Abstract
Background
Vector arthropods control arbovirus replication and spread through antiviral innate immune responses including RNA interference (RNAi) pathways. Arbovirus infections have been shown to induce the exogenous small interfering RNA (siRNA) and Piwi-interacting RNA (piRNA) pathways, but direct antiviral activity by these host responses in mosquito cells has only been demonstrated against a limited number of positive-strand RNA arboviruses. For bunyaviruses in general, the relative contribution of small RNA pathways in antiviral defences is unknown.
Methodology/Principal Findings
The genus Orthobunyavirus in the Bunyaviridae family harbours a diverse range of mosquito-, midge- and tick-borne arboviruses. We hypothesized that differences in the antiviral RNAi response in vector versus non-vector cells may exist and that could influence viral host range. Using Aedes aegypti-derived mosquito cells, mosquito-borne orthobunyaviruses and midge-borne orthobunyaviruses we showed that bunyavirus infection commonly induced the production of small RNAs and the effects of the small RNA pathways on individual viruses differ in specific vector-arbovirus interactions.
Conclusions/Significance
These findings have important implications for our understanding of antiviral RNAi pathways and orthobunyavirus-vector interactions and tropism.
Author Summary
A number of orthobunyaviruses such as Oropouche virus, La Crosse virus and Schmallenberg virus are important global human or animal pathogens transmitted by arthropod vectors. Further understanding of the antiviral control mechanisms in arthropod vectors is key to developing novel prevention strategies based on preventing transmission. Antiviral small RNA pathways such as the exogenous siRNA and piRNA pathways have been shown to mediate antiviral activity against positive-strand RNA arboviruses, but information about their activities against negative-strand RNA arboviruses is critically lacking. Here we show that in Aedes aegypti-derived mosquito cells, the antiviral responses to mosquito-borne orthobunyaviruses is largely mediated by both siRNA and piRNA pathways, whereas the piRNA pathway plays only a minor role in controlling midge-borne orthobunyaviruses. This suggests that vector specificity is in part controlled by antiviral responses that depend on the host species. These findings contribute significantly to our understanding of arbovirus-vector interactions.
Introduction
Orthobunyaviruses are endemic in tropical and subtropical regions worldwide and are transmitted by mosquitoes, midges, ticks or other arthropods. The Orthobunyavirus genus within the Bunyaviridae family comprises at least 30 viruses that can cause disease in humans, including Oropouche virus (OROV; febrile illness), La Crosse virus (LACV; encephalitis) and Ngari virus (haemorrhagic fever) [1]. In addition, infection by orthobunyaviruses such as Cache Valley virus (CVV) and Schmallenberg virus (SBV) can lead to disease in animals [2].
Bunyamwera virus (BUNV) is the prototype virus of both the Orthobunyavirus genus and the family. Like most viruses in the genus, the BUNV genome possesses a tripartite, single-stranded negative sense RNA genome, in which the small (S) segment encodes the nucleocapsid (N) protein and the nonstructural protein NSs in overlapping reading frames, the medium (M) segment encodes a viral glycoprotein precursor (in the order Gn-NSm-Gc) for two envelope glycoproteins Gn, Gc and a nonstructural protein NSm, and the large (L) segment encodes the RNA-dependent RNA polymerase. This genome structure is generally reflected by most orthobunyaviruses with some differences for example in the presence or length of NSs [1].
BUNV was originally isolated from Aedes spec. mosquitoes in the Semliki Forest in Uganda in 1943 [3] but has since also been found in Culex spec. and Mansonia spec. (see [4] and references on BUNV therein) and Ochlerotatus spec. [5]. BUNV infections cause febrile illness and (rarely) encephalitis in humans in Sub-Saharan Africa, in particular in Nigeria and the Central African Republic, with wild rodents likely to be serving as amplifying reservoir [6]. Cache Valley virus (CVV) belongs to the Bunyamwera serogroup and is enzootic throughout North and South America [7, 8]. It was first isolated from Culiseta inornata mosquitoes in the Cache Valley in Utah, United States of America in 1956 [8], and has since been shown to be transmitted by mosquitoes of the Culiseta, Anopheles, Aedes, Culex and Ochleratatus genera [9, 10]. A small number of CVV infections in humans have been reported [11–13], where infection rarely leads to serious disease. In ruminants, including sheep and cattle, CVV causes spontaneous abortions and multiple congenital malformations [14–16]. Large mammals including deer, horses and sheep are known to serve as amplifying hosts [6].
In addition to mosquito-borne orthobunyaviruses some members of the genus are exclusively transmitted by biting midges (e.g. SBV, OROV and Sathuperi virus [SATV]) [17–22], ticks (Tete group viruses Bahig and Matruh) [23] or are mosquito/insect-specific [24, 25].
SBV and SATV are closely related orthobunyaviruses of the Simbu serogroup. SBV was first discovered in 2011 in Germany and the Netherlands [26] and infections have since been reported in many European countries [27]. Infections can lead to reduced milk yield, fever, fetal malformations and abortions in ruminants (primarily sheep, goats and cattle) [26, 28]; human infections have not been reported. SATV was isolated from mosquitoes in India in 1957 [29], and was later detected in cattle and biting midges in Nigeria [30, 31]. More recently, SATV was detected in Japan in 1999 [32]. To date little information is available on its pathogenicity in ruminants [22]. Both SBV and SATV are transmitted by Culicoides sp. biting midges [22, 31, 33–35].
The exogenous small interfering (exo-si) RNA and Piwi-interacting (pi)RNA pathways have previously been described as important mosquito antiviral responses limiting the replication of positive-strand RNA flaviviruses and togaviruses [36, 37]. The activity of the exo-siRNA pathway is mediated by two key proteins, the endoribonuclease Dicer-2 (Dcr2) and Argonaute-2 (Ago2). Dcr2 cleaves long virus-derived double-stranded RNAs (dsRNAs) into 21 nucleotides (nt) long small interfering RNA (viRNA) duplexes. These viRNAs are then incorporated into the multiprotein RNA-induced silencing complex (RISC), where presumably one strand is retained (guide strand) by Ago2 to detect, bind and catalyze the degradation of complementary single-stranded (ss)RNA such as viral mRNA. Indeed, 21 nt viRNAs were produced in mosquitoes or mosquito-derived cell lines upon infection with several arboviruses, for example flaviviruses (dengue virus, DENV; West Nile virus, WNV), alphaviruses of the Togaviridae family (chikungunya virus, CHIKV; Semliki Forest virus, SFV; Sindbis virus, SINV; o’nyong’nyong virus, ONNV), bunyaviruses (Rift Valley fever virus, RVFV; and SBV) as well as reoviruses (bluetongue virus, BTV) [38–42]. Silencing experiments in mosquitoes have confirmed the antiviral activity of the exo-siRNA pathway against DENV, SINV, CHIKV and ONNV in vivo [42–46].
In addition to the exo-siRNA pathway, the piRNA pathway has been shown to be a contributor to antiviral immunity in mosquitoes or derived cells [38, 47]. In Drosophila, the piRNA pathway is involved in the epigenetic regulation of the expression of transposable elements (TE) in the germline, and thus preserves the genome integrity [48–51]. However, PIWI proteins have also been detected in somatic cells [49, 52–54]. The production of transposon-specific piRNAs is complex and relies on proteins of PIWI clade, in particular Piwi, Aubergine (Aub) and Argonaute-3 (Ago3). piRNAs are believed to be generated via a primary processing pathway and a secondary ping-pong amplification loop [51, 52]. Primary piRNAs have a 5’ uridine (U1) bias. Secondary piRNAs have a 10 nt overlap with primary piRNAs and contain an adenine at position 10 (A10 bias). Mature piRNAs are generally 26–32 nt in length. The piRNA pathway is functional in mosquito germline and somatic cells [38, 39, 47, 55–57]. Interestingly, in mosquitoes a loss of Aub and a diversification of Piwi proteins has occurred [58]. This gene diversification has been linked with a gain of function of the piRNA pathway and a new role in antiviral immunity since the pathway has also been found to target a number of mosquito-borne viruses including DENV, CHIKV, SFV and RVFV [38, 39, 47]. Further, in the Ae. aegypti-derived Aag2 cell line an antiviral effect of Piwi4 against SFV has been directly demonstrated [47]. Recently it was shown that Ago3 and Piwi5 (and Piwi6 to a lesser extent) are needed for the generation of SINV and DENV specific piRNAs in Aag2 cells [59, 60]; however, the viral RNA substrate that induces this pathway is unknown. Importantly, the respective contribution of the two small RNA pathways to immune defenses against negative-sense RNA arboviruses has not been studied. In short, little is known about the interactions of viruses and vectors, and antiviral responses of vectors, which may govern viral infection, dissemination and transmission.
In this study we investigated the antiviral activities of the mosquito exo-siRNA and piRNA pathway against two mosquito- and three midge-borne orthobunyaviruses. Using reporter BUNV and SBV that express Nano luciferase we compared these responses in vector-virus and non-vector-virus interactions. We performed small RNA sequencing and showed that mosquito as well as midge-borne viruses produce virus-specific siRNAs and piRNAs in mosquito cells. Interestingly we found that silencing of Ago2 and Piwi4 in Aag2 cells led to increased viral replication of mosquito-borne orthobunyaviruses, in contrast to midge-borne orthobunyaviruses where only Ago2 silencing increased virus replication. Additionally, silencing of other piRNA pathway members (Piwi5, 6 and Ago3) affected virus replication differently depending on whether the virus was mosquito- or midge borne. These findings indicate that RNAi pathways play a crucial role in the control of orthobunyavirus replication; however, the piRNA pathway in Aag2 cells seems to be adapted to specific virus-vector combinations and may have important consequences for arbovirus tropism and vector specificity.
Results
Induction of small RNA production by orthobunyaviruses in mosquito and midge cells
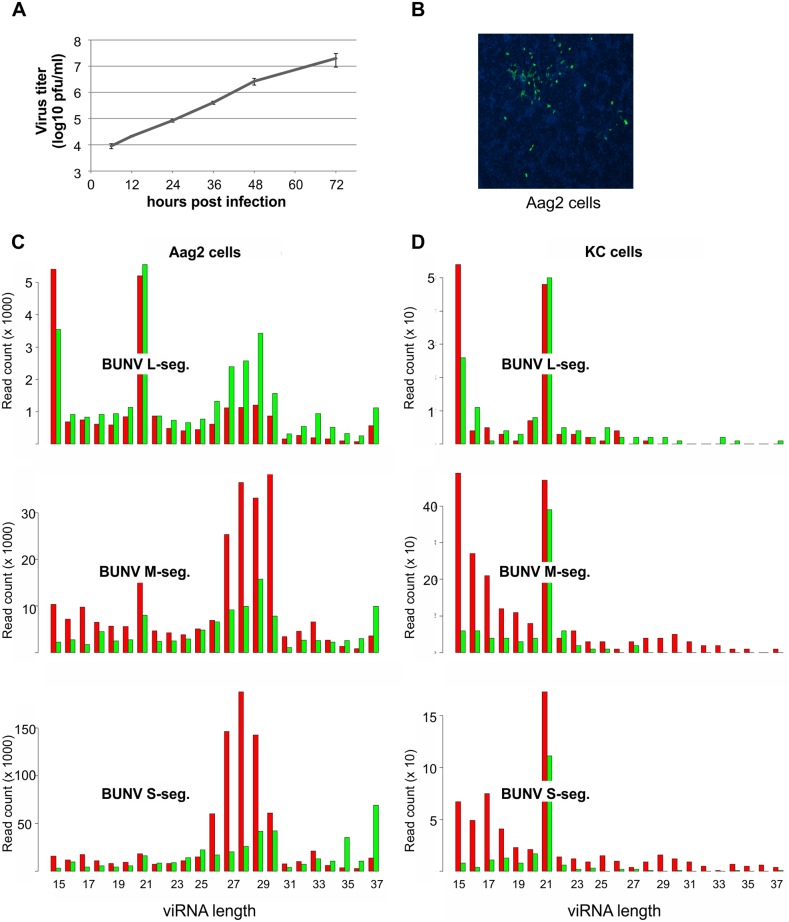
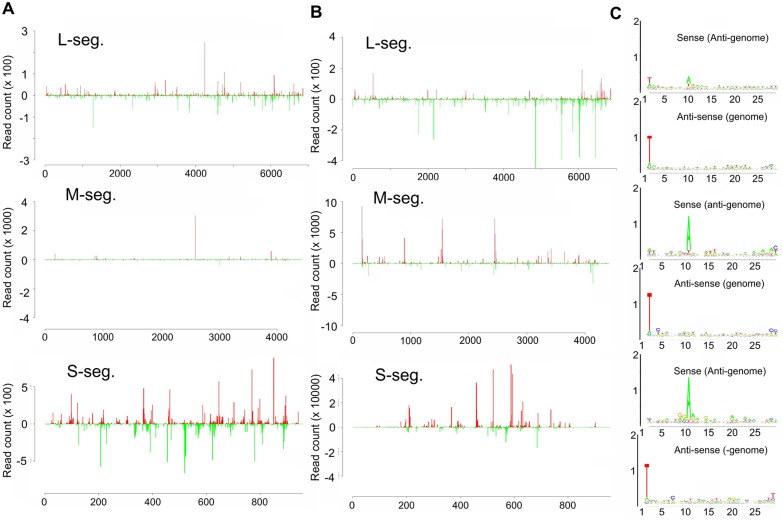
Our previous work has shown that small RNAs with viRNA and piRNA characteristics are produced in non-vector mosquito cells infected with the midge-borne SBV [40] (S1A Fig). To determine if this is similar for a mosquito-borne virus, Ae. aegypti-derived Aag2 cells were infected with BUNV (Fig 1A and 1B) and small RNAs were isolated, sequenced and mapped to the virus genome and antigenome. As shown in Figs 1C and 2A, 21 nt long small RNAs were produced from all three segments which mapped along the genome and antigenome in a cold and hot spot pattern. For the L segment, these 21 nts viRNAs were the major small RNA species produced. Moreover, small RNAs of 24–30 nts with piRNA-specific features (A10, U1 bias), which mapped across the genome and antigenome in a hot and cold spot pattern (Fig 2B), were produced for all three segments (Fig 2C); however, they were the major small RNA species only for M and S segments with a bias for small RNAs mapping to the antigenome. In contrast, 24–30 nt small RNAs mapping to the L segment had a bias for the genome (Fig 1C). Similar results were obtained in BUNV-infected Ae. albopictus-derived U4.4 cells (S2 Fig).
Fig 1. BUNV growth and BUNV-specific viRNA production in Aag2 cells.
(A) BUNV growth curve following infection of Aag2 cells at MOI 1, determined by plaque assay. (B) Immunofluorescence of BUNV-infected Aag2 cells (MOI 0.01) at 48 hours p.i., using BUNV-N specific primary antibody and Alexa 488-conjugated anti-rabbit-IgG secondary antibody (green). Nucleus stained by Dapi (blue). (C) BUNV-specific small RNAs produced in infected Aag2 cells at 24 hours p.i. against L, M and S segments. The y axis indicates read frequency; the x axis indicates the length of the small RNAs (nt). Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome. (D) BUNV-specific small RNAs produced in infected KC cells at 48 hours p.i. against L, M and S segments. The y axis indicates read frequency; the x axis indicates the length of the small RNAs. Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome.
Fig 2. BUNV-specific small RNA and piRNA-like molecules produced in infected Aag2 cells at 24 hours p.i.
(A) Frequency distribution of 21 nt small RNAs along the BUNV antigenome from the 5` to 3` (positive numbers and red), genome (negative numbers and green) from 3` to 5`. (B) Frequency distribution of 24–30 nt small RNAs along the BUNV antigenome from the 5` to 3` (positive numbers and red), genome (negative numbers and green) from 3` to 5` (C) Relative nucleotide frequency and conservation per position of 24–30 nt small RNAs mapping to the genome or antigenome of BUNV S, M and L segments. Sequence is represented as DNA.
To determine if similar small RNAs are produced for BUNV and SBV in midge cells; experiments were repeated with infected C. sonorensis KC cells. As previously reported no piRNA-like molecules could be detected for SBV in KC cells, in contrast, 21 nt small RNAs were mapped to the genome and antigenome of all three segments [40] (S1B Fig). Similar results were observed for BUNV small RNAs in infected KC cells (Fig 1D). Although small RNAs of 24–28 nts could be detected in KC cells, they lacked the piRNA-specific features (A10, U1 bias and 10 nts overlap of sense and antisense small RNAs) (Fig 1D).
Overall, our data showed that BUNV infection induced small RNA patterns comparable with SBV [40] in mosquito-derived cells as well as Culicoides-derived cells. In addition, differences in small RNA patterns were found between mosquito and midge cells.
Generation of BUNV reporter virus expressing Nano luciferase
Previously, luciferase expressing alphaviruses were employed to investigate the antiviral RNAi response in arthropod cells [47, 61, 62]. To obtain similar tools for bunyaviruses, Nano luciferase (NL) expressing BUNV (BUNV-NL) and SBV (SBV-NL) were constructed in the course of this study. BUNV NSm protein, which is dispensable in viral replication in tissue culture, consists of the ectodomain, transmembrane, cytoplasmic domain and type-II transmembrane domain that also serves as internal Gc signal peptide [63, 64]. For BUNV-NL, the 62 residues of the NSm cytoplasmic domain (residues 395 to 455) was replaced by the NL coding region between residues 394 and 456, resulting in a chimeric NSm-NL fusion protein (S3A Fig). NSm-NL has a similar molecular weight to that of N protein (28.67 versus 26.67 kDa) and was indistinguishable from the N protein band in the protein profile of the reporter virus (S3B Fig). BUNV-NL exhibited smaller plaque size than the wildtype virus (S3C Fig). The reporter virus could readily infect Aag2 cells, similar to wildtype virus (S3D Fig). The SBV-NL reporter virus was constructed in the same way as BUNV-NL and successful infection of Aag2 cells, comparable to wildtype SBV, was verified by immunostaining (S3D Fig).
piRNA and miRNA pathways differentially affect midge- and mosquito-borne virus infections in Aag2 cells
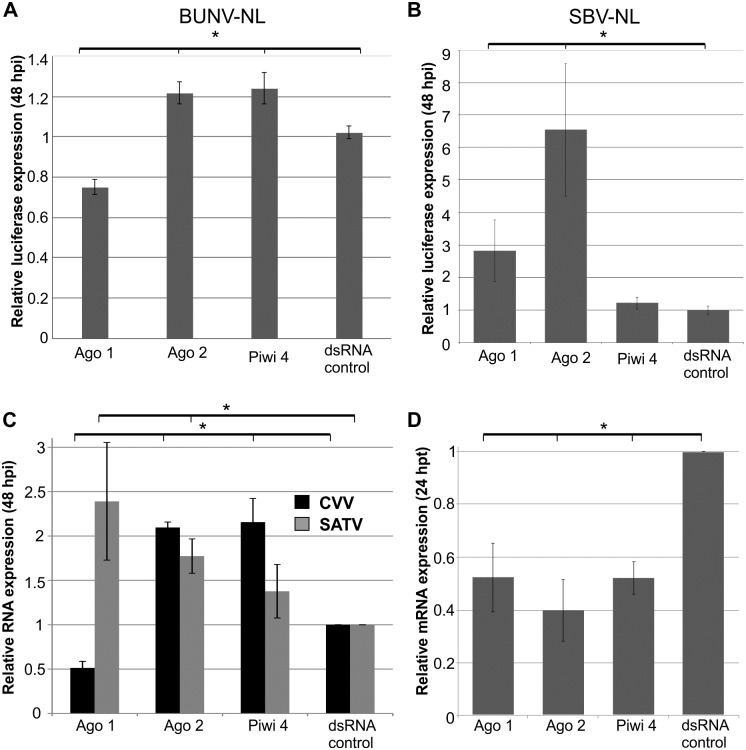
To assess the antiviral role of the small RNAi pathways in BUNV and SBV infected Aag2 mosquito cells, knockdown experiments were performed. Transcripts of the different RNAi pathway key effectors (Ago2, exo-siRNA pathway; Ago1, miRNA pathway; Piwi4-6 and Ago3, piRNA pathway) were silenced by transfection of sequence-specific dsRNA. The effect of the silencing was evaluated by BUNV-NL or SBV-NL infection at 24 hours post-transfection (p.t.) at low MOI (0.01) and subsequent luciferase detection at 48 hours p.i. (Figs 3A, 3B, 4A and 4B); dsRNA specific to eGFP was used as negative control.
Fig 3. Effects of silencing exo-siRNA, miRNA and piRNA pathway key proteins on different orthobunyavirus infections in Aag2 cells.
Silencing of key transcripts by transfection of sequence-specific dsRNA (Ago1, Ago2, Piwi4 or eGFP as negative control) into Aag2 cells, followed by infection at MOI 0.01 at 24 hours p.t.; BUNV-NL (A), SBV-NL (B) or CVV, SATV (C). Cells were lysed at 48 hours p.i. and luciferase activity determined (A, B) or RNA isolated, followed by cDNA synthesis and virus specific qRT-PCR (C). Knockdown of RNAi transcripts was verified by qRT-PCR (D). S7 was used as internal control in qRT-PCR assays. All data were normalized to cells transfected with control dsRNA. Graphs represent three independent experiments performed in triplicate for the luciferase assays and three independent experiments for the qRT-PCR assays. * p≤0.05 student t-test.
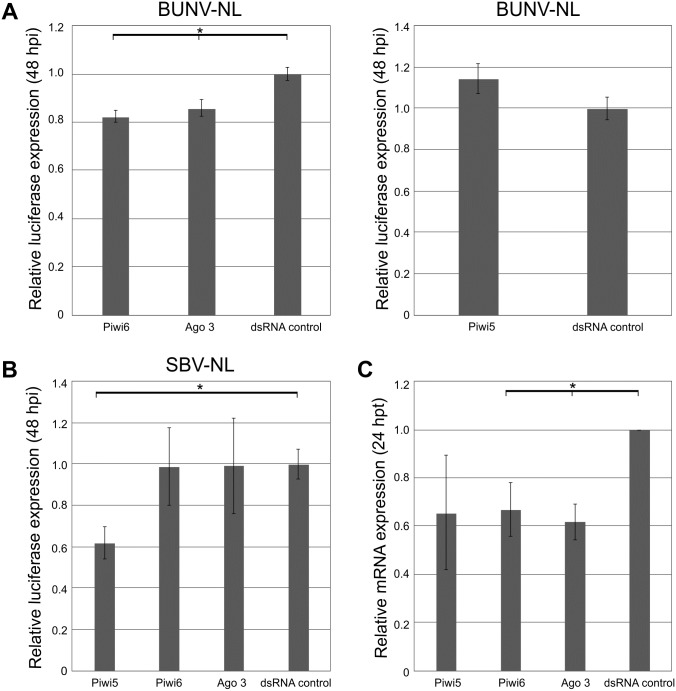
Fig 4. Effects of silencing Piwi5, Piwi6 and Ago3 on BUNV-NL and SBV-NL replication in Aag2 cells.
Silencing of transcripts by transfection of sequence-specific dsRNA (Piwi5, Piwi6, Ago3 or eGFP as negative control) into Aag2 cells, followed by infection at MOI 0.01 at 24 hours p.t.; BUNV-NL (A), SBV-NL (B). Cells were lysed at 48 hours p.i. and luciferase activity determined (A-B). Knockdown of RNAi transcripts was verified by qRT-PCR (C). S7 was used as internal control in qRT-PCR assays. All data were normalized to cells transfected with control dsRNA. Graphs represent three independent experiments performed in triplicate for the luciferase assays and three independent experiments for the qRT-PCR assays. * p≤0.05 student t-test.
Silencing of Ago2 led to an increase in luciferase expression for both BUNV-NL and SBV-NL (Fig 3A and 3B). In contrast, silencing of Piwi4 had no effect on SBV-NL, but resulted in an increase of BUNV-NL replication. Interestingly, silencing expression of other piRNA pathway related genes resulted in a significant decrease of BUNV-NL (Piwi6 and Ago3 knockdowns) and SBV-NL (Piwi5 knockdown) replication (Fig 4A and 4B). A decrease of luciferase expression was also observed for BUNV-NL following Ago1 silencing, compared to an increase of luciferase expression for SBV-NL (Fig 3A and 3B). These results suggested differences in the ability of the piRNA and miRNA pathway to interact with BUNV and SBV.
To determine if this was virus-specific or due to vector versus non-vector arbovirus interaction, silencing experiments were repeated with the mosquito-borne Cache Valley orthobunyavirus (CVV) as well as the midge-borne Sathuperi (SATV) orthobunyaviruses after their ability to grow in Aag2 cells was verified (S4A Fig).
Similar to BUNV, silencing of Ago2 or Piwi4 promoted CVV replication, whereas Ago1 knockdown reduced CVV replication (Fig 3C). Moreover, CVV-infected, Piwi4-silenced Aag2 cells showed cytopathic effects not observed for any of the other knockdown experiments with this virus (S4B Fig). For midge-borne SATV, an increase in replication was observed in cells silenced for Ago2 and Ago1. However, no significant effect on SATV replication was observed in cells treated with Piwi4 dsRNA (Fig 3C). Successful silencing of transcripts by sequence specific dsRNAs was verified by quantitative RT-PCR (Figs 3D and 4C).
In short, Ago2 silencing led to a consistent increase in virus replication for mosquito and midge-borne orthobunyaviruses, supporting an antiviral activity of the exo-siRNA pathway. Similar effects were observed for Piwi4 silencing in the case of mosquito-borne viruses, but not midge-borne viruses.
Silencing of Ago1 resulted in a decrease of replication of the tested mosquito-borne viruses, suggesting an importance of the miRNA pathway for a successful infection in Aag2 cells for these viruses. In contrast, silencing of Ago1 resulted in an increase of SBV and SATV, albeit only slightly significant.
Discussion
The RNAi response is a major antiviral response in arthropods against arbovirus infection. The activated exo-siRNA pathway plays a role in a variety of organisms, including mosquitoes and the model insect D. melanogaster. In contrast, the antiviral activity of the piRNA pathway and the production of viral specific piRNA molecules have been restricted to mosquitoes, especially Aedes spp. Besides, interactions between the miRNA pathway and viruses have been reported in several organisms [65, 66], acting either pro- or antiviral. This can be by expression changes of vector/host miRNAs or viral encoded miRNAs which can either directly target the virus or have host/vector targets, resulting in changes of the cell environment. Previous research has often used D. melanogaster to investigate the interaction between the insect RNAi response and different viruses including arboviruses; however little is known about the specificity of the antiviral response in vector- or non-vector arbovirus interactions. Knockdown experiments of key proteins of the exo-siRNA, miRNA and piRNA pathway in mosquito cells and orthobunyavirus infection, either mosquito- or midge-borne, supports the broad antiviral activity of the exo-siRNA pathway, based on an observed increase in virus replication upon Ago2 silencing for all viruses used. In contrast, the miRNA pathway seemed to be only important for virus replication in the case of a virus-vector match. miRNA expression is often species or even tissue specific and some miRNA-arbovirus interactions have been reported [67]. Whether a similar interaction is important for BUNV and CVV infection in mosquitoes has to be investigated in the future.
Infections with both mosquito-borne and midge-borne viruses were able to induce viral specific piRNAs in the used mosquito cell line. However, the antiviral activity of the piRNA pathway was only confirmed for the mosquito-borne viruses in the Piwi4 knockdown cells. Interestingly knockdown of other piRNA pathway members indicated that they may have pro-viral activities, knockdown of Piwi6 and Ago3 had replication suppressive effects on the mosquito-borne BUNV while knockdown of these proteins had no effect on the midge-borne SBV, whereas knockdown of Piwi5 reduced SBV replication but not BUNV replication. The meaning and biological relevance of these observations is not yet clear. Co-silencing/infection experiments and sequencing of virus-derived small RNAs from such cells may give clues to how different Piwi proteins interact, their role in piRNA-like small RNA production and potentially a hierarchy of protein effector and regulatory functions within this pathway. Perhaps, similar to other RNA binding proteins some of these proteins are important for promoting virus replication though this would require in depth analysis of RNA protein interactions and/or protein-protein interaction studies. Little is known about the piRNA pathway in mosquitoes, the production of virus specific piRNAs and how/if they target the virus. Recently, it has been shown that Piwi5 (and to a lesser extent Piwi6) and Ago3 are needed for virus-derived piRNA production [59, 60]; however, no antiviral activity has previously been linked to these transcripts. In contrast, Piwi4 seems not to be involved in virus-derived piRNA production [59]; however knockdown of Piwi4 can result in antiviral activity for mosquito-borne viruses, like SFV [47]. Similar antiviral activity of Piwi4 is observed for the mosquito-borne orthobunyaviruses: CVV and BUNV, but not for midge-borne orthobunyaviruses (SBV and SATV). This is especially striking as both the mosquito-borne BUNV as well as midge-borne SBV produce similar amounts of virus-specific piRNA molecules in infected Aag2 cells. This would suggest that the antiviral activity of Piwi4 is species specific and either acts as an effector protein downstream of the virus specific-piRNA production or through a different as yet unidentified pathway. Interestingly, no viral specific piRNAs have been reported in midges so far; although this has only been investigated in the C. sonorensis-derived KC cell line and not whole midges [40].
Interestingly, it has been shown that infection of mosquitoes and mosquito-derived cultured cells with arboviruses can lead to generation of virus derived cDNA forms, which are important for mosquito tolerance to virus infection and survival [68]. These DNA forms have the potential to become the template for small RNA production and could therefore determine the action of RNAi pathways on acute arbovirus infection. If the generation of cDNA forms in mosquitoes and derived cells is mosquito-borne virus specific (not occurring with e.g. midge borne viruses) is not known, but could explain the observed species specific antiviral activity of Piwi4.
Overall, these results show the importance to investigate the antiviral RNAi response in vector cells to understand the complex interaction between virus-vector interplay and its effect on tropism.
Materials and Methods
Cell lines
BSR-T7/5 cells [derived from the BSR clone of baby hamster kidney cells-21 [BHK-21] and stably expressing T7 RNA polymerase [69]; a kind gift of Dr. K.K. Conzelmann, Max-von-Pettenkofer Institut, Munich, Germany] were maintained in Glasgow minimal essential medium (GMEM) supplemented with 10% tryptose phosphate broth (TPB), 10% fetal calf serum (FCS), and 1 mg/mL geneticin. BHK-21 cells were maintained in GMEM supplemented with 10% TPB, 10% newborn calf serum (NCS) and 1% penicillin/streptomycin (P/S). Sheep choroid plexus cells (CPT-Tert) [70] were grown in Iscove's modified Dulbecco's media (IMDM) supplemented with 10% FCS and 1% P/S. Vero E6 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) and 10% FCS. BSR-T7/5, BHK-21, CPT-Tert and Vero E6 cells were grown at 37°C and 5% CO2. Ae. aegypti-derived Aag2 cells were maintained in L-15 medium supplemented with 10% TBP, 10% FCS and 1% P/S at 28°C.
Plasmids
Plasmids to generate full-length BUNV antigenome RNA transcripts [pT7riboBUNL(+), pTVT7RBUNM(+), and pT7riboBUNS(+)] have been described previously [71, 72]. pTVT7RBUNM-NL was generated by PCR-directed internal deletion to replace the coding region of the NSm cytoplasmic tail (residues 395 to 456) with that of Nano luciferase (NL). A five amino acid linker, GASGA, was inserted between the NSm transmembrane domain (TMD) and the N-terminus of NL. To facilitate the cloning of NL, a unique KpnI restriction enzyme site was introduced at nt 1419 in the BUNV M segment cDNA (S3A Fig). The plasmids, pUCSBVST7, pUCSBVMT7 and pUCSBVLT7, used to rescue SBV have been described previously [73]. pUCSBVT7-NL was generated through the introduction of two unique restriction sites, MIuI and XhoI, in the SBV M segment (provided by M. Varela; University of Glasgow). The restrictions sites were used to delete 90 nt of NSm (corresponding to nt 1235–1325 in JX853180.1) and NL was subsequently cloned in the deletion site. NL is a small luciferase subunit (19 kDa) from the deep sea shrimp Oplophorus gracilirostris with significantly increased luminescence expression and signal half-life as well as specific activity in mammalian cells compared to both Firefly and Renilla luciferases. NL uses a novel imidazopyrazinone substrate (furimazine) [74].
Virus rescue by reverse genetics and plaque assay
Rescue experiments were performed as essentially described previously, with a modification [75]. Briefly, BSR-T7/5 cells were transfected with a mixture of plasmids comprising 0.5 μg each of pT7riboBUNL(+), pT7riboBUNS(+) and either TVT7RBUNM(+) or TVT7RBUNM-NLuc cDNA. At 4 hours p.t., 2 ml growth medium was added and incubation continued for 5–11 days at 33°C until cytopathic effect (CPE) was evident. Virus titre was determined by plaque assay on BSR-T7/5 cells. Cells were fixed in 4% formaldehyde and plaques stained in 0.01% toluidine blue. The rescue of SBV-NL was performed using pUCSBVST7, pUCSBVMT7-NL and pUCSBVLT7 as described for BUNV but with the exception that 1 μg of each plasmid was used and that the plaque assay was performed using BHK-21 cells.
Preparation of virus working stocks
BUNV, BUNV-NL, SBV, SBV-NL, CVV and SATV stocks were grown in BHK-21 cells. Cells were infected with viruses at a multiplicity of infection (MOI) of 0.01 PFU/cell and incubated at 33°C. Virus-containing cell supernatant was harvested when CPE was evident (usually 2–4 days p.i.), cleared by centrifugation and stored at -80°C. Virus titres of BUNV and CVV were determined by plaque assay on BHK-21 cells and those of SBV and SATV on CPT-Tert cells.
Metabolic radiolabelling and immunoprecipitation
Procedures for metabolic radiolabelling and immunoprecipitation of BUNV proteins were described previously [76]. Briefly, at 24 hours p.i., BSR-T7/5 cells were labelled with [35S]methionine (50 Ci) for 2 hours and then lysed, on ice, in 300 μl of non-denaturing RIPA buffer (50 mM Tris-HCl [pH7.4], 1% Triton X-100, 300 mM NaCl, 5 mM EDTA) containing a cocktail of protease inhibitors (Roche). For immunoprecipitation assays, BUNV viral proteins were immunoprecipitated with anti-BUNV antibody, a rabbit antisera raised against purified BUNV [77] that had been conjugated to magnetic Protein A-Dynabeads (Life Technologies). Viral proteins were analysed by SDS-PAGE under reducing conditions.
Plaque assays in mammalian cells
The plaque phenotype of BUNV and BUNV-NL in BSR cells (routinely grown in GMEM supplemented with 10% foetal calf serum at 37°C in a 5% CO2 incubator) was investigated by plaque assay. Cells were seeded into 12-well plates at a density of 1.2 x 105 cells/well and left to adhere overnight. Cells were infected with BUNV or BUNV-NL at a MOI of 0.05 and cells were fixed with 4% formaldehyde-PBS and stained with 0.1% crystal violet blue solution at 3 days post infection.
Virus growth assays in mosquito cells
Growth of BUNV, SBV, CVV and SATV in Aag2 cells was assessed. Briefly, Aag2 cells were seeded into 24-well plates at a density of 1.7 x 105 cells/well and left to adhere overnight. Cells were then infected with viruses at MOI 1 (BUNV, CVV) or MOI 0.01 (SBV, SATV) and culture supernatant was harvested at different time points p.i. Viral titres were determined by plaque assays on BHK-21 cells (BUNV, CVV) or CPT-Tert cells (SBV, SATV).
Immunofluorescence assays
Aag2 cells were seeded into 24-well glass bottom plates at a density of 1.7 x 105 cells/well and infected with BUNV, BUNV-NL, SBV or SBV-NL at a MOI of 0.01 for 48 hours. Cells were fixed and stained with anti-BUNV N or–SBV N antibody [78, 79]. Goat anti-rabbit Alexa Fluor 488 antibody (Molecular Probes) was used to detect primary antibodies and cells were mounted using Vectashield mounting media containing DAPI (Vectorlabs). Images were taken using the EVOS FL Cell Imaging System.
Small RNA deep sequencing
Small RNA sequencing of BUNV-infected Aag2 and U4.4 cells was carried out by Edinburgh Genomics (University of Edinburgh) using the Illumina HighSeq 2000 platform, as previously described [40]. In short, 2.6 x 106 Aag2 cells/well were seeded into a 6-well plate and left to adhere overnight. Cells were infected with BUNV at a MOI of 10. At 24 hours p.i., RNA was isolated from individual wells using 1 ml TRIzol (Life Technologies) followed by purification, sequencing, analysis and mapping of virus specific small RNAs using viRome [80]. Small RNA sequences were submitted to the European Nucleotide Archive (accession number PRJEB15203). Data for SBV are referenced under [40]. The actual number of reads for each experiment is shown S1 Table.
Silencing of RNAi pathway components
Silencing of Ae. aegypti Ago2, Piwi4, and Ago1 was performed in Aag2 cells using dsRNAs as previously described [81]. dsRNA targeting eGFP was used as control. Silencing of transcripts was confirmed by qRT-PCR using Fast SYBR Green Master Mix (Applied Biosystems) and the corresponding primers [S2 Table; [81]]; on an ABI 7500 Fast real-time PCR instrument. Ae. aegypti S7 ribosomal transcript was used as housekeeping gene for relative quantification as previously described [81]. At 24 hours p.t., cells were infected with BUNV-NL, SBV-NL, CVV or SATV at a MOI of 0.01. At 48 hours p.i. cells were either lysed in passive lysis buffer and NL luminescence was measured (BUNV-NL and SBV-NL) or RNA was isolated using TRIzol (CVV and SATV), followed by cDNA production and qRT-PCR analysis. cDNA synthesis was performed using random hexamer primers (Promega) and SuperScript III reverse transcriptase (Life Technologies). qRT-PCR was performed using Fast SYBR Green Master Mix using corresponding primers (S2 Table). Ae. aegypti S7 ribosomal transcript was used as housekeeping gene.
Supporting Information
Size distribution of SBV-specific small RNAs in Ae. aegypti-derived Aag2 cells at 48 hours p.i. (A) or C. sonorensis KC cells (B) at 24 hours p.i. The y-axis indicates read frequency; the x-axis indicates the length of the small RNAs (nt). Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome of L, M and S segments.
(TIF)
(A) Size distribution of BUNV-specific small RNAs. The y axis indicates read frequency; the x axis indicates the length of the small RNAs (nt). Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome. (B) Relative nucleotide frequency and conservation per position of 24–30 nt small RNAs mapping to the genome or antigenome of BUNV S, M and L segments. Sequence is represented as DNA. (C) Frequency distribution of the 21 nt and 24–30 nt BUNV-specific small RNAs across the antigenome (positive numbers and red) from the 5` to 3` and genome (negative numbers and green) from the 3` to 5`.
(TIF)
(A) Construction of TVT7BUNM-NL in which the coding region of the NSm cytoplasmic tail (residues 395 to 455) was replaced by that of Nano luciferase (NL); BUN-NL GPC was cleaved into Gn, Gc, and NSm-NL chimeric protein. The fused NL is shown in orange, signal peptide (sp) in the grey box and transmembrane domain (TM) in the black box. The amino acid positions at the boundary of each protein are marked on top of Wt BUNV GPC. (B) Comparison of protein profiles of BUNV and BUNV-NL. BSR-T7/5 cells were infected with BUNV and BUNV-NL at MOI of 0.5 and labelled with [35S]methionine at 24 hours p.i for 20 hours. Viral proteins were precipitated with anti-BUNV antibody and analysed by 12.5% SDS-PAGE tris-glycine gel under reducing conditions. Positions of viral proteins are indicated. (C) Comparison of plaque phenotypes of BUNV and BUNV-NL on Vero E6 cells. Cells were fixed with 4% formaldehyde-PBS and stained with 0.1% crystal violet blue solution. (D) Immunofluorescence of Aag2 cells infected with either BUNV-NL or SBV-NL at MOI 0.01 at 48 hours p.i. Anti-BUNV or anti-SBV N primary antibody, followed by an anti-rabbit Alexa Fluor 488-conjugated secondary antibody (green) and nucleic acid staining with Dapi (blue) was used.
(TIF)
(A) Aag2 cells were infected with CVV (MOI 1), SBV, or SATV (MOI 0.01) and culture supernatants were harvested at different time points p.i. as indicated. Viral titres were determined by plaque assays on BHK-21 cells (CVV) or CPT-Tert cells (SBV, SATV). Graphs represent one experiment performed in triplicate. Error bars represent standard errors of the means (SE). (B) Aag2 cells were transfected with dsRNA either specifically to Piwi4 or eGFP as control, followed by CVV infection at 24 hours p.t. Images of cells shown were taken at 48 hours p.i using the EVOS FL Cell Imaging System.
(TIF)
The actual numbers of small RNA reads obtained in each experiment performed and analysed in this study are outlined in the table. The total number of reads against each virus segment as well as the number of reads of each size for each repeat is shown.
(XLSX)
(DOC)
Acknowledgments
The Authors want to thank the colleagues mentioned in the text for useful reagents.
Data Availability
Small RNA sequences were submitted to the European Nucleotide Archive (accession number PRJEB15203).
Funding Statement
This work was funded by UK Medical Research Council (MC_UU_12014) (AK, ES), Wellcome Trust (099220/B/12/Z) (RME), UK Biotechnology and Biological Sciences Research Council (BBSRC BB/J004324/1) (MW), Swedish Research Council (grant no. 2012–6555) (ALB) and the ERA-Net EMIDA RiftVectors project (contract no. 219235) (AK). Edinburgh Genomics is partly supported by UK MRC (MR/K001744/1), UK BBSRC (BB/J004243/1) and UK NERC (R8/H10/56) (MW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Elliott RM. Orthobunyaviruses: recent genetic and structural insights. Nature reviews Microbiology. 2014;12(10):673–85. Epub 2014/09/10. 10.1038/nrmicro3332 [DOI] [PubMed] [Google Scholar]
- 2.Hubalek Z, Rudolf I, Nowotny N. Arboviruses pathogenic for domestic and wild animals. Adv Virus Res. 2014;89:201–75. 10.1016/B978-0-12-800172-1.00005-7 [DOI] [PubMed] [Google Scholar]
- 3.Smithburn KC, Haddow AJ, Mahaffy AF. A neurotropic virus isolated from Aedes mosquitoes caught in the Semliki forest. Am J Trop Med Hyg. 1946;26:189–208. [DOI] [PubMed] [Google Scholar]
- 4.Berge TO. International Catalogue of Arboviruses: Including Certain Other Viruses of Vertebrates In: American Committee on Arthropod Borne Viruses Subcommittee on Information Exchange CfDC, editor. 2 ed: Public Health Service; 1975. [Google Scholar]
- 5.Tauro LB, Batallan GP, Rivarola ME, Visintin A, Berron CI, Sousa EC Jr., et al. Detection of Orthobunyavirus in mosquitoes collected in Argentina. Med Vet Entomol. 2015;29(3):338–43. 10.1111/mve.12121 [DOI] [PubMed] [Google Scholar]
- 6.Wertheim HFL, Horby P, John P. Woodall JP. Atlas of Human Infectious Diseases: Wiley-Blackwell; 2012. [Google Scholar]
- 7.Calisher CH, Francy DB, Smith GC, Muth DJ, Lazuick JS, Karabatsos N, et al. Distribution of Bunyamwera serogroup viruses in North America, 1956–1984. Am J Trop Med Hyg. 1986;35(2):429–43. [DOI] [PubMed] [Google Scholar]
- 8.Grimstad PR. Cache Valley virus In: Service MW, editor. Encyclopedia of Arthropod-transmitted Infections of Man and Domestic Animals. New York: CABI Publishing; 2001. p. 101–4. [Google Scholar]
- 9.Andreadis TG, Armstrong PM, Anderson JF, Main AJ. Spatial-temporal analysis of Cache Valley virus (Bunyaviridae: Orthobunyavirus) infection in anopheline and culicine mosquitoes (Diptera: Culicidae) in the northeastern United States, 1997–2012. Vector Borne Zoonotic Dis. 2014;14(10):763–73. 10.1089/vbz.2014.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ngo KA, Maffei JG, Dupuis AP 2nd, Kauffman EB, Backenson PB, Kramer LD. Isolation of Bunyamwera serogroup viruses (Bunyaviridae, Orthobunyavirus) in New York state. J Med Entomol. 2006;43(5):1004–9. [DOI] [PubMed] [Google Scholar]
- 11.Campbell GL, Mataczynski JD, Reisdorf ES, Powell JW, Martin DA, Lambert AJ, et al. Second human case of Cache Valley virus disease. Emerg Infect Dis. 2006;12(5):854–6. 10.3201/eid1205.051625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sexton DJ, Rollin PE, Breitschwerdt EB, Corey GR, Myers SA, Dumais MR, et al. Life-threatening Cache Valley virus infection. N Engl J Med. 1997;336(8):547–9. 10.1056/NEJM199702203360804 [DOI] [PubMed] [Google Scholar]
- 13.Nguyen NL, Zhao G, Hull R, Shelly MA, Wong SJ, Wu G, et al. Cache valley virus in a patient diagnosed with aseptic meningitis. J Clin Microbiol. 2013;51(6):1966–9. 10.1128/JCM.00252-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Concha-Bermejillo de la A. Cache Valley Virus is a cause of fetal malformation and pregnancy loss in sheep. Small Ruminant Res. 2003;49:1–9. [Google Scholar]
- 15.Edwards JF, Livingston CW, Chung SI, Collisson EC. Ovine arthrogryposis and central nervous system malformations associated with in utero Cache Valley virus infection: spontaneous disease. Vet Pathol. 1989;26(1):33–9. [DOI] [PubMed] [Google Scholar]
- 16.Chung SI, Livingston CW Jr., Edwards JF, Gauer BB, Collisson EW. Congenital malformations in sheep resulting from in utero inoculation of Cache Valley virus. Am J Vet Res. 1990;51(10):1645–8. [PubMed] [Google Scholar]
- 17.Wernike K, Jost H, Becker N, Schmidt-Chanasit J, Beer M. Lack of evidence for the presence of Schmallenberg virus in mosquitoes in Germany, 2011. Parasit Vectors. 2014;7:402 10.1186/1756-3305-7-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elbers AR, Meiswinkel R, van Weezep E, Kooi EA, van der Poel WH. Schmallenberg Virus in Culicoides Biting Midges in the Netherlands in 2012. Transbound Emerg Dis. 2015;62(3):339–42. 10.1111/tbed.12128 [DOI] [PubMed] [Google Scholar]
- 19.Larska M, Lechowski L, Grochowska M, Zmudzinski JF. Detection of the Schmallenberg virus in nulliparous Culicoides obsoletus/scoticus complex and C. punctatus—the possibility of transovarial virus transmission in the midge population and of a new vector. Vet Microbiol. 2013;166(3–4):467–73. 10.1016/j.vetmic.2013.07.015 [DOI] [PubMed] [Google Scholar]
- 20.Mercer DR, Castillo-Pizango MJ. Changes in relative species compositions of biting midges (Diptera: Ceratopogonidae) and an outbreak of Oropouche virus in Iquitos, Peru. J Med Entomol. 2005;42(4):554–8. [DOI] [PubMed] [Google Scholar]
- 21.Mercer DR, Spinelli GR, Watts DM, Tesh RB. Biting rates and developmental substrates for biting midges (Diptera: Ceratopogonidae) in Iquitos, Peru. J Med Entomol. 2003;40(6):807–12. [DOI] [PubMed] [Google Scholar]
- 22.Kato T, Shirafuji H, Tanaka S, Sato M, Yamakawa M, Tsuda T, et al. Bovine Arboviruses in Culicoides Biting Midges and Sentinel Cattle in Southern Japan from 2003 to 2013. Transbound Emerg Dis. 2015. [DOI] [PubMed] [Google Scholar]
- 23.Hubalek Z, Rudolf I. Tick-borne viruses in Europe. Parasitol Res. 2012;111(1):9–36. Epub 2012/04/25. 10.1007/s00436-012-2910-1 [DOI] [PubMed] [Google Scholar]
- 24.Marklewitz M, Zirkel F, Kurth A, Drosten C, Junglen S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(24):7536–41. 10.1073/pnas.1502036112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marklewitz M, Zirkel F, Rwego IB, Heidemann H, Trippner P, Kurth A, et al. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J Virol. 2013;87(23):12850–65. 10.1128/JVI.01862-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffmann B, Scheuch M, Hoper D, Jungblut R, Holsteg M, Schirrmeier H, et al. Novel orthobunyavirus in cattle, europe, 2011. Emerg Infect Dis. 2012;18(3):469–72. Epub 2012/03/02. 10.3201/eid1803.111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lievaart-Peterson K, Luttikholt S, Peperkamp K, Van den Brom R, Vellema P. Schmallenberg disease in sheep or goats: Past, present and future. Vet Microbiol. 2015;181(1–2):147–53. 10.1016/j.vetmic.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 28.van den Brom R, Luttikholt SJ, Lievaart-Peterson K, Peperkamp NH, Mars MH, van der Poel WH, et al. Epizootic of ovine congenital malformations associated with Schmallenberg virus infection. Tijdschr Diergeneeskd. 2012;137(2):106–11. [PubMed] [Google Scholar]
- 29.Dandawate CN, Rajagopalan PK, Pavri KM, Work TH. Virus isolations from mosquitoes collected in North Arcot district, Madras state, and Chittoor district, Andhra Pradesh between November 1955 and October 1957. Indian J Med Res. 1969;57(8):1420–6. [PubMed] [Google Scholar]
- 30.Causey OR, Kemp GE, Causey CE, Lee VH. Isolations of Simbu-group viruses in Ibadan, Nigeria 1964–69, including the new types Sango, Shamonda, Sabo and Shuni. Ann Trop Med Parasitol. 1972;66(3):357–62. [DOI] [PubMed] [Google Scholar]
- 31.Lee VH. Isolation of viruses from field populations of culicoides (Diptera: Ceratopogonidae) in Nigeria. J Med Entomol. 1979;16(1):76–9. [DOI] [PubMed] [Google Scholar]
- 32.Yanase T, Fukutomi T, Yoshida K, Kato T, Ohashi S, Yamakawa M, et al. The emergence in Japan of Sathuperi virus, a tropical Simbu serogroup virus of the genus Orthobunyavirus. Arch Virol. 2004;149(5):1007–13. 10.1007/s00705-003-0266-7 [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen LD, Kristensen B, Kirkeby C, Rasmussen TB, Belsham GJ, Bodker R, et al. Culicoids as vectors of schmallenberg virus. Emerg Infect Dis. 2012;18(7):1204–6. Epub 2012/06/20. 10.3201/eid1807.120385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbers AR, Meiswinkel R, van Weezep E, Sloet van Oldruitenborgh-Oosterbaan MM, Kooi EA. Schmallenberg virus in Culicoides spp. biting midges, the Netherlands, 2011. Emerg Infect Dis. 2013;19(1):106–9. 10.3201/eid1901.121054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Regge N, Deblauwe I, De Deken R, Vantieghem P, Madder M, Geysen D, et al. Detection of Schmallenberg virus in different Culicoides spp. by real-time RT-PCR. Transbound Emerg Dis. 2012;59(6):471–5. 10.1111/tbed.12000 [DOI] [PubMed] [Google Scholar]
- 36.Blair CD, Olson KE. The role of RNA interference (RNAi) in arbovirus-vector interactions. Viruses. 2015;7(2):820–43. 10.3390/v7020820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donald CL, Kohl A, Schnettler E. New insights into control of arbovirus replication and spread by insect RNA interference pathways. Insects. 2012;3(2):511–31. 10.3390/insects3020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8(1):e1002470 Epub 2012/01/14. 10.1371/journal.ppat.1002470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leger P, Lara E, Jagla B, Sismeiro O, Mansuroglu Z, Coppee JY, et al. Dicer-2- and Piwi-mediated RNA interference in Rift Valley fever virus-infected mosquito cells. J Virol. 2013;87(3):1631–48. 10.1128/JVI.02795-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnettler E, Ratinier M, Watson M, Shaw AE, McFarlane M, Varela M, et al. RNA interference targets arbovirus replication in culicoides cells. J Virol. 2013;87(5):2441–54. Epub 2012/12/28. 10.1128/JVI.02848-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4(10):e848 Epub 2010/11/05. 10.1371/journal.pntd.0000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19938–43. Epub 2008/12/03. 10.1073/pnas.0803408105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17240–5. 10.1073/pnas.0406983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5(7):e1000502 Epub 2009/07/07. 10.1371/journal.ppat.1000502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 2009;9:49 10.1186/1471-2180-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, et al. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiol. 2008;8:47 10.1186/1471-2180-8-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnettler E, Donald CL, Human S, Watson M, Siu RW, McFarlane M, et al. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J Gen Virol. 2013;94(Pt 7):1680–9. Epub 2013/04/06. 10.1099/vir.0.053850-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–4. Epub 2006/07/01. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- 49.Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, et al. The genetic makeup of the Drosophila piRNA pathway. Mol Cell. 2013;50(5):762–77. 10.1016/j.molcel.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12(1):45–55. 10.1016/j.devcel.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 51.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315(5818):1587–90. Epub 2007/02/27. 10.1126/science.1140494 [DOI] [PubMed] [Google Scholar]
- 52.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. Epub 2007/03/10. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- 53.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137(3):509–21. 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–35. 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vodovar N, Bronkhorst AW, van Cleef KW, Miesen P, Blanc H, van Rij RP, et al. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One. 2012;7(1):e30861 Epub 2012/02/01. 10.1371/journal.pone.0030861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.George P, Jensen S, Pogorelcnik R, Lee J, Xing Y, Brasset E, et al. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin. 2015;8:50 10.1186/s13072-015-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castellano L, Rizzi E, Krell J, Di Cristina M, Galizi R, Mori A, et al. The germline of the malaria mosquito produces abundant miRNAs, endo-siRNAs, piRNAs and 29-nt small RNAs. BMC Genomics. 2015;16:100 10.1186/s12864-015-1257-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell CL, Black WCt, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9(1):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miesen P, Girardi E, van Rij RP. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic acids research. 2015;43(13):6545–56. 10.1093/nar/gkv590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miesen P, Ivens A, Buck AH, van Rij RP. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl Trop Dis. 2016;10(2):e0004452 10.1371/journal.pntd.0004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siu RW, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol. 2011;85(6):2907–17. Epub 2010/12/31. 10.1128/JVI.02052-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Attarzadeh-Yazdi G, Fragkoudis R, Chi Y, Siu RW, Ulper L, Barry G, et al. Cell-to-cell spread of the RNA interference response suppresses Semliki Forest virus (SFV) infection of mosquito cell cultures and cannot be antagonized by SFV. J Virol. 2009;83(11):5735–48. 10.1128/JVI.02440-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi X, Botting CH, Li P, Niglas M, Brennan B, Shirran SL, et al. Bunyamwera orthobunyavirus glycoprotein precursor is processed by cellular signal peptidase and signal peptide peptidase. Proceedings of the National Academy of Sciences. 2016;113(31):8825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi X, Kohl A, Léonard V, Li P, McLees A, Elliott R. Requirement of the N-terminal region of the orthobunyavirus non-structural protein NSm for virus assembly and morphogenesis. J Virol. 2006;80(16):8089–99. 10.1128/JVI.00579-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flor TB, Blom B. Pathogens Use and Abuse MicroRNAs to Deceive the Immune System. Int J Mol Sci. 2016;17(4):538 10.3390/ijms17040538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma N, Singh SK. Implications of non-coding RNAs in viral infections. Rev Med Virol. 2016;26(5):356–68. 10.1002/rmv.1893 [DOI] [PubMed] [Google Scholar]
- 67.Asgari S. Role of microRNAs in arbovirus/vector interactions. Viruses. 2014;6(9):3514–34. 10.3390/v6093514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goic B, Stapleford KA, Frangeul L, Doucet AJ, Gausson V, Blanc H, et al. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat Commun. 2016;7:12410 10.1038/ncomms12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73(1):251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arnaud F, Black SG, Murphy L, Griffiths DJ, Neil SJ, Spencer TE, et al. Interplay between ovine bone marrow stromal cell antigen 2/tetherin and endogenous retroviruses. J Virol. 2010;84(9):4415–25. 10.1128/JVI.00029-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(26):15400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(2):664–9. 10.1073/pnas.98.2.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, et al. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLos Pathogens. 2013;9(1):e1003133 Epub 2013/01/18. 10.1371/journal.ppat.1003133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7(11):1848–57. 10.1021/cb3002478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330(2):493–500. 10.1016/j.virol.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 76.Shi X, Elliott RM. Analysis of N-linked glycosylation of Hantaan virus glycoproteins and the role of oligosaccharide side chains in protein folding and intracellular trafficking. J Virol. 2004;78(10):5414–22. 10.1128/JVI.78.10.5414-5422.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watret GE, Pringle CR, Elliott RM. Synthesis of Bunyavirus-specific Proteins in a Continuous Cell Line (XTC-2) Derived from Xenopus laevis. Journal of General Virology. 1985;66(3):473–82. [DOI] [PubMed] [Google Scholar]
- 78.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, et al. Schmallenberg Virus Pathogenesis, Tropism and Interaction with the Innate Immune System of the Host. PLoS Pathog. 2013;9(1):e1003133 10.1371/journal.ppat.1003133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber F, Dunn EF, Bridgen A, Elliott RM. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology. 2001;281(1):67–74. 10.1006/viro.2000.0774 [DOI] [PubMed] [Google Scholar]
- 80.Watson M, Schnettler E, Kohl A. viRome: an R package for the visualization and analysis of viral small RNA sequence datasets. Bioinformatics. 2013;29(15):1902–3. 10.1093/bioinformatics/btt297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McFarlane M, Arias-Goeta C, Martin E, O'Hara Z, Lulla A, Mousson L, et al. Characterization of Aedes aegypti Innate-Immune Pathways that Limit Chikungunya Virus Replication. PLoS Negl Trop Dis. 2014;8(7):e2994 10.1371/journal.pntd.0002994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Size distribution of SBV-specific small RNAs in Ae. aegypti-derived Aag2 cells at 48 hours p.i. (A) or C. sonorensis KC cells (B) at 24 hours p.i. The y-axis indicates read frequency; the x-axis indicates the length of the small RNAs (nt). Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome of L, M and S segments.
(TIF)
(A) Size distribution of BUNV-specific small RNAs. The y axis indicates read frequency; the x axis indicates the length of the small RNAs (nt). Red indicates the small RNAs mapping to the antigenome and green the small RNAs mapping to the genome. (B) Relative nucleotide frequency and conservation per position of 24–30 nt small RNAs mapping to the genome or antigenome of BUNV S, M and L segments. Sequence is represented as DNA. (C) Frequency distribution of the 21 nt and 24–30 nt BUNV-specific small RNAs across the antigenome (positive numbers and red) from the 5` to 3` and genome (negative numbers and green) from the 3` to 5`.
(TIF)
(A) Construction of TVT7BUNM-NL in which the coding region of the NSm cytoplasmic tail (residues 395 to 455) was replaced by that of Nano luciferase (NL); BUN-NL GPC was cleaved into Gn, Gc, and NSm-NL chimeric protein. The fused NL is shown in orange, signal peptide (sp) in the grey box and transmembrane domain (TM) in the black box. The amino acid positions at the boundary of each protein are marked on top of Wt BUNV GPC. (B) Comparison of protein profiles of BUNV and BUNV-NL. BSR-T7/5 cells were infected with BUNV and BUNV-NL at MOI of 0.5 and labelled with [35S]methionine at 24 hours p.i for 20 hours. Viral proteins were precipitated with anti-BUNV antibody and analysed by 12.5% SDS-PAGE tris-glycine gel under reducing conditions. Positions of viral proteins are indicated. (C) Comparison of plaque phenotypes of BUNV and BUNV-NL on Vero E6 cells. Cells were fixed with 4% formaldehyde-PBS and stained with 0.1% crystal violet blue solution. (D) Immunofluorescence of Aag2 cells infected with either BUNV-NL or SBV-NL at MOI 0.01 at 48 hours p.i. Anti-BUNV or anti-SBV N primary antibody, followed by an anti-rabbit Alexa Fluor 488-conjugated secondary antibody (green) and nucleic acid staining with Dapi (blue) was used.
(TIF)
(A) Aag2 cells were infected with CVV (MOI 1), SBV, or SATV (MOI 0.01) and culture supernatants were harvested at different time points p.i. as indicated. Viral titres were determined by plaque assays on BHK-21 cells (CVV) or CPT-Tert cells (SBV, SATV). Graphs represent one experiment performed in triplicate. Error bars represent standard errors of the means (SE). (B) Aag2 cells were transfected with dsRNA either specifically to Piwi4 or eGFP as control, followed by CVV infection at 24 hours p.t. Images of cells shown were taken at 48 hours p.i using the EVOS FL Cell Imaging System.
(TIF)
The actual numbers of small RNA reads obtained in each experiment performed and analysed in this study are outlined in the table. The total number of reads against each virus segment as well as the number of reads of each size for each repeat is shown.
(XLSX)
(DOC)
Data Availability Statement
Small RNA sequences were submitted to the European Nucleotide Archive (accession number PRJEB15203).