Abstract
In eukaryotes, formation of short duplexes between the U3 small nucleolar RNA (snoRNA) and the precursor rRNA (pre-rRNA) at multiple sites is a prerequisite for three endonucleolytic cleavages that initiate small subunit biogenesis by releasing the 18S rRNA precursor from the pre-rRNA. The most likely role of these RNA duplexes is to guide the U3 snoRNA and its associated proteins, designated the small subunit processome, to the target cleavage sites on the pre-rRNA. Studies by others in Saccharomyces cerevisiae have identified the proteins Mpp10p, Imp3p, and Imp4p as candidates to mediate U3–pre-rRNA interactions. We report here that Imp3p and Imp4p appear to stabilize an otherwise unstable duplex between the U3 snoRNA hinge region and complementary bases in the external transcribed spacer of the pre-rRNA. In addition, Imp4p, but not Imp3p, seems to rearrange the U3 box A stem structure to expose the site that base-pairs with the 5′ end of the 18S rRNA, thereby mediating duplex formation at a second site. By mediating formation of both essential U3–pre-rRNA duplexes, Imp3p and Imp4p may help the small subunit processome to dock onto the pre-rRNA, an event indispensable for ribosome biogenesis and hence for cell growth.
Keywords: ribosome biogenesis, U3 snoRNA, RNA-binding proteins, annealing
Eukaryotic ribosome biogenesis is a dynamic process involving the assembly and disassembly of RNA–protein and RNA–RNA complexes. Considering the temporal complexity of this process and its importance to cell growth (reviewed in refs. 1 and 2), cofactors are expected to regulate the formation and dissociation of key transient RNA–protein and RNA–RNA interactions. Even though RNA duplexes can form spontaneously, cells have evolved proteins to stimulate hybridization for a variety of reasons: the hybrid is unstable, the rate of duplex formation is too slow, or structural rearrangements in one or both RNA partners are necessary to unmask the site of hybridization. Identifying proteins with putative RNA annealing activity is challenging because, unlike RNA helicases, such proteins share neither distinctive sequence elements nor a common mechanism (such as the use of the ATP cofactor) by which their activity can be monitored. Our studies center on two transient RNA–RNA interactions needed for the endonucleolytic cleavages that release the 18S precursor from the precursor rRNA (pre-rRNA) and thereby initiate biogenesis of the ribosomal small subunit (SSU). The cleavages depend on the U3 small nucleolar (sno) RNA (snoRNA) and its 28 or more associated proteins, a large ribonucleoprotein (RNP) designated the small subunit processome (SSUP) (3).
Ribosome biogenesis begins with transcription of the prerRNA. The pre-rRNA contains the small subunit rRNA (18S) and two of the three large subunit rRNAs (the 5.8S and the 25S) that are separated by two internal transcribed spacers (ITS1 and ITS2) and flanked by two external transcribed spacers (5′ ETS and 3′ ETS) (Fig. 1). A series of highly coordinated events is necessary to produce mature ribosomal subunits: the pre-rRNA has to be modified (by pseudouridylation and methylation), cleaved (by exo- and endonucleases), folded, and assembled with at least 79 ribosomal proteins (reviewed in ref. 4). After transcription of the pre-rRNA, the SSUP mediates the release of the 18S rRNA precursor by three endonucleolytic cleavages of the pre-rRNA at sites A0, A1, and A2 (3). These endonucleolytic cleavages require base-pairing interactions between the U3 snoRNA and the pre-rRNA (5–10). The expected role of these U3–pre-rRNA duplexes is to guide the SSUP and the as-yet-unidentified endoribonuclease(s) to the cleavage sites, just as duplex formation between the pre-rRNA and the snoRNA guides the pseudouridylation and methylation snoRNPs to their target sites of modification (1).
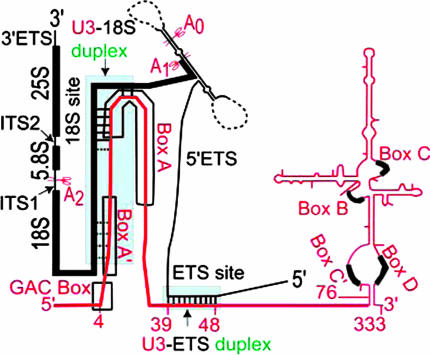
Fig. 1.
Schematic of the pre-rRNA and the U3 snoRNA [secondary structure drawing of the U3 snoRNA (red) base-paired to the pre-rRNA (black)]. Conserved sequence elements of the 5′ domain are boxed (GAC box and boxes A and A′). The secondary structure of the U3 snoRNA was derived from chemical protection experiments (37); missing regions of the secondary structure of the pre-rRNA fragment are dashed. Helix 1a (nucleotides 4–39) and helix 1b′ (nucleotides 47–63) of the U3 snoRNA are not shown because formation of the U3 snoRNA–pre-rRNA base-pairing interactions is expected to replace these structural elements (37, 38). At the duplex sites (green), genetically verified (solid lines) and expected (dotted lines) interactions are shown.
The pre-rRNA processing pathways and most of the genes identified to play a role in ribosome biogenesis in the yeast Saccharomyces cerevisiae have counterparts in higher eukaryotes. Moreover, the potential for forming multiple base-pairing sites between the pre-rRNA and the U3 snoRNA is conserved (11–13). Hence, S. cerevisiae is the model organism for our studies. The U3 snoRNA contains conserved sequence elements (designated the GAC box and boxes A, A′,B,C,C′, and D) and can be divided into three regions: the 5′ domain with the GAC box and boxes A and A′; the hinge region; and the 3′ domain with boxes B, C, C′, and D, which classifies the U3 snoRNA as a box C/D snoRNA (Fig. 1). Both U3–pre-rRNA duplexes occur in the U3 snoRNA 5′ domain and the hinge region (nucleotides 1–76): nucleotides 39–48 of the U3 snoRNA interact with the complementary site of the 5′ ETS of the pre-rRNA [designated the U3–ETS duplex (7)], and nucleotides 16–22 of the U3 snoRNA interact with the complementary site in the 18S portion of the pre-rRNA [designated the U3–18S duplex (9, 10)]. The base-pairing sites in the 5′ ETS and the 5′ terminus of the 18S portion of the pre-rRNA are designated the ETS site and the 18S site, respectively. By analogy with premRNA splicing (14), RNA editing (15), and viral RNA replication (16), proteins are expected to stimulate formation of these U3–pre-rRNA duplexes and thereby provide a site for controlling initiation of SSU biogenesis. For example, the abundant nucleolar protein nucleolin may help to recruit the SSUP to the pre-rRNA (17, 18), possibly via RNA annealing activities; however, only DNA annealing activity was shown (19).
Of the 28 SSUP proteins (3), Imp3p, Imp4p, and Mpp10p are promising candidates to mediate the U3–pre-rRNA interactions. Affinity purification, immunoprecipitation, and two-hybrid studies suggest that Mpp10p, Imp3p, and Imp4p form a heterotrimer in vivo (20–23). The studies imply that the heterotrimer interacts with the 5′ domain and hinge region of the U3 snoRNA, but they fail to distinguish between direct binding to the U3 snoRNA and protein-mediated interactions.
Our studies focus on Imp3p and Imp4p, both of which have RNA-binding modules and higher sequence conservation throughout the eukaryotic kingdom than Mpp10p (20, 23, 24). The central portion of Imp3p has sequence homology to an RNA-binding motif of ribosomal protein S4 that forms a winged helix (20, 25, 26). Imp4p is a member of the Imp4 superfamily of proteins, four of which (Rfp1p, Rfp2p, Ssf1/2p, and Brx1p) are involved in large-subunit biogenesis (27–31). Phylogenetic analyses and mutagenesis studies indicate that a σ70 motif (a putative helix–turn–helix structure) in the superfamily contributes to binding homopolymeric RNA sequences; however, the RNA-binding target and the function of these proteins in ribosome biogenesis were previously unknown (27).
To determine whether the candidate proteins (Imp3p and Imp4p) bind to RNA (the U3 snoRNA or the pre-rRNA), mediate formation of the essential U3-pre-rRNA hybrids, or both, we carried out in vitro studies. To overcome protein insolubility, we identified refolding conditions that produced Imp3p and Imp4p in a biologically functional state. Both proteins bind independently to the 5′ domain and the adjacent hinge region of the U3 snoRNA, whereas protein binding was not observed to fragments of the pre-rRNA and the 3′ domain of the U3 snoRNA. We discovered that either protein mediates formation of the U3–ETS duplex, apparently by increasing the stability of the otherwise unstable hybrid. In contrast, only Imp4p is shown to promote formation of the U3–18S duplex by rearranging the box A stem to expose the relevant nucleotides, thereby permitting hybridization. This protein-dependent U3–pre-rRNA hybridization may facilitate duplex formation between the 5′ domain and the 5′ hinge of the U3 snoRNA and the pre-rRNA in vivo, thereby recruiting the SSUP to its pre-rRNA substrate.
Methods
DNA template construction, RNA synthesis, nitrocellulose filter-binding assays, electromobility-shift assays, and RNase protections assays are in Supporting Methods, which is published as supporting information on the PNAS web site.
Cloning, Expression, and Purification of Recombinant Imp3p and Imp4p. Genes encoding the Imp3p and Imp4p proteins in S. cerevisiae were amplified from ORFs in plasmids provided by S. J. Baserga (Yale University, New Haven, CT) (20). Imp3p was cloned into pET21d (Novagen) by using NcoI and NotI restriction sites (New England Biolabs), and Imp4p was cloned into pET21a (Novagen) by using NdeI and NotI restriction sites (New England Biolabs). Proteins were expressed at 37°C for 4 h in Escherichia coli BL21(DE3) cells supplemented with a vector coding for the rare AGA and AGG tRNAArg codons. After cell breakage, inclusion bodies were solubilized in buffer A (20 mM Tris, pH 8.0/50 mM MgCl2/5 mM 2-mercaptoethanol) containing 6 M urea. Denatured proteins were loaded onto a CMSepharose resin (Amersham Pharmacia) in buffer B [50 mM 2-morpholinoethanesulfonate acid (Mes), pH 5.2/5 mM 2-mercaptoethanol/6 M urea] with 100 mM NaCl and recovered with gradient elution by using buffer B with 300–400 mM NaCl. After dilution, both proteins were renatured by overnight incubation in a refolding buffer at 4°C. Optimal refolding conditions were selected from the protein-folding screen (Hampton Research, Riverside, CA) (33), based on protein solubility and the ability to migrate as a single species on nondenaturing PAGE. Imp3p was diluted to ≈0.1 mg/ml in refolding buffer (55 mM Mes, pH 6.5/264 mM NaCl/11 mM KCl/550 mM guanidine·HCl) that contained 0.055% (wt/vol) polyethylene glycol 3350/2.2 mM MgCl2/2.2 mM CaCl2/440 mM sucrose. Imp4p was diluted to ≈0.1 mg/ml in refolding buffer that contained 1.1 mM EDTA/550 mM l-arginine. Before reverse-phase chromatography, the salt concentration of each protein solution was adjusted to be equivalent to that of buffer A containing 1 M urea/2 M NaCl. To remove improperly folded proteins and RNase contamination, each protein was loaded onto a butyl-Sepharose resin (Amersham Pharmacia) in buffer A containing 1 M urea/2 M NaCl and recovered with gradient elution by using buffer A containing 1 M urea/0.5 M NaCl. Imp3p was stored at 30 mg/ml in buffer A containing 0.08 mM Tween 20 (Sigma). Imp4p was stored at 16 mg/ml in 50 mM Mes, pH 6.5/50 mM MgCl2/5 mM 2-mercaptoethanol/8 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) (Calbiochem). The concentrations of Tween 20 and CHAPS were equal to the critical micelle concentration. Both proteins migrated as single bands on nondenaturing PAGE, consistent with the homogeneous conformation of a folded protein (Fig. 5, which is published as supporting information on the PNAS web site).
Cloning, Expression, and Purification of an Imp4p Fragment (Imp4-Core) and Point Mutants Thereof. Imp4-core (amino acids 87–275 of Imp4p) was amplified from an ORF coding for WT Imp4p and cloned into pProExHTa (Life Technologies, Gaithersburg, MD) by using EcoRI and HindIII restriction sites (New England Biolabs) (primer sequences are in Supporting Methods). This protein was expressed at 15°C for 16 h in E. coli BL21(DE3) cells supplemented with a vector coding for rare tRNAArg codons, as before (32). After cell breakage in 20 mM Tris, pH 8.0/200 mM NaCl/0.08 mM Tween 20, inclusion bodies were solubilized in buffer C (50 mM Mes, pH 6.5/200 mM (NH4)2SO4/0.08 mM Tween 20) containing 2 M urea. The solubilized protein was purified via Talon resin (Clontech) by following the manufacturer's instructions. Protein was stored at a concentration of 5 mg/ml in buffer C. Point mutants of Imp4-core (R119A, R201A, R220A, and R253A) were made with QuikChange (Stratagene) by using Imp4-core as the template and expressed and purified as described for Imp4-core.
Duplex Formation Assays. The ability of proteins to stimulate annealing of complementary RNA strands was assayed as follows: RNP complexes containing Imp3p, Imp4p, or control proteins at various concentrations (0.5–5 μM) and refolded unlabeled minimal U3 snoRNA-binding site [10 nM minimal U3 snoRNA-binding site for Imp3p and Imp4p (U3 MINI) (nucleotides 4–50)] were mixed in the binding buffer [20 mM Tris, pH 8.0/30 mM NH4Cl/100 mM KCl/0.5 mM MgCl2/1 mM DTT/4% (vol/vol) glycerol/0.1% (wt/vol) Nonidet P-40/0.2 units/μl RNasin (Promega)]. After a 5-min incubation at room temperature, radiolabeled (1,000 cpm) complementary strand comprising the relevant sites of the pre-rRNA [the 10-mer 5′ ETS site (nucleotides 470–479) or the 17-mer 18S site (nucleotides 706–722)] at 10 nM final concentration was added to the mixture and incubated for an additional 15 min. Base-paired RNP complexes were resolved on 6% nondenaturing PAGE for 35 min at 100 V at 4°C. Proteinase K treatment included a 10-min incubation of the preformed ternary complex with 2 μl of stop buffer (0.03 mg/ml tRNA/0.3 mg/ml proteinase K/0.67% SDS/0.02% bromophenol blue/0.02% xylene cyanole) at 37°C.
Results
To identify and investigate the role of Imp3p and Imp4p in ribosome biogenesis, both proteins from S. cerevisiae were expressed in E. coli as inclusion bodies, purified, and renatured (see Methods). Nondenaturing PAGE (Fig. 5), resistance to proteolytic cleavage (data not shown), and specific binding to RNA (see below and Fig. 2) indicated that the proteins were refolded to a biologically relevant state. To study the region of sequence conservation shared among the Imp4p superfamily, which includes a σ70 motif expected to interact with the U3 snoRNA (27), we cloned Imp4-core, a truncation mutant that contains amino acids 87–275 of Imp4p. The Imp4-core protein and point mutants thereof were expressed and extracted from inclusion bodies under nondenaturing conditions.
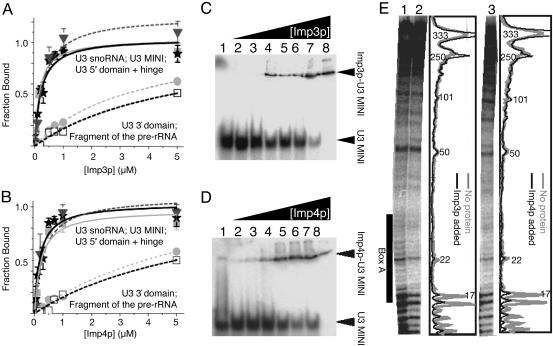
Fig. 2.
Imp3p and Imp4p bind to U3 MINI. Nitrocellulose filter-binding assays of Imp3p (A) and Imp4p (B) binding to the full-length U3 snoRNA (square), the 3′ domain (nucleotides 71–333; circle), U3 MINI (nucleotides 4–50; triangle) and a pre-rRNA fragment containing the ETS site, the 18S site, and the intervening sequence (open square); and electromobility-shift assays of Imp3p (A) and Imp4p (B) binding to the 5′ domain plus hinge region (nucleotides 1–76; star). Electromobility-shift assays of nucleotides 1–76 with varying amounts of Imp3p (C) and Imp4p (D); lanes 1–8 have 0, 0.05, 0.1, 0.2, 0.5, 0.7, 1, and 5 μM of protein, respectively. (E) Footprinting assays of the U3 snoRNA without protein (lane 1), with 5 μM Imp3p (lane 2), and with 5 μM Imp4p (lane 3) using 0.4 milliunits/μl RNase V1. Graphical quantitations are shown to the right of lanes 2 and 3.
Imp3p and Imp4p Bind Directly to a 5′ Portion of the U3 snoRNA. To test for direct RNA binding, we used nitrocellulose filter-binding assays to measure apparent dissociation constants (Kd) for these proteins in complex with parts of the pre-rRNA, the full-length U3 snoRNA, and fragments thereof (Table 1 and Fig. 2). Protein binding to the U3 snoRNA (nucleotides 1–333) and to the 5′ domain plus hinge region (nucleotides 1–76) was indistinguishable (Fig. 2 A and B). In contrast, saturatable binding was not observed for either the 3′ domain of the U3 snoRNA (nucleotides 71–333) or fragments of the pre-rRNA that include the ETS site, the 18S site, or both sites and the intervening sequence (Table 1 and Fig. 2). RNase protection assays show that binding of either protein to the U3 snoRNA preferentially inhibits endonucleolytic cleavage of the 5′ domain plus hinge region by the double-strand-specific RNase V1, especially nucleotides in the box A/A′ stem structure (Fig. 2E). Competition experiments further support direct binding between the 5′ domain plus hinge region of the U3 snoRNA (nucleotides 1–76) and either protein. The unlabeled 5′ domain plus hinge region and the full-length U3 snoRNA compete equally well for binding to a preformed complex of the full-length U3 snoRNA and Imp3p or Imp4p (Fig. 6, which is published as supporting information on the PNAS web site). In line with specific binding, complexes of the U3 snoRNA and either protein were insensitive to competition with tRNA (up to a concentration of 3 μM) or salt (up to a concentration of 0.5 M KCl). A similar resistance to salt concentration was observed for complexes purified from cells (34).
Table 1. Apparent dissociation constants (Kd).
| RNA constructs | Imp3p, nM | Imp4p, nM |
|---|---|---|
| U3 snoRNA (1–333) | 164 ± 35 | 226 ± 66 |
| 5′ domain plus hinge region of the U3 snoRNA (1–76) | 218 ± 62* | 194 ± 59* |
| 3′ domain of the U3 snoRNA (71–333) | NB | NB |
| U3 MINI (4–50) | 316 ± 70 | 315 ± 60 |
| U3 MINIAS1† (4–50) | 290 ± 11 | 793 ± 10 |
| Hinge of the U3 snoRNA (39–62) | NB | NB |
| 5′ domain of the U3 snoRNA (1–36) | NB | NB |
| Fragment of the pre-rRNA (469–722) | NB | NB |
| ETS site of the pre-rRNA (470–479) | NB | NB |
Reported values are the average and standard deviation of at least three independent measurements. Nucleotide numbers are given in parentheses. NB, no binding detected or binding did not saturate.
Binding constants determined by electromobility-shift assays; other binding constants determined by nitrocellulose filter-binding assays
U3 MINIAS1 is a mutant of U3 MINI with compensatory base changes to allow pairing with the antisense ETS site
Studies of a series of U3 snoRNA truncations defined U3 MINI: a minimal binding site (nucleotides 4–50) with virtually the entire 5′ domain and the 5′ part of the hinge region. Further 5′ or 3′ truncations of U3 MINI did not result in saturatable binding when using 500-fold excess of protein (Table 1). Binding to U3 MINI is typified by Kd values of 316 ± 70 nM for Imp3p and 315 ± 60 nM for Imp4p. Similar results were obtained for binding to the full-length U3 snoRNA with Kd values of 164 ± 35 nM for Imp3p and 226 ± 66 nM for Imp4p. Our in vitro studies of truncated RNA constructs demonstrate that recognition of the 5′ domain and the 5′ part of the hinge region is sufficient for binding of Imp3p and Imp4p to the U3 snoRNA. Complexes between U3 MINI and Imp3p or Imp4p migrated as single bands on nondenaturing PAGE (Figs. 2 C and D), indicating that the RNP is a homogeneous complex. Similar results were obtained for protein binding to the full-length U3 snoRNA (data not shown). Determining similar dissociation constants by these two complementary assays, mobility shift and filter binding, increases our confidence that a bona fide interaction is being measured between protein and RNA (Table 1 and Fig. 2).
Two R→A Mutations of Imp4-Core Impair RNA Binding. The highly charged N- and/or C-terminal regions of Imp4p are not essential for RNA binding, because Imp4p and Imp4-core bind to U3 MINI with similar affinities, Kd values of 315 ± 60 nM and 255 ± 50 nM, respectively (data not shown). To identify possible protein–RNA contacts, conserved arginines were targeted for alanine-scanning mutagenesis, because arginines are expected to be surface residues and are found with unusual frequency at RNA–protein interfaces (35). Of the four conserved arginine residues of Imp4-core, two R→A mutations retain RNA binding: the R119A and R201A mutants bound to U3 MINI with Kd values of 883 ± 15 nM and 308 ± 24 nM, respectively. In contrast, binding was not observed between U3 MINI and either the R220A mutant or the R253A mutant. Notably, R253 resides in the putative helix–turn–helix motif, suggesting an RNA–protein contact.
Imp3p and Imp4p Mediate Formation of the U3–ETS Duplex. U3 MINI participates in two functional base-pairing interactions: the U3–ETS duplex is between the U3 hinge (nucleotides 39–49) and the ETS site of the pre-rRNA (nucleotides 470–479); the U3–18S duplex is between the box A/A′ stem of U3 (nucleotides 6–23) and the 18S site, a region near the 5′ terminus of the 18S portion of the pre-rRNA. To test whether Imp3p, Imp4p, or both mediates formation of the U3–ETS duplex, we performed annealing assays with U3 MINI and the radiolabeled ETS site using nondenaturing PAGE. A ternary complex forms in the presence of protein (Imp3p and Imp4p), U3 MINI, and the ETS site (Fig. 3 A and B, lanes 3–6); in contrast, the ternary complex was not detected either in the absence of protein (lane 2) or when noncomplementary RNA sequences (antisense ETS) were used (lane 7). In the latter case, the ternary complex can be partially restored by introducing compensatory mutations in the base-pairing site of U3 MINI (lane 8). Moreover, neither protein mediates formation of the U3–ETS duplex using a U3 construct containing the 5′ hinge region but lacking the 5′ domain (Fig. 7, which is published as supporting information on the PNAS web site).
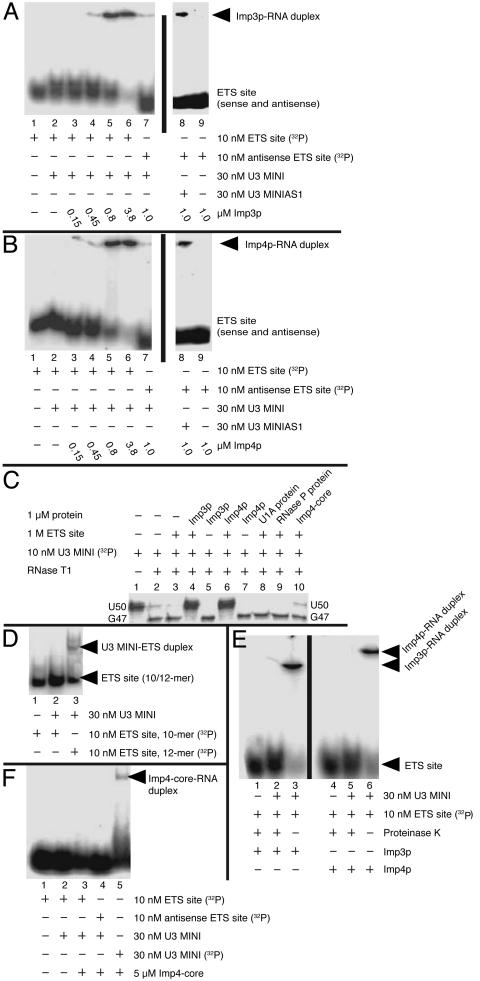
Fig. 3.
Imp3p and Imp4p mediate formation of the U3–ETS duplex. Electromobility shifts on nondenaturing PAGE of Imp3p (A) and Imp4p (B) were used to detect duplex formation between the 32P-labeled ETS site and the U3 hinge region of U3 MINI. U3 MINIAS1 is a mutant of U3 MINI with compensatory base changes to allow pairing with the antisense ETS site. (C) T1 protection assays were used to verify duplex formation and its dependence on protein [only the base-pairing site of U3 MINI is shown (nucleotides 45–50)]. Differences in the radioactivity reflect the extent of cleavage by T1. (D) Protein-independent duplex formation was observed by electromobility shift only when the base-pairing site was extended from 10 to 12 bp; to resolve the ETS site from the duplex, a 10% instead of a 6% nondenaturing PAGE was used. (E) Protein association is required to maintain the U3–ETS duplex; proteinase K treatment of a preformed complex among protein, U3 MINI, and the ETS site resulted in duplex dissociation of 32P-labeled ETS and U3 MINI. (F) Nondenaturing PAGE shows that the Imp4 core protein binds weakly to U3 MINI but does not promote formation of the U3–ETS duplex.
To verify protein-dependent formation of the U3–ETS duplex, we performed nuclease protection experiments with RNase T1, which cleaves single-stranded regions of RNA on the 3′ side of G nucleotides (Fig. 3C). Formation of the U3–ETS duplex is expected to protect only G47 of U3 MINI from RNase T1 digestion. G47 of U3 MINI is cleaved in the absence and presence of Imp3p or Imp4p (Fig. 3C, lanes 3, 5, and 7, respectively), demonstrating that G47 is accessible even after forming a complex with protein. In contrast, cleavage at G47 is inhibited in the presence of protein, U3 MINI and the ETS site, consistent with protein-dependent formation of the U3-ETS duplex (Fig. 3C, lanes 4 and 6).
Next, we investigated whether mediating formation of the U3–ETS duplex is specific. Imp4p-core fails to support hybridization even though it retains RNA binding, indicating that part or all of the regions truncated from Imp4p (the N terminus, the C terminus, or both) is required for duplex formation (Fig. 3F). To test whether other proteins could promote formation of this duplex, we assayed two well characterized RNA-binding proteins: the U1A protein and the RNase P protein. Neither protein is part of the SSUP (3); however, each binds in vitro to U3 MINI (Kd values of ≈0.1 μM and ≈0.3 μM, respectively). Importantly, neither the U1A protein nor the RNase P protein was able to inhibit the T1-dependent cleavage of G47 (Fig. 3C, lanes 8 and 9, respectively). The data argue that Imp3p and Imp4p use a specific mechanism to enable formation of the U3–ETS duplex.
Protein Stabilizes the Otherwise Unstable U3–ETS Duplex. To assess how protein binding changes duplex stability, we performed proteinase K treatment after assembly of the ternary complex (ETS site, U3 MINI, and Imp3p or Imp4p). The duplex dissociated after treatment, demonstrating that the presence of protein is necessary to maintain a stable hybrid (Fig. 3E). We extended the hybrid by two base pairs to test the effect of duplex stability on hybridization. In this case, spontaneous hybridization was observed; however, protein is required for >50% duplex formation (compare Fig. 3 D and E).
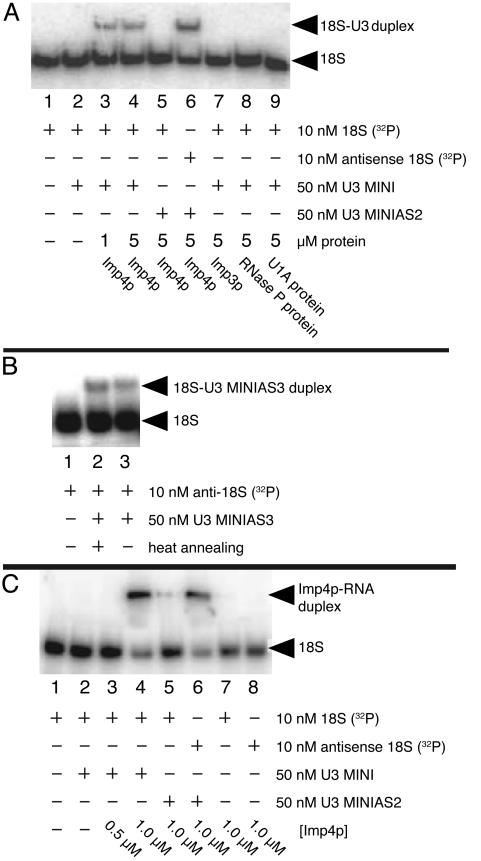
Imp4p Mediates Formation of the U3–18S Duplex. Unlike the U3 snoRNA nucleotides in the U3–ETS duplex that are expected to be single-stranded before hybridization and thus accessible, most of the corresponding U3 snoRNA nucleotides in the U3–18S duplex form base pairs in the box A stem before duplex formation (Fig. 1 A). A trans-acting factor is therefore expected to disrupt this stem to make its nucleotides accessible for hybridization. Interestingly, in vitro formation of the U3–18S duplex, as mimicked by U3 MINI and the 18S site, is detected only in the presence of Imp4p (Fig. 4A, lanes 3 and 4). After removal of Imp4p by proteinase K treatment, the duplex does not fully dissociate. Formation of this duplex was not detected when other proteins were added (Imp3p, the RNase P protein, or the U1A protein) or when noncomplementary sequences were used at the base-pairing site (Fig. 4A, lanes 7, 8, 9, and 5, respectively); in the presence of Imp4p, duplex formation was restored when compensatory mutations were made in U3 MINI (Fig. 4A, lane 6). To test whether proteinase K destabilizes the duplex or the duplex has limited stability once the protein is removed, we surveyed the percentage of duplex formation using variants of U3 MINI designed to disrupt the box A stem structure and to pair with the antisense of the 18S site. Consistent with a limited duplex stability, an equivalent percentage of duplex was observed with proteinase K treatment (Fig. 4A, lanes 3, 4, and 6) and without proteinase K treatment using 18S and U3 variants that permit hybridization in the absence of protein (Fig. 4B, lanes 2 and 3). Annealing assays using nondenaturing PAGE confirm that Imp4p promotes formation of the U3–18S duplex (Fig. 4C). The data argue that Imp4p disrupts the box A stem, via chaperonic activity, to expose the 18S base-pairing site in box A, thereby enabling formation of the U3–18S duplex.
Fig. 4.
Imp4p mediates formation of the U3–18S duplex as detected by nondenaturing PAGE. (A) Protein and 32P-labeled 18S site were incubated with either U3 MINI or U3 MINIAS2. U3 MINIAS2 is a mutant in which the nucleotides involved in pairing with the 18S site are antisense and the corresponding nucleotides involved in the box A/A′ stem are mutated to maintain the WT stem structure. Before the assay, protein was removed with proteinase K treatment. (B) Formation of the U3–18S duplex is spontaneous only when the box A/A′ stem structure is disrupted by mutation (U3 MINIAS3). (C) Nondenaturing PAGE verifies duplex formation between 32P-labeled 18S site and U3 MINI.
Discussion
A major quest in the field of ribosome biogenesis is to determine the role played by the numerous essential nonribosomal proteins. An emerging view is that many of these proteins mediate the dynamic rearrangements associated with the assembly and disassembly of RNA–protein and RNA–RNA complexes critical during eukaryotic ribosome biogenesis.
Our studies show that Imp3p and Imp4p mediate two U3–pre-rRNA interactions (Figs. 3 and 4) that are critical for ribosome biogenesis (5–10). Our in vitro studies argue that Imp3p and Imp4p are primary binders to U3 MINI (nucleotides 4–50) where both U3–pre-rRNA duplexes occur (Figs. 1 and 2). Because Imp3p and Imp4p are part of the SSUP and have the same minimal binding site, simultaneous binding to the U3 snoRNA is expected but has not yet been demonstrated. Imp4p mediates the formation of the transient interactions between the 5′ domain of U3 snoRNA and the part of the pre-rRNA that becomes the 5′ terminus of the mature 18S rRNA. In contrast, both proteins mediate formation of the transient interaction between the U3 snoRNA hinge and the ETS site of the prerRNA, which is removed by cleavage during processing.
Several lines of evidence argue that Imp3p and Imp4p use a specific mechanism to facilitate the essential U3–pre-rRNA interactions. Proteins that bind nonspecifically to U3 MINI and the pre-rRNA (U1A protein and RNase P protein) do not promote formation of either the U3–ETS or the U3–18S duplex. Moreover, Imp4-core binds to the U3 snoRNA but fails to promote formation of the U3–ETS duplex. We observed direct binding to the U3 snoRNA but not to pre-rRNA fragments, ruling out a matchmaker mechanism (36), which requires simultaneous binding to both RNAs. Chaperone activity is not necessary for duplex formation at the ETS site, because the part of the U3 snoRNA involved in this interaction is accessible for endonucleolytic cleavage either in the presence or absence of Imp3p and Imp4p (Fig. 3C). We propose that both proteins target the 5′ domain and the hinge of the U3 snoRNA and enable duplex formation by stabilizing the U3–ETS interaction. The charged terminal regions of Imp4p are required to mediate formation of the U3–ETS duplex, because Imp4-core lacking these regions binds to the U3 MINI but fails to anneal the U3–ETS duplex (Fig. 3F). These charged residues can stabilize duplex formation by decreasing charge repulsion that arises during hybridization of polyanionic RNAs. A similar mechanism was proposed for the charged RS domain of the splicing factor U2AF65 that helps recruit the U2 small nuclear RNP to the branch point of the intron in pre-mRNA (14).
Unlike the U3–ETS duplex, a structural change is necessary to form the U3–18S duplex, because the relevant nucleotides of the U3 snoRNA are base-paired and therefore inaccessible. We hypothesize that Imp4p mediates base-pair disruption in the box A stem to permit duplex formation at the U3–18S site. In the case of the U3–ETS duplex, removal of Imp3p and Imp4p is sufficient to disengage the apparently unstable U3–ETS duplex. In contrast, the U3–18S duplex remains after removal of Imp4p. Formation of the U3–18S duplex is incompatible with the participation of these 18S nucleotides in a central and universal RNA tertiary interaction (pseudoknot) found in mature 18S rRNA; the U3–18S duplex thus prevents premature formation of this tertiary interaction. By mediating formation of the U3–ETS and the U3–18S duplexes (Figs. 3 and 4), Imp3p and Imp4p may help recruit the SSUP to the pre-rRNA (3, 22, 23).
In vivo data suggest that the numerous transient RNA–RNA and RNA–protein interactions during ribosome biogenesis occur in a strict temporal order (4, 30). To achieve this order, specific steps in this process must be regulated. Imp3p and Imp4p are the first proteins shown to facilitate formation of a particular RNA–RNA interaction that is a key step in ribosome biogenesis. It is intriguing to think that temporal control of ribosome biogenesis may be achieved via regulation of factors like Imp3p and Imp4p. Our studies identify an RNA target and possible function for a member of the Imp4 superfamily of proteins, five of which have been implicated in ribosome biogenesis (27–31). A possible role of other proteins in the Imp4 superfamily is to mediate formation of specific RNA duplexes, thereby providing sites for regulation.
Supplementary Material
Acknowledgments
We thank Matthew Plantinga for technical help and helpful discussions, Yuen-Ling Chan for invaluable advice and helpful discussions, Anna Brady for technical assistance, and Susan Baserga (Yale University, New Haven, CT) and Barbara Golden (Purdue University, West Lafayette, IN) for providing reagents. This work was supported by grants (to C.C.C.) from the National Institutes of Health (GM59782) and by an award to the University of Chicago's Division of Biological Sciences under the Research Resources Program for Medical Schools of the Howard Hughes Medical Institute.
Author contributions: T.G. and C.C.C. designed research; and T.G. performed research.
Abbreviations: sno, small nucleolar; RNP, ribonucleoprotein; SSU, small subunit; SSUP, SSU processome; pre-rRNA, precursor rRNA; U3 MINI, minimal U3 snoRNA-binding site for Imp3p and Imp4p; ETS, external transcribed spacer; Mes, 2-morpholinoethanesulfonic acid.
References
- 1.Venema, J. & Tollervey, D. (1999) Annu. Rev. Genet. 33, 261-311. [DOI] [PubMed] [Google Scholar]
- 2.Kressler, D., Linder, P. & de La Cruz, J. (1999) Mol. Cell. Biol. 19, 7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragon, F., Gallagher, J. E., Compagnone-Post, P. A., Mitchell, B. M., Porwancher, K. A., Wehner, K. A., Wormsley, S., Settlage, R. E., Shabanowitz, J., Osheim, Y., et al. (2002) Nature 417, 967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fatica, A. & Tollervey, D. (2002) Curr. Opin. Cell Biol. 14, 313-318. [DOI] [PubMed] [Google Scholar]
- 5.Hughes, J. M. & Ares, M., Jr. (1991) EMBO J. 10, 4231-4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame, M. & Tollervey, D. (1992) EMBO J. 11, 1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltrame, M. & Tollervey, D. (1995) EMBO J. 14, 4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltrame, M., Henry, Y. & Tollervey, D. (1994) Nucleic Acids Res. 22, 5139-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes, J. M. (1996) J. Mol. Biol. 259, 645-654. [DOI] [PubMed] [Google Scholar]
- 10.Sharma, K. & Tollervey, D. (1999) Mol. Cell. Biol. 19, 6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borovjagin, A. V. & Gerbi, S. A. (1999) J. Mol. Biol. 286, 1347-1363. [DOI] [PubMed] [Google Scholar]
- 12.Borovjagin, A. V. & Gerbi, S. A. (2000) J. Mol. Biol. 300, 57-74. [DOI] [PubMed] [Google Scholar]
- 13.Borovjagin, A. V. & Gerbi, S. A. (2004) RNA 10, 942-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valcarcel, J., Gaur, R. K., Singh, R. & Green, M. R. (1996) Science 273, 1706-1709. [DOI] [PubMed] [Google Scholar]
- 15.Muller, U. F., Lambert, L. & Goringer, H. U. (2001) EMBO J. 20, 1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rein, A., Henderson, L. E. & Levin, J. G. (1998) Trends Biochem. Sci. 23, 297-301. [DOI] [PubMed] [Google Scholar]
- 17.Ginisty, H., Amalric, F. & Bouvet, P. (1998) EMBO J. 17, 1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginisty, H., Serin, G., Ghisolfi-Nieto, L., Roger, B., Libante, V., Amalric, F. & Bouvet, P. (2000) J. Biol. Chem. 275, 18845-18850. [DOI] [PubMed] [Google Scholar]
- 19.Hanakahi, L. A., Bu, Z. & Maizels, N. (2000) Biochemistry 39, 15493-15499. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. J. & Baserga, S. J. (1999) Mol. Cell. Biol. 19, 5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wormsley, S., Samarsky, D. A., Fournier, M. J. & Baserga, S. J. (2001) RNA 7, 904-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehner, K. A., Gallagher, J. E. & Baserga, S. J. (2002) Mol. Cell. Biol. 22, 7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granneman, S., Gallagher, J. E., Vogelzangs, J., Horstman, W., van Venrooij, W. J., Baserga, S. J. & Pruijn, G. J. (2003) Nucleic Acids Res. 31, 1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunbar, D. A., Wormsley, S., Agentis, T. M. & Baserga, S. J. (1997) Mol. Cell. Biol. 17, 5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies, C., Gerstner, R. B., Draper, D. E., Ramakrishnan, V. & White, S. W. (1998) EMBO J. 17, 4545-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodersen, D. E., Clemons, W. M., Jr., Carter, A. P., Wimberly, B. T. & Ramakrishnan, V. (2002) J. Mol. Biol. 316, 725-768. [DOI] [PubMed] [Google Scholar]
- 27.Wehner, K. A. & Baserga, S. J. (2002) Mol. Cell 9, 329-339. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhaber, F., Wechselberger, C. & Kreil, G. (2001) Trends Biochem. Sci. 26, 345-347. [DOI] [PubMed] [Google Scholar]
- 29.Kaser, A., Bogengruber, E., Hallegger, M., Doppler, E., Lepperdinger, G., Jantsch, M., Breitenbach, M. & Kreil, G. (2001) Biol. Chem. 382, 1637-1647. [DOI] [PubMed] [Google Scholar]
- 30.Fatica, A., Cronshaw, A. D., Dlakic, M. & Tollervey, D. (2002) Mol. Cell 9, 341-351. [DOI] [PubMed] [Google Scholar]
- 31.Bogengruber, E., Briza, P., Doppler, E., Wimmer, H., Koller, L., Fasiolo, F., Senger, B., Hegemann, J. H. & Breitenbach, M. (2003) FEM Yeast Res. 3, 35-43. [DOI] [PubMed] [Google Scholar]
- 32.Brinkmann, U., Mattes, R. E. & Buckel, P. (1989) Gene 85, 109-114. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong, N., de Lencastre, A. & Gouaux, E. (1999) Protein Sci. 8, 1475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker, K. A. & Steitz, J. A. (1987) Mol. Cell. Biol. 7, 2899-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allers, J. & Shamoo, Y. (2001) J. Mol. Biol. 311, 75-86. [DOI] [PubMed] [Google Scholar]
- 36.Sancar, A. & Hearst, J. E. (1993) Science 259, 1415-1420. [DOI] [PubMed] [Google Scholar]
- 37.Mereau, A., Fournier, R., Gregoire, A., Mougin, A., Fabrizio, P., Lührmann, R. & Branlant, C. (1997) J. Mol. Biol. 273, 552-571. [DOI] [PubMed] [Google Scholar]
- 38.Antal, M., Mougin, A., Kis, M., Boros, E., Steger, G., Jakab, G., Solymosy, F. & Branlant, C. (2000) Nucleic Acids Res. 28, 2959-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.