libraries were sequenced on a PacBio RS using the recommended protocol for large insert libraries (C2 sequencing chemistry, magnetic bead loading, stage start enabled, and 2-hour acquisitions). Sequence was obtained from 4 to 8 SMRT cells per genome, resulting in an average depth of coverage of approximately 150-fold. Genomes were assembled de novo using the hierarchical genome assembly process (HGAP), and sequences were polished using Quiver to obtain a final consensus assembly (53). Both modules are part of the SMRT Analysis package (version 1.3). Methylation detection was carried out using the base modification analysis protocol “RS_Modification_and_Motif_Analysis.1” as part of the SMRT Analysis software package (Pacific Biosciences, Menlo Park, CA), and methylation signals were visualized using SMRT View. Methyltransferase prediction was carried out using SEQWARE (54).
Genome annotation, distance matrix calculation, and RMS gene presence/absence analyses.
All draft genomes, including those publicly available, were assembled and annotated as a group. Briefly, assembly of paired-end reads was done using ABySS 1.5.2 (55) and annotated using Prokka (56) before comparative genomic analyses were done. Genomic distances were determined using the Web server Genome-to-Genome Distance Calculator (GGDC) (http://ggdc.dsmz.de/distcalc2.php) as published previously (57, 58). Dendroscope was used to generate a maximum-likelihood tree based on the genome distance matrix (60). REBASE was used to infer RMS gene presence/absence for each isolate and to identify methylation motifs (31). Mobile element analysis was completed with PHAST (http://phast.wishartlab.com/) (61).
Mauve alignment.
Closed genomes were aligned using Mauve 2.3.1 with default settings (62).
MUMmer and NUCmer analysis.
Whole-genome nucleotide and protein alignments were completed in all pairwise combinations using MUMmer 3.0 (63) with default settings.
Accession number(s).
The sequences obtained from SMRT sequencing were deposited in GenBank with accession numbers NC_021837.1, NC_021823.1, NZ_CP007688.1, NZ_CP007689.1, NZ_CP007687.1, NZ_CP007684.1, NZ_CP007685.1, NZ_CP007686.1, NC_021826.1, NC_021824.1, NC_021838.1, NC_021839.1, NC_021827.1, and NC_021840.1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cheryl L. Tarr and the team at the Enteric Diseases Laboratory Branch at the Centers for Disease Control and Prevention for their generous contribution of bacterial isolates to this study as well as invaluable input into the preparation of the manuscript. We thank Susana Wang and Yan Guo for library preparation and sequencing. We also thank Richard Roberts for methyltransferase prediction analysis and deposition into REBASE.
Funding was provided to B.C.W and the 100K Pathogen Genome Project (FDA grant no. 5U01FD003572-04).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02091-16.
REFERENCES
- 1.Hernandez-Milian A, Payeras-Cifre A. 2014. What is new in listeriosis? Biomed Res Int 2014:358051. doi: 10.1155/2014/358051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cossart P, Lebreton A. 2014. A trip in the “New Microbiology” with the bacterial pathogen Listeria monocytogenes. FEBS Lett 588:2437–2445. doi: 10.1016/j.febslet.2014.05.051. [DOI] [PubMed] [Google Scholar]
- 3.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect 16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 5.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couve E, de Daruvar A, Dehoux P, Domann E, Dominguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, Garcia-del Portillo F, Garrido P, Gautier L, Goebel W, Gomez-Lopez N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Perez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, et al. . 2001. Comparative genomics of Listeria species. Science 294:849–852. [DOI] [PubMed] [Google Scholar]
- 6.Kuenne C, Voget S, Pischimarov J, Oehm S, Goesmann A, Daniel R, Hain T, Chakraborty T. 2010. Comparative analysis of plasmids in the genus Listeria. PLoS One 5:e12511. doi: 10.1371/journal.pone.0012511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.den Bakker HC, Desjardins CA, Griggs AD, Peters JE, Zeng Q, Young SK, Kodira CD, Yandava C, Hepburn TA, Haas BJ, Birren BW, Wiedmann M. 2013. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS One 8:e67511. doi: 10.1371/journal.pone.0067511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. 2010. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics 11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenz LL, Portnoy DA. 2002. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol Microbiol 45:1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 10.Lindback T, Secic I, Rorvik LM. 2011. A contingency locus in prfA in a Listeria monocytogenes subgroup allows reactivation of the PrfA virulence regulator during infection in mice. Appl Environ Microbiol 77:3478–3483. doi: 10.1128/AEM.02708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, Le Monnier A, Brisse S. 2008. A new perspective on Listeria monocytogenes evolution. PLoS Pathog 4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JH, Karamychev VN, Kozyavkin SA, Mills D, Pavlov AR, Pavlova NV, Polouchine NN, Richardson PM, Shakhova VV, Slesarev AI, Weimer B, O'Sullivan DJ. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. doi: 10.1186/1471-2164-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klaenhammer T, Altermann E, Arigoni F, Bolotin A, Breidt F, Broadbent J, Cano R, Chaillou S, Deutscher J, Gasson M, van de Guchte M, Guzzo J, Hartke A, Hawkins T, Hols P, Hutkins R, Kleerebezem M, Kok J, Kuipers O, Lubbers M, Maguin E, McKay L, Mills D, Nauta A, Overbeek R, Pel H, Pridmore D, Saier M, van Sinderen D, Sorokin A, Steele J, O'Sullivan D, de Vos W, Weimer B, Zagorec M, Siezen R. 2002. Discovering lactic acid bacteria by genomics. Antonie Van Leeuwenhoek 82:29–58. doi: 10.1023/A:1020638309912. [DOI] [PubMed] [Google Scholar]
- 15.Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, Grimmond SM, Smith AL, Barenkamp SJ, Korlach J, Bakaletz LO, Jennings MP. 2015. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun 6:7828. doi: 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC, Jennings MP, Oggioni MR. 2014. A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5:5055. doi: 10.1038/ncomms6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blakeway LV, Power PM, Jen FE, Worboys SR, Boitano M, Clark TA, Korlach J, Bakaletz LO, Jennings MP, Peak IR, Seib KL. 2014. ModM DNA methyltransferase methylome analysis reveals a potential role for Moraxella catarrhalis phasevarions in otitis media. FASEB J 28:5197–5207. doi: 10.1096/fj.14-256578. [DOI] [PubMed] [Google Scholar]
- 18.Le Bourgeois P, Lautier M, van den Berghe L, Gasson MJ, Ritzenthaler P. 1995. Physical and genetic map of the Lactococcus lactis subsp. cremoris MG1363 chromosome: comparison with that of Lactococcus lactis subsp. lactis IL 1403 reveals a large genome inversion. J Bacteriol 177:2840–2850. doi: 10.1128/jb.177.10.2840-2850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guinane CM, Kent RM, Norberg S, Hill C, Fitzgerald GF, Stanton C, Ross RP. 2011. Host specific diversity in Lactobacillus johnsonii as evidenced by a major chromosomal inversion and phage resistance mechanisms. PLoS One 6:e18740. doi: 10.1371/journal.pone.0018740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dybvig K, Sitaraman R, French CT. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc Natl Acad Sci U S A 95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Low DA, Casadesus J. 2008. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr Opin Microbiol 11:106–112. doi: 10.1016/j.mib.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez D, Kozdon JB, McAdams HH, Shapiro L, Collier J. 2014. The functions of DNA methylation by CcrM in Caulobacter crescentus: a global approach. Nucleic Acids Res doi: 10.1093/nar/gkt1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weimer BC, Storey DB, Elkins CA, Baker RC, Markwell P, Chambliss D, Edlund S, Kaufman JH. 2016. Defining the food microbiome for authentication, safety, and process management. IBM J Res Dev doi: 10.1147/JRD.2016.2582598. [DOI] [Google Scholar]
- 24.Chen P, Jeannotte R, Weimer BC. 2014. Exploring bacterial epigenomics in the next-generation sequencing era: a new approach for an emerging frontier. Trends Microbiol 22:292–300. doi: 10.1016/j.tim.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Casadesus J, Low D. 2006. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev 70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis BM, Chao MC, Waldor MK. 2013. Entering the era of bacterial epigenomics with single molecule real time DNA sequencing. Curr Opin Microbiol 16:192–198. doi: 10.1016/j.mib.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta Y, Abe K, Kobayashi I. 2010. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res 38:2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. doi: 10.1186/1471-2164-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng W, Kathariou S. 1997. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl Environ Microbiol 63:3085–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yildirim S, Elhanafi D, Lin W, Hitchins AD, Siletzky RM, Kathariou S. 2010. Conservation of genomic localization and sequence content of Sau3AI-like restriction-modification gene cassettes among Listeria monocytogenes epidemic clone I and selected strains of serotype 1/2a. Appl Environ Microbiol 76:5577–5584. doi: 10.1128/AEM.00648-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loenen WA, Dryden DT, Raleigh EA, Wilson GG, Murray NE. 2014. Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res 42:3–19. doi: 10.1093/nar/gkt990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Chen S, Vergin KL, Giovannoni SJ, Chan SW, DeMott MS, Taghizadeh K, Cordero OX, Cutler M, Timberlake S, Alm EJ, Polz MF, Pinhassi J, Deng Z, Dedon PC. 2011. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc Natl Acad Sci U S A 108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiedmann M, Bruce JL, Keating C, Johnson AE, McDonough PL, Batt CA. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect Immun 65:2707–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng X, Desai PT, den Bakker HC, Mikoleit M, Tolar B, Trees E, Hendriksen RS, Frye JG, Porwollik S, Weimer BC, Wiedmann M, Weinstock GM, Fields PI, McClelland M. 2014. Genomic epidemiology of Salmonella enterica serotype Enteritidis based on population structure of prevalent lineages. Emerg Infect Dis 20:1481–1489. doi: 10.3201/eid2009.131095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A 101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill CW, Harnish BW. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A 78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivatsan A, Tehranchi A, MacAlpine DM, Wang JD. 2010. Co-orientation of replication and transcription preserves genome integrity. PLoS Genet 6:e1000810. doi: 10.1371/journal.pgen.1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisen JA, Heidelberg JF, White O, Salzberg SL. 2000. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol 1:RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naito Y, Naito T, Kobayashi I. 1998. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol Chem 379:429–436. doi: 10.1515/bchm.1998.379.4-5.429. [DOI] [PubMed] [Google Scholar]
- 42.Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, Peterson J, White O, Nelson WC, Nierman W, Beanan MJ, Brinkac LM, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Haft DH, Selengut J, Van Aken S, Khouri H, Fedorova N, Forberger H, Tran B, Kathariou S, Wonderling LD, Uhlich GA, Bayles DO, Luchansky JB, Fraser CM. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res 32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schirm M, Kalmokoff M, Aubry A, Thibault P, Sandoz M, Logan SM. 2004. Flagellin from Listeria monocytogenes is glycosylated with beta-O-linked N-acetylglucosamine. J Bacteriol 186:6721–6727. doi: 10.1128/JB.186.20.6721-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. 2001. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev 14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts AJ, Williams SK, Wiedmann M, Nightingale KK. 2009. Some Listeria monocytogenes outbreak strains demonstrate significantly reduced invasion, inlA transcript levels, and swarming motility in vitro. Appl Environ Microbiol 75:5647–5658. doi: 10.1128/AEM.00367-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novichkov PS, Wolf YI, Dubchak I, Koonin EV. 2009. Trends in prokaryotic evolution revealed by comparison of closely related bacterial and archaeal genomes. J Bacteriol 191:65–73. doi: 10.1128/JB.01237-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verghese B, Lok M, Wen J, Alessandria V, Chen Y, Kathariou S, Knabel S. 2011. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl Environ Microbiol 77:3279–3292. doi: 10.1128/AEM.00546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elsinghorst EA. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol 236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 50.Shah J, Desai PT, Weimer BC. 2014. Genetic mechanisms underlying the pathogenicity of cold-stressed Salmonella enterica serovar Typhimurium in cultured intestinal epithelial cells. Appl Environ Microbiol 80:6943–6953. doi: 10.1128/AEM.01994-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shah J, Desai PT, Chen D, Stevens JR, Weimer BC. 2013. Preadaptation to cold stress in Salmonella enterica serovar Typhimurium increases survival during subsequent acid stress exposure. Appl Environ Microbiol 79:7281–7289. doi: 10.1128/AEM.02621-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong N, Ng W, Lee V, Kelly L, Weimer BC. 2013. Production and analysis of high molecular weight genomic DNA for NGS pipelines using Agilent DNA extraction kit (p/n 200600). Agilent Technologies Inc., Santa Clara, CA. [Google Scholar]
- 53.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 54.O'Connor BD, Merriman B, Nelson SF. 2010. SeqWare Query Engine: storing and searching sequence data in the cloud. BMC Bioinformatics 11(Suppl 12):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 57.Auch AF, von Jan M, Klenk HP, Goker M. 2010. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reference deleted.
- 60.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weis AM, Storey DB, Taff CC, Townsend AK, Huang BC, Kong NT, Clothier KA, Spinner A, Byrne BA, Weimer BC. 2016. Genomic comparison of Campylobacter spp. and their potential for zoonotic transmission between birds, primates, and livestock. Appl Environ Microbiol 82:7165–7175. doi: 10.1128/AEM.01746-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.