Abstract
Polarized membrane trafficking during plant cytokinesis and cell expansion are critical for plant morphogenesis, yet very little is known about the molecular mechanisms that guide this process. Dynamin and dynamin-related proteins are large GTP binding proteins that are involved in membrane trafficking. Here, we show that two functionally redundant members of the Arabidopsis dynamin-related protein family, ADL1A and ADL1E, are essential for polar cell expansion and cell plate biogenesis. adl1A-2 adl1E-1 double mutants show defects in cell plate assembly, cell wall formation, and plasma membrane recycling. Using a functional green fluorescent protein fusion protein, we show that the distribution of ADL1A is dynamic and that the protein is localized asymmetrically to the plasma membrane of newly formed and mature root cells. We propose that ADL1-mediated membrane recycling is essential for plasma membrane formation and maintenance in plants.
INTRODUCTION
Dynamin and dynamin-related proteins constitute a structurally related but functionally diverse family of large GTP binding proteins (for review, see van der Bliek, 1999; McNiven et al., 2000) that share a conserved N-terminal GTPase domain and a C-terminal GTPase effector domain. A wide range of cellular processes involve dynamin and dynamin-related proteins. Dynamin has been shown to constrict membranes and to facilitate the release of vesicles from the plasma membrane (PM) and the Golgi apparatus. In addition, dynamins and dynamin-related proteins function at other stages of the secretory pathway (reviewed by van der Bliek, 1999; McNiven et al., 2000), in actin cytoskeletal dynamics (Ochoa et al., 2000; Lee and De Camilli, 2002; Orth et al., 2002), and in the maintenance of mitochondrial and peroxisome morphology and inheritance (Otsuga et al., 1998; Smirnova et al., 1998; Hoepfner et al., 2001).
Three dynamin-related protein subfamilies have been identified in Arabidopsis. One of these, which consists of the Arabidopsis Dynamin-Like Protein6 (ADL6) gene product, is highly related to mammalian dynamin I and has been shown to be required for trans-Golgi–to–vacuole trafficking (Jin et al., 2001). Two other Arabidopsis dynamin subfamilies, ADL1 and ADL2, appear to be unique to plants (van der Bliek, 1999). These dynamin-related proteins lack the pleckstrin and Pro-rich domains characteristic of dynamin I and are likely to perform plant-specific functions. ADL2A and ADL2B constitute the ADL2 subfamily. ADL2A is targeted to plastids (Kang Shin et al., 1998), suggesting that it may function in chloroplast/plastid biogenesis and maintenance. ADL2B localizes to the constriction site of dividing mitochondria, and expression of a dominant-negative form resulted in the fusion and tubulation of mitochondria, indicating its involvement in mitochondria division (Arimura and Tsutsumi, 2002).
The largest plant-specific dynamin subfamily consists of the 68-kD dynamin-related proteins: the ADL1 multigene family in Arabidopsis and phragmoplastin in soybean. The ADL1 gene family is composed of five highly related members, ADL1A to ADL1E (Kang et al., 2001). Localization studies have shown that phragmoplastin and members of the ADL1 family are associated with the developing cell plate during somatic and syncytial plant cytokinesis (Gu and Verma, 1996, 1997; Lauber et al., 1997; Kang et al., 2001; Otegui et al., 2001). Like other dynamins and dynamin-related proteins (Nakayama et al., 1993; Hinshaw and Schmid, 1995; Zhang et al., 2000; Kim et al., 2001; Klockow et al., 2002), phragmoplastin and ADL1A have been shown to polymerize into helical structures (Zhang et al., 2000; Otegui et al., 2001). ADL1A rings were found to encircle constricted regions of “nonconventional” syncytial-type cell plates during endosperm cellularization in developing Arabidopsis seeds (Otegui et al., 2001); however, the function of these structures is unknown.
Previously, we isolated complete loss-of-function adl1A mutants (Kang et al., 2001). Analysis of these mutants confirmed that ADL1A is critical for normal embryogenesis, seedling development, and reproduction. Here, we describe the further characterization of ADL1A and another member of the ADL1 family, ADL1E, and demonstrate that these dynamin-related proteins are required for the de novo assembly and maintenance of the PM during cell plate formation and cell expansion.
RESULTS
Expression of ADL1A and ADL1E Reporter Fusions in Transgenic Plants
We demonstrated previously that Arabidopsis plants express multiple ADL1 isoforms (Kang et al., 2001). Development of adl1A mutant seedlings was found to be dependent on exogenous metabolizable sugars (e.g., sucrose or glucose). However, ADL1A was not essential for further vegetative growth once the sucrose-rescued adl1A mutants became photosynthetically active. These observations suggested that another functionally redundant member of the ADL1 family could compensate for the loss of ADL1A. Consistent with this finding, RNA gel blot analysis demonstrated that ADL1A, ADL1C, and ADL1E, but not ADL1B and ADL1D, were expressed at similar levels throughout the plant, including roots, leaves, stems, and flowers (data not shown). However, further studies indicated that adl1C plants have a unique phenotype and that the gene is expressed in a cell type–specific manner distinct from the expression of ADL1A and ADL1E. These results are reported elsewhere (Kang et al., 2003).
To examine the cell- and tissue-specific expression patterns of ADL1A and ADL1E, we generated transgenic plants that express translational fusion constructs containing either the ADL1A or ADL1E promoter and the bacterial β-glucuronidase (GUS) reporter gene UID. The ADL1A and ADL1E promoter sequences used in the GUS fusion constructs were sufficient to control the expression of ADL1A (Kang et al., 2001) and ADL1E (data not shown) genomic DNA for molecular complementation experiments. Progeny from eight independent ADL1-GUS transgenic plant lines were used for histochemical analysis of GUS activity (Figure 1). Untransformed lines and transgenic lines harboring GUS but lacking the ADL1 promoters did not show any GUS activity (data not shown).
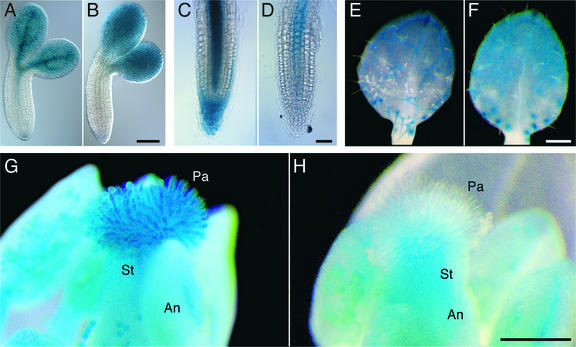
Figure 1.
Histochemical Localization of ADL1A-GUS and ADL1E-GUS Reporter Gene Expression.
(A) and (B) Torpedo-stage embryos.
(C) and (D) Three-day-old seedling root tips.
(E) and (F) Primary leaves of 7-day-old seedlings showing three-branched trichomes.
(G) and (H) Mature flowers (stage 14).
Processing of ADL1A-GUS ([A], [C], [E], and [G]) and ADL1E-GUS ([B], [D], [F], and [H]) tissue samples for the analysis of GUS activity at each developmental stage was performed in a pair-wise fashion. An, anther; Pa, stigmatic papillae cells; St, stigma. Bars = 100 μm in (B) and (D) and 250 μm in (F) and (H).
ADL1A-GUS and ADL1E-GUS showed overlapping but distinct expression profiles during embryogenesis and plant development (Figure 1). As early as the heart stage of development, ADL1A-GUS activity was detected in the suspensor and hypophysis (data not shown). At later stages of development, strong GUS staining was observed in ADL1A-GUS embryos, extending from the cotyledons into the hypocotyl procambium (Figure 1A). The ADL1A-GUS stain in the embryonic axis became progressively weaker toward the root tip, reappearing in the hypophysis. By contrast, ADL1E-GUS staining was observed in the cotyledons and weakly in the root tip (Figure 1B). After germination, however, both ADL1A-GUS and ADL1E-GUS were expressed in the root provascular tissue (Figures 1C and 1D). Strong ADL1A-GUS expression also was observed in the lateral and columella root cap cells (Figure 1C), which are derived from the hypophysis (Dolan et al., 1993), whereas no staining was noted for ADL1E-GUS in the root cap (Figure 1D). Throughout early flower development, we observed weak GUS staining in the anthers and styles of ADL1A-GUS and ADL1E-GUS transgenic plants. However, the specialized epidermal cells of the stigma, known as papillae, exhibited strong ADL1A-GUS expression (Figure 1G). These cells serve as attachment sites for pollen and are required for successful fertilization (Thorsness et al., 1993). As floral structures mature, the stigmatic papillae expand anisotropically and reach full length by the time of pollen sac dehiscence. ADL1A-GUS expression correlated with papillae elongation. GUS activity was observed only just before and during papillae elongation. No GUS staining was detected in the papillae after flower maturation and bud opening (data not shown). In contrast to ADL1A expression, ADL1E-GUS expression was not detected in expanding or mature papillae (Figure 1H).
Defective Anisotropic Expansion of adl1A Stigmatic Papillae
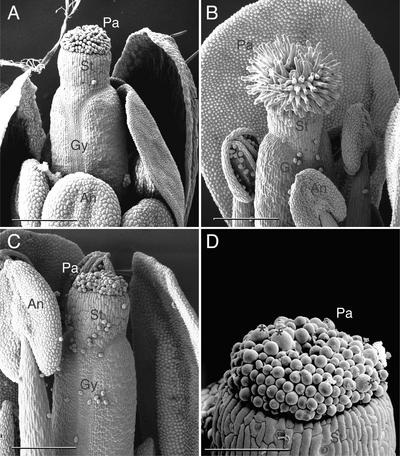
Although sucrose-rescued adl1A mutants display normal vegetative growth, they are highly infertile as a result of a maternal sporophytic defect in the mutant flowers that severely inhibits fertilization (Kang et al., 2001). We reasoned that the maternal defect could result from problems in the mutant adl1A-2 carpel that would disrupt normal pollen tube germination and/or growth to the ovule. Thus, we reexamined gynoecium development in wild-type and adl1A-2 flowers by scanning electron microscopy. Based on morphological criteria, the development of Arabidopsis flowers is divided into 20 stages, beginning with the appearance of the floral primordium and ending with seed release (Smyth et al., 1990). Stigmatic papillae are observed first in stage-11 or -12 unopened flowers, in which the sepals completely enclose the other three floral organs (Figure 2A). As the flower buds mature and undergo anthesis, individual papillae expand anisotropically (Figure 2B). Mature wild-type papillae have a flask-shaped appearance and are receptive to pollen grains. By contrast, the papillae of adl1A-2 flowers failed to elongate (Figure 2C) and frequently displayed abnormal isotropic expansion, giving rise to large, bloated cells (Figure 2D).
Figure 2.
Scanning Electron Microscopy Analysis of Wild-Type and adl1A-2 Flowers.
(A) Dissected stage-12 wild-type flower. Anthers have not dehisced at this stage.
(B) Late stage-15 wild-type flower exhibiting dehisced anthers. The gynoecium has begun to elongate rapidly at this stage, presumably as a result of successful self-fertilization.
(C) Infertile late stage-16 adl1A-2 flower. The gynoecium has failed to lengthen.
(D) Close-up view of adl1A-2 mutant papillae cells showing abnormal isotropically expanded cells (asterisks).
An, anther; Gy, gynoecium; Pa, stigmatic papillae cells; St, stigma. Bars = 250 μm in (A) to (C) and 100 μm in (D).
Before elongation, wild-type and adl1A-2 mutant papillae were densely cytoplasmic and appeared morphologically similar by thin section transmission electron microscopy (data not shown). As the flowers matured, wild-type and adl1A-2 papillae cells became more vacuolate and the number of Golgi stacks increased significantly. The vacuoles in wild-type papillae were consistently larger and occupied a greater proportion of the cell volume than in the adl1A-2 mutant.
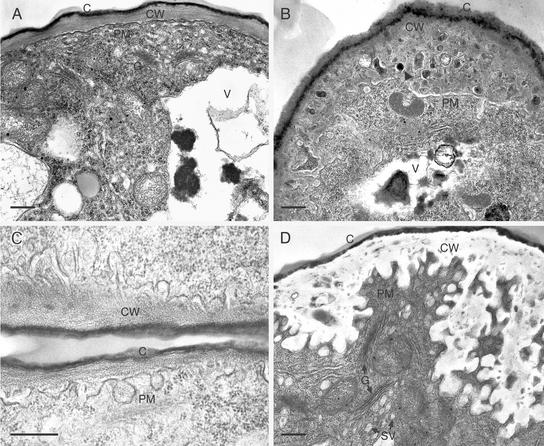
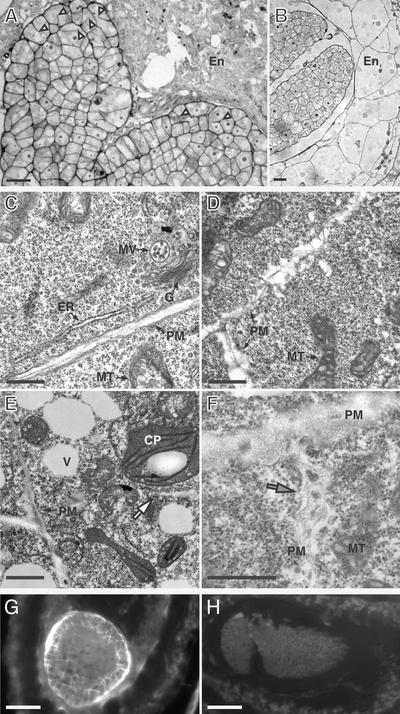
The most prominent difference between wild-type and adl1A-2 papillae cells was in the appearance of highly elaborated PM infoldings during mutant flower maturation (Figure 3). Wild-type cells were devoid of abnormal ingrowths, with the PM being appressed to the cell wall throughout papillae expansion (Figure 3A). Minor infoldings and thickened cell walls were observed in the youngest mutant flowers examined (stage 11) and were located primarily near the distal region of the adl1A papillae (Figures 3B and 3C). By the time that flowers had opened and reached stage 15 of development, the ingrowths were found along the entire length of the cell, extending deep into the cytoplasm (Figure 3D). The PM elaborations at the base of the mutant papillar cells, near their junction with the style, were not as extensive as those observed at the distal tip of the mutant cells. In addition to the PM defects, the electron-dense cell wall material, which was present on the extracellular face of the PM in the papillae of young adl1A-2 flowers (Figure 3B), was absent in the mature mutant papillae (Figure 3D), suggesting that the wall material had been degraded or that its rate of synthesis had been reduced severely. Dark inclusions were present within the cell wall matrices of young adl1A-2 papillae (Figure 3B). Numerous Golgi stacks and possible secretory vesicles were observed near the PM of adl1A-2 papillae during flower maturation (Figure 3D). Thus, the accumulation of PM most likely represents a net gain of membrane in cells lacking ADL1A.
Figure 3.
Transmission Electron Micrographs of Developing Wild-Type and adl1A-2 Papillae.
Longitudinal sections of developing and mature papillae cells are shown.
(A) Stigmatic papillae of a wild-type stage-15 flower.
(B) and (C) Stigmatic papillae of early stage-11 adl1A-2 flowers. A dark inclusion within the cell wall matrix is indicated by the arrowhead in (B).
(D) Stigmatic papillae of a late stage-15 adl1A-2 flower showing PM invaginations.
C, cuticle; CW, cell wall; G, Golgi stack; SV, putative secretory vesicle; V, vacuole. Bars = 5 μm in (A) and (B) and 500 nm in (C) and (D).
adl1A Affects Leaf Trichome Biogenesis
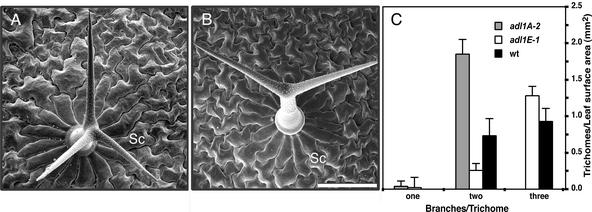
In addition to the effect on the polarized growth of papillae, adl1A-2 plants display defects in the development of leaf trichomes (leaf hairs). Arabidopsis leaf trichomes are unicellular structures that initially expand out of epidermal precursors and then form three or four branches (Figure 4A). Initiation and expansion of the third branch are suppressed completely in adl1A mutant trichomes (Figure 4B). No defects were observed in the formation and length of the trichome stalk, the orientation of the initial two branches, the surrounding socket cell morphology, or the total leaf trichome density (Figure 4C).
Figure 4.
Scanning Electron Microscopy Analysis of Wild-Type and adl1A Leaf Trichomes.
(A) and (B) Comparison of trichome branching in wild-type (A) and homozygous adl1A-2 (B) siblings from an adl1A-2/ADL1A heterozygous plant. Sc, socket cell. Bar = 100 μm.
(C) Trichome density and the number of branches per trichome were measured on the fourth leaves of wild-type and mutant plants grown under identical conditions. Data are means ± sd of three leaf samples (>125 trichomes per leaf).
The stages of trichome morphogenesis are approximately distributed (i.e., youngest to oldest) along the leaf surface in a base-to-tip direction (Hülskamp et al., 1994). Consistent with a role for ADL1A in trichome morphogenesis, GUS activity was detected in developing leaf trichomes and socket cells of ADL1A-GUS plants. ADL1A-GUS staining of trichomes was observed only during the early stages of development and not in mature leaf trichomes (Figure 1E). ADL1E-GUS staining of developing trichomes was restricted to the socket cells of expanding trichomes (Figure 1F). These results suggest that ADL1A function may be more critical than ADL1E function for trichome expansion and branching.
Identification and Characterization of an adl1E Loss-of-Function Mutant
In addition to displaying overlapping promoter expression patterns, ADL1A and ADL1E share significant amino acid sequence similarity (>80%) (Kang et al., 2001). To examine the role of ADL1E, we isolated an adl1E::T-DNA insertion line (adl1E-1) containing a T-DNA insert in the 14th exon (Figure 5). Homozygous adl1E-1 plants showed no discernible phenotypes and were completely fertile. Immunoblot analysis of total Arabidopsis seedling extracts from wild-type and adl1E-1 mutants using an anti-ADL1 GTPase domain–specific antibody (α-GTPase) (Kang et al., 2001) demonstrated that ADL1E encodes a 70-kD dynamin-related polypeptide and that adl1E-1 was a complete loss-of-function mutant. As shown in Figure 6A, α-GTPase recognized the 68-kD ADL1A polypeptide, which is absent in homozygous adl1A-2 mutants, and a 70-kD polypeptide in total protein extracts prepared from wild-type seedlings. Neither the 70-kD polypeptide nor any shorter ADL1E truncation products was detected in homozygous adl1E-1 plants (data not shown). AtCDC48 (Rancour et al., 2002) was used on immunoblots as a protein loading control.
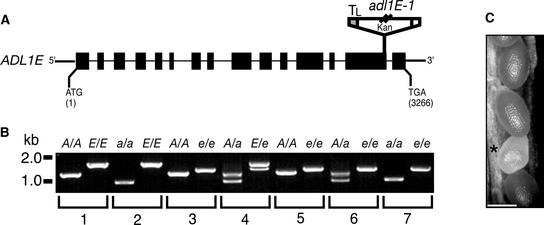
Figure 5.
Homozygous adl1A; E Mutants Display a Synthetic Embryo-Lethal Phenotype.
(A) Scheme of the deduced exon/intron structure of ADL1E. Black boxes and lines (drawn to scale) represent exons and introns, respectively. ATG and TGA signify the positions of the translation initiation and termination codons, respectively. The position and orientation of the T-DNA insert (not drawn to scale) in adl1E-1 is shown. Kan, T-DNA neomycin phosphotransferase selectable gene marker; TL, T-DNA left border.
(B) Genotypic analysis of the wild type and the adl1A-2, adl1E-1, and adl1A; E mutants. Total DNA was prepared from leaves (lanes 1 to 4) or isolated embryos (lanes 5 to 7) and analyzed by PCR using ADL1A–specific (A), adl1A-2–specific (a), ADL1E–specific (E), and adl1E-1–specific (e) primer sets. PCR products in wild-type [A/A; E/E] (lane 1), adl1A-2 [a/a; E/E] (lane 2), adl1E-1 [A/A; e/e] (lane 3), ADL1A/adl1A-2; ADL1E-1/adl1E-1 [A/a; E/e] (lane 4), ADL1A/ADL1A; adl1E-1/adl1E-1 [A/A; e/e] (lane 5), ADL1A/adl1A-2; adl1E-1/adl1E-1 [A/a; e/e] (lane 6), and adl1A-2/adl1A-2; adl1E-1/adl1E-1 [a/a; e/e] (lane 7) were generated from plants and embryos isolated from self-fertilized heterozygous ADL1A/adl1A-2; ADL1E-1/adl1E-1 plants.
(C) Portion of an immature silique (10 to 12 days after flowering) from an ADL1A/adl1A-2; adl1E-1/adl1E-1 plant. Genotypic analysis of genomic DNA prepared from six independent embryos isolated from the pale developing seeds (asterisk) indicated that they were homozygous adl1A; E double mutants, as shown in lane 7 of (B). Bar = 500 μm.
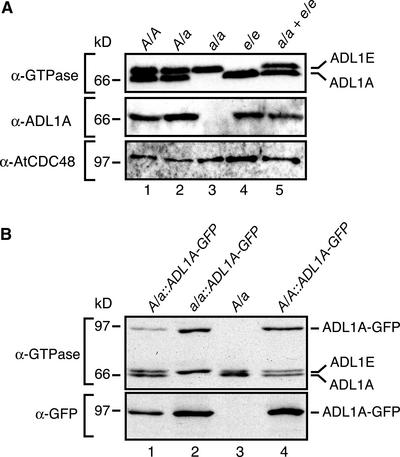
Figure 6.
Analysis of ADL1A, ADL1E, and ADL1A-mGFP5 Expression in Wild-Type and Mutant Plants.
(A) Immunoblot analysis of total protein prepared from rosette leaves of wild-type (lane 1), heterozygous adl1A-2 (lane 2), homozygous adl1A-2 (lane 3), and homozygous adl1E-1 (lane 4) plants. The relative mobilities of the ADL1 proteins in lanes 3 and 4 were confirmed by analysis of a protein mixture containing both homozygous adl1A-2 and homozygous adl1E-1 protein extracts (lane 5). The immunoblots were probed using α-GTPase, α-ADL1A, or α-AtCDC48 antibody. Abbreviations are as in Figure 5.
(B) Immunoblot analysis of transgenic lines expressing ADL1A-mGFP5. Total protein was prepared from rosette leaves of transgenic heterozygous adl1A-2 (lane 1), homozygous adl1A-2 (lane 2), and wild-type (lane 4) plants and untransformed heterozygous adl1A-2 plants (lane 3). The immunoblots were probed using α-GTPase or α-GFP antibody.
ADL1E Is Associated with the Cell Plate during Cytokinesis
To compare the subcellular distribution of ADL1A and ADL1E, we analyzed their localization by indirect immunofluorescence microscopy. Because ADL1E-specific antibodies were unavailable, we examined the localization of ADL1E in developing adl1A-2 null mutant roots using the α-GTPase antiserum (Kang et al., 2001), which detects ADL1A and ADL1E (Figure 6A) but not ADL1C (Kang et al., 2003). Expression of the 68-kD polypeptides encoded by ADL1B and ADL1D was not detected by the α-GTPase antiserum in homozygous adl1A-2 protein extracts (Figure 6A, lane 3), as expected based on the lack of detectable ADL1B and ADL1D mRNA in wild-type plants (Kang et al., 2003). Localization studies were performed in developing roots because of their high mitotic activity and amenability to processing for immunofluorescence microscopy. As shown previously (Kang et al., 2001), affinity-purified anti-ADL1A antibodies (α-ADL1A) strongly labeled the division plane in wild-type cells during cytokinesis (Figure 7A). In homozygous adl1A-2 roots, we observed only weak background staining, as expected for a loss-of-function mutant (data not shown). Like ADL1A, ADL1E was found to be associated with the cell plate in dividing adl1A-2 root cells immunolabeled with the α-GTPase antiserum (Figure 7B). ADL1A and ADL1E also were found by immunofluorescence microscopy to be associated with punctate subcellular structures. However, in certain cells, ADL1A and ADL1E were associated with the cell surface (Figures 7A, 7B, 7H, and 7I). Further analysis of the localization of ADL1A will be described below. The overlapping distribution of ADL1A and ADL1E in dividing and nondividing cells lends further support to our hypothesis that these proteins are functionally redundant.
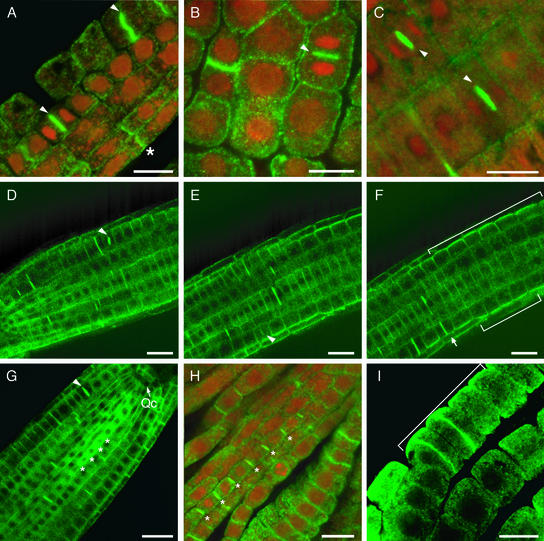
Figure 7.
Polarized Localization of ADL1A and ADL1A-mGFP5 in Arabidopsis Roots.
(A) Procambial root transition cells stained with affinity-purified α-ADL1A.
(B) Lateral root primordium of a homozygous adl1A-2 mutant seedling immunolabeled with affinity-purified GTPase-specific antibodies.
(C) ADL1A-mGFP5 in root tissue in chemically fixed homozygous adl1A-2 root cells.
(D) to (F) Three frames from the beginning (D), middle (E), and end (F) of the movie (see supplemental data online) showing the localization of ADL1A-mGFP5 in live homozygous adl1A-2 root cells. The arrow in (F) denotes the outer tangential surface of a dividing epidermal cell.
(G) ADL1A-mGFP5 localization in the root vascular procambium of a homozygous adl1A-2 seedling.
(H) Immunolocalization of ADL1A with α-ADL1A in the root vascular procambium.
(I) Immunolocalization of ADL1A with α-ADL1A in the outer tangential surface of epidermal cells in the root elongation zone.
ADL1 proteins are shown in green. Nuclei in (A) and (B) are shown in red. (A) to (G) show expanding cell plates in dividing root cells labeled with arrowheads. Asterisks in (A), (G), and (H) indicate ADL1 protein localization in transverse root cell surfaces. Brackets in (F) and (I) indicate the outer tangential surfaces of epidermal cells in the root elongation zone. Qc, quiescent center. Bars = 10 μm in (A), (B), (C), (H), and (I) and 20 μm in (D) to (G).
Defective Embryo Development of adl1A-2 adl1E-1 Double Mutants
To examine the genetic interaction between ADL1A and ADL1E, we generated a line that was heterozygous for adl1A-2 and homozygous for adl1E-1 (ADL1A/adl1A; adl1E/adl1E). Upon self-fertilization, no homozygous adl1A/adl1A adl1E/adl1E (adl1A; E) double mutant seedlings were recovered. Morphological and PCR genotype analysis of the developing seeds from ADL1A/adl1A; adl1E/adl1E plants confirmed that 25% of the progeny were homozygous embryo-lethal adl1A; E mutants (Figure 5B). Double mutant progeny were discernible from the phenotypically normal siblings by visual inspection of developing seeds (Figure 5C). Homozygous adl1A; E embryos were contained in pale developing seeds, which shriveled upon desiccation. Molecular complementation of the embryo-lethal defect of adl1A; E double mutants with genomic copies of ADL1A and ADL1E demonstrated that the phenotype was specific to the T-DNA–induced disruption of ADL1A and ADL1E (data not shown).
We compared the development of adl1A; E and phenotypically normal (ADL1A/adl1A; adl1E/adl1E or ADL1A/ADL1A; adl1E/adl1E) sibling embryos in the same silique to determine at what stage during development adl1A; E mutant embryos arrested (Figure 8). The development of adl1A; E embryos first deviated from normal during the late globular stage (Figures 8A and 8F). Mutant cells above the hypophysis, which give rise to the vascular procambium as well as to cortical cell layers, failed to expand anisotropically along the apical-basal embryo axis (Figure 8G, arrow), giving the mutant embryos a flattened and bloated appearance. At the heart stage, when wild-type embryos begin to assume their normal body plan by coordinated cell division and elongation, the mutant embryo phenotype became progressively more obvious (Figures 8G to 8I). The ordered cell files noted in wild-type embryos (Figures 8B to 8E) became increasingly disorganized in the adl1A; E embryos as they continued to develop. Unlike other embryo-defective mutants, such as knolle and keule, mature adl1A; E embryos were highly vacuolated (Figure 8J) and failed to germinate at maturity.
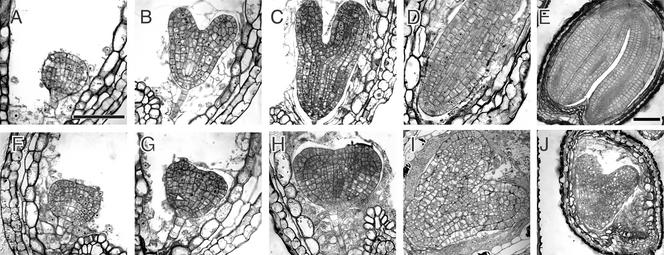
Figure 8.
Development of Homozygous adl1A; E Mutant Embryos.
Bright-field micrographs of histological sections of morphologically wild-type ([A] to [E]) and abnormal homozygous adl1A; E ([F] to [J]) seeds in the siliques of self-fertilized ADL1A/adl1A; adl1E/adl1E plants at various times during development. Embryos in (A) and (F), (B) and (G), (C) and (H), and (D) and (I) are sibling pairs from the same silique. Developmental stages relative to the wild type (Goldberg et al., 1994) are as follows: (A) and (F), globular; (B) and (G), heart; (C) and (H), torpedo; (D) and (I), walking stick; (E) and (J), mature embryo. Mutant embryos were indistinguishable from wild-type embryos before the globular stage. The arrow in (G) indicates mutant cells that failed to expand anisotropically. Bar in (A) = 50 μm for (A) to (D) and (F) to (I); bar in (E) = 100 μm for (E) and (J).
Loss of ADL1A and ADL1E Affects Cytokinesis and PM Morphology during Embryogenesis
adl1A; E embryos showed defects in cell plate formation. Analysis of plastic sections of developing adl1A; E embryos during the heart to late torpedo stages of development by bright-field microscopy revealed multinucleated cytokinesis-defective cells with partial and askew cell wall stubs (Figure 9A) in the rapidly dividing and expanding cotyledon protoderm (Mansfield and Briarty, 1991). As in other cytokinesis-defective mutants (Söllner et al., 2002), the cell wall stubs of adl1A; E protodermal cells were attached predominantly to one side of the cell with no stub on the opposing side. Cellularization of the syncytial endosperm also was affected in adl1A; E seeds. We have shown previously that ADL1A is associated with developing syncytial-type cell plates of endosperm (Otegui et al., 2001). Cellularization of the syncytial Arabidopsis endosperm was initiated at ∼60 h after fertilization (late globular stage) and was fully cellularized by the torpedo stage (Mansfield and Briarty, 1990) (Figures 8C and 9B). By contrast, the endosperms of seeds containing torpedo- and walking stick–stage adl1A; E embryos were highly amorphous (Figures 8H, 8I, and 9A), suggesting that syncytial-type cell plate formation was affected in the mutants.
Figure 9.
Cellular Morphology of Developing adl1A; E Embryos.
(A) and (B) Histological sections of developing mutant (A) and wild-type (B) seeds containing walking stick–stage embryos and endosperm. Arrowheads in (A) indicate cytokinesis-defective protodermal cells with cell wall stubs.
(C) to (F) Transmission electron micrographs of phenotypically wild-type (C) and mutant torpedo– (D) and walking stick–stage (E) and (F) embryonic cells. The white arrow in (E) indicates a translucent cell wall stub. The mutant PM is highly irregular and encloses a thickened diffuse cell wall that contains dense inclusions (gray arrow in [F]).
(G) and (H) Transverse sections of adl1A; E (G) and phenotypically wild-type (H) sibling embryos stained with the callose binding dye aniline blue.
CP, chloroplast; En, endosperm; ER, endoplasmic reticulum; G, Golgi apparatus; MT, mitochondria; MV, multivesicular body; PM, plasma membrane; V, vacuole. Bars = 10 μm in (A) and (B), 200 nm in (B) to (E), and 40 μm in (G) and (H).
Transmission electron microscopy analysis of adl1A; E embryo cells showed cellular changes similar to those observed in adl1A mutant papillae (Figures 9D to 9F). At the torpedo stage, the PM of phenotypically normal sibling embryo cells was flat (Figure 9C), whereas the PM of mutant cells was irregular. The PM, cell wall stubs, and new cross walls of adl1A; E were broad and translucent (Figures 9D to 9F). As shown in Figure 9G, the abnormal cell walls of adl1A; E embryos were characterized by the ectopic deposition of callose. When sections of mutant and wild-type embryos were stained with the UV light fluorescent callose binding dye aniline blue, we observed strong fluorescence in the cell walls of adl1A; E embryos (Figure 9G) but not wild-type embryos (Figure 9H). Consistent with the mutant phenotype, callose accumulation was highest in the cotyledon protoderm (Figure 9G). No defects in mitochondria, chloroplast, nuclei, or ER morphology were detected in the cells of heart- and torpedo-stage mutant embryos. These observation suggest that adl1A; E mutant embryos have defects in PM and cell wall formation and maintenance.
Polarized Localization of ADL1A in the PM of Root Cells
To further examine the role of ADL1A in de novo PM biogenesis and maintenance, we generated transgenic plants expressing an ADL1A-mGFP5 fusion protein. Previous studies have shown that overexpression of an N-terminal green fluorescent protein (GFP)–phragmoplastin fusion protein resulted in abnormal cell plate formation in transgenic tobacco suspension-cultured cells (Gu and Verma, 1997). Therefore, ADL1A-mGFP5 was expressed under the control of its native promoter in homozygous adl1A plants. The ADL1A-mGFP5 construct rescued all of the phenotypic defects associated with homozygous adl1A-2 mutants, demonstrating that the ADL1A-mGFP5 fusion protein was functional (data not shown). Immunoblot analysis of protein extracts prepared from ADL1A-mGFP5 seedlings confirmed that the 96-kD fusion protein was intact and expressed at levels comparable to those of native ADL1A in wild-type plants (Figure 6B). The distribution of ADL1A-mGFP5 in chemically fixed roots (Figure 7C) was very similar or identical to that observed for ADL1A (Figures 7A, 7H, and 7I) by whole-mount in situ immunostaining. ADL1A-mGFP5 was found to be associated predominantly with the developing cell plate during cytokinesis (Figures 7C to 7G; see also supplemental data online). In addition, ADL1A-mGFP5 expression was observed in all cells of the root cap, the root transition zone (also referred to as the distal elongation zone), and the elongation zone (Figures 7D to 7G; see also supplemental data online). No signal was observed in the root quiescent center (Figure 7G, arrow). Consistent with the expression profile of ADL1A-GUS (Figure 1A), the highest level of ADL1A-mGFP5 and ADL1A expression was in the vascular procambium (Figures 7G and 7H).
ADL1A and its soybean homolog, phragmoplastin, were shown previously to associate with small punctate intracellular structures in nondividing cells processed for indirect immunofluorescence microscopy (Gu and Verma, 1996; Kang et al., 2001) (Figures 7A, 7H, and 7I). By contrast, ADL1A-mGFP5 fluorescence strongly labeled the cell surface and exhibited diffuse cytoplasmic staining in most living root cells (Figures 7D to 7G; see also supplemental data online), suggesting that the distribution of ADL1A is affected by chemical fixation. Indeed, we observed a significant reduction in the intensity of cell surface–associated ADL1A-mGFP5 and the appearance of bright punctate intracellular structures similar to those observed by indirect immunofluorescence microscopy in ADL1A-mGFP5 seedlings fixed with 4% (w/v) paraformaldehyde (Figure 7C). Consistent with these results, ADL1C and ADL1E were identified recently in purified Arabidopsis leaf cell PM fractions by two-dimensional gel electrophoresis and mass spectroscopic analysis (Santoni et al., 2000; M.R. Sussman, personal communication).
The distribution of PM-associated ADL1A-mGFP5 in living root cells was found to be nonuniform and varied depending on root cell identity. In the cortex and endodermis, ADL1A-mGFP5 was localized preferentially to the transverse cell surfaces (Figures 7D to 7F; see also supplemental data online). Very strong labeling of the transverse surfaces of cell files within the vascular procambium of the root transition zone was observed in living cells (Figure 7G) and fixed cells processed for immunofluorescence microscopy (Figures 7A and 7H). In the epidermal cell files of the elongation zone, ADL1A-mGFP5 and ADL1A labeling was associated asymmetrically with the outer tangential cell surface (Figures 7F and 7I; see also supplemental data online)
Time-lapse imaging of growing roots that express ADL1A-mGFP5 revealed additional information about the dynamic distribution of ADL1A during cell plate development and new cell surface maturation. In dividing root cells, cell plates were observed frequently to be asymmetric, as described previously (Cutler and Ehrhardt, 2002), making initial contact with one side of the parental cell and then extending to the opposite side (see supplemental data online). ADL1A-mGFP5–specific fluorescence extended the length of the cell plate and was strongest at the leading edge(s). After fusion with the parental PM, the level of ADL1A-mGFP5 in the new cell surface gradually declined to basal levels (∼30 to 40 min after the completion of cytokinesis). In dividing epidermal root cells within the expansion zone, we observed a significant decrease in ADL1A-mGFP5 signal in the outer tangential surface PM during cell plate assembly (Figure 7F, arrow; see also supplemental data online), suggesting that the localization of ADL1A is dynamic and that it is redistributed during division to the developing cell plate. The polarized cell surface distribution of ADL1A, its persistence in newly formed cell surfaces, and the defects observed in the PM of rapidly expanding adl1A papillae suggest a role for ADL1A in membrane recycling.
DISCUSSION
ADL1A has been localized to conventional cytokinetic and syncytial-type endosperm cell plates (Kang et al., 2001; Otegui et al., 2001). Here, we show that ADL1A and another member of the ADL1 protein family, ADL1E, are functionally redundant and are essential for cell plate assembly. Cytokinesis-defective adl1A; E embryonic cells are multinucleate, often with cell wall stubs, indicative of arrested cell plate formation (Nacry et al., 2000). The cell stubs observed in the adl1A; E embryos likely are caused by a disruption in the assembly of asymmetrically oriented cell plates that we and others have observed (Cutler and Ehrhardt, 2002) (see supplemental data online). Endosperm cellularization, which is mediated by syncytial-type cell plates (Otegui et al., 2001), also is disrupted in mutant seeds. In addition to cytokinesis defects, cells of adl1A; E embryos fail to expand anisotropically and have distorted PM and cell walls. In marked contrast to knolle, keule, and several other recently identified Arabidopsis seedling-defective cytokinesis mutants (Assaad et al., 1996; Lukowitz et al., 1996; Söllner et al., 2002), adl1A; E mutant seeds fail to germinate. The defects observed closely match those in cyt1, an embryo-lethal cytokinesis-defective Arabidopsis mutant, and in embryos treated with the herbicide 2,6-dichlorobenzonitrile, both of which affect cellulose biosynthesis (Vaughn et al., 1996; Nickle and Meinke, 1998; Lukowitz et al., 2001). Therefore, adl1A; E embryo lethality may be related to more general problems in cell wall and PM integrity than defects in cell plate biogenesis. Similar defects in cell wall and PM structure were observed in adl1C mutant pollen grains, which collapse and arrest during desiccation (Kang et al., 2003). Together, these results support a vital role for the ADL1 family in PM maintenance and cell wall integrity in various developmental processes in Arabidopsis.
ADL1-Mediated Membrane Dynamics Is Required for Cell Expansion
During flower development, adl1A stigmatic papillae failed to expand anisotropically and accumulate PM in the mutant cells (Figures 3B to 3D). Morphometric studies have estimated that ≥75% of the total membrane incorporated into the PM of an expanding cell or cell plate during cytokinesis is recycled (Samuels and Bisalputra, 1990; Battey et al., 1999; Otegui et al., 2001). The phenotypes of adl1A papillae and adl1A; E embryos are consistent with a role for ADL1A in endocytic membrane recycling from the rapidly expanding cell surface and the developing cell plate in wild-type cells.
Membrane invaginations, similar to those observed in adl1A mutants, accumulate in the neurons and epithelial cells of the Drosophila mutant shibirets, a temperature-sensitive dynamin mutant required for the clathrin-mediated and non-clathrin-dependent endocytic process (McNiven et al., 2000). Clathrin-coated vesicles have been shown to be associated with the developing cell plate and PM in plants (Samuels et al., 1995; Holstein, 2002). ADL1A, however, lacks the critical pleckstrin homology and SH3 domain binding Pro-rich domains required for the interaction of mammalian dynamins with components of the clathrin-dependent vesicle-budding machinery. It remains to be determined whether ADL1A functions directly in membrane recovery from the PM and, if so, whether this process is a clathrin-dependent or clathrin-independent process. Other plant dynamin-like proteins, including a cell plate–associated plant dynamin that is related closely to animal dynamin I (Verma, 2001), may be required for clathrin-dependent membrane recycling.
How defects in ADL1A-mediated PM dynamics affect polarized plant cell expansion is not understood. The direction of plant cell expansion, which is driven by turgor pressure, is thought to be determined by the localized deposition of membrane and cell wall material, via fusion of exocytic vesicles, and the loosening of cell wall polymers (Kropf et al., 1998). As suggested previously, exocytosis and endocytosis in plant cells are highly interdependent processes (Battey et al., 1999). The abnormal proliferation of the PM in the absence of ADL1A could disrupt the targeting and fusion of exocytic vesicles to the cell surface, thereby disrupting cell wall biosynthesis, which is necessary for polarized expansion. In addition, the loss of ADL1 function may affect either directly or indirectly the organization of the cortical microtubule and/or actin microfilaments, which also play a major role in plant cell shape determination (Kropf et al., 1998). In this regard, it is interesting that adl1A trichomes (Figure 4B) phenotypically resemble those of the Arabidopsis trichome-branching mutant zwichel, which fails to form the distal pointing branch. ZWICHEL encodes a kinesin-like Ca2+/calmodulin-regulated motor protein that may function in intracellular membrane transport and/or in the reorganization of the microtubule cytoskeleton required for the establishment and polarized expansion of trichome branches (Oppenheimer et al., 1997). However, recent studies in mammalian cells have demonstrated that dynamins link cellular membranes to the actin cytoskeleton and regulate a variety of actin-dependent membrane processes, including membrane protrusion, tubulation, and endocytosis (Ochoa et al., 2000; Lee and De Camilli, 2002; Orth et al., 2002). Consistent with this finding, we have demonstrated that the asymmetric localization of ADL1A-mGFP5 in root cells was disrupted by an actin antagonist (B.-H. Kang and S.Y. Bednarek, unpublished data). Further work is necessary to establish the role of ADL1 proteins in microtubule- and/or actin microfilament-dependent membrane processes.
The progressive loss of morphologically normal cell wall material observed during adl1A papillae maturation likely is a consequence of defective ADL1A-mediated PM dynamics (cf. Figures 3C and 3D). Failure to recycle, localize, or regulate hydrolytic enzymes involved in cell wall biosynthesis, such as KORRIGAN, a PM-associated endo-1,4-β-d-glucanase (Nicol et al., 1998) required for normal cell wall biosynthesis and cell plate formation (Nicol et al., 1998; Zuo et al., 2000), could lead to the degradation of the existing extracellular matrix. Callose deposition at the cell surface and in the developing cell plate also is affected in adl1 mutants. Similar to the cyt1 mutation, which affects N-linked glycosylation and thereby cell wall biosynthesis (Nickle and Meinke, 1998; Lukowitz et al., 2001), the accumulation of callose in the adl1A; E embryos may be an indirect “stress” response to the loss or improper synthesis of the extracellular matrix. However, callose not only accumulates in response to defects or injury to the cell wall; it also is synthesized normally during the process of cell plate biogenesis.
Callose deposition within the developing somatic and syncytial-type cell plates precedes cellulose biosynthesis and is thought to help drive the spreading and stabilization of the cell plate membrane system (Samuels et al., 1995; Otegui and Staehelin, 2000). As the cell plate matures and fuses to the parental cell surface, callose is replaced gradually by cellulose. Callose synthase is localized at the cell plate, and its activity is regulated by the ADL1A homolog phragmoplastin (Hong et al., 2001; Verma, 2001). These data suggest that ADL1A may control the activity of the callose synthase complex within the cell plate and PM, thereby affecting cell plate maturation and cell wall synthesis. Indeed, one possible role for the ADL1A rings that constrict membrane tubules in the syncytial-type cell plate (Otegui et al., 2001) is to regulate the localized deposition of callose within the developing tubular network. Defects in the proper localization and activity of callose synthase complexes would be expected to block syncytial cell plate maturation and thus endosperm cellularization, as we have observed in adl1A; E seeds.
A Role for ADL1A in Asymmetric PM Protein Localization?
In nondividing, subcortical, root transition zone cells, ADL1A was localized asymmetrically in the transverse surfaces (i.e., surfaces that do not undergo significant expansion after their formation). Based on its persistence in these surfaces, we propose that ADL1A-mediated membrane dynamics also is essential for the maintenance of the proper protein and lipid compositions of biochemically distinct PM subdomains in polarized plant cells. During cytokinesis, exocytic membrane trafficking appears to be polarized toward the division plane, resulting in the localization of cell cycle–specific and constitutively synthesized PM proteins and cell wall precursors to the cell plate (for review, see Bednarek and Falbel, 2002), including PIN1 (Geldner et al., 2001). PIN1, a PM protein required for the vectoral transport of auxin through the plant, is associated asymmetrically with the basal (toward the shoot) end of cells in the vascular tissue. Therefore, PIN1 would need to be removed from the apical surface of the new daughter cell after cytokinesis and delivered to the basal side to maintain its polar localization. Consistent with this notion, polar localization of PIN1 has been shown to be maintained by actin-dependent membrane trafficking between the PM and an endocytic compartment, a process that is blocked by the polar auxin transport inhibitor naphthylphthalmic acid (Geldner et al., 2001). Biochemical studies using immobilized naphthylphthalmic acid affinity chromatography recently implicated ADL1A and several other proteins involved in vesicular trafficking between the trans-Golgi, the PM, and the endocytic compartment (Murphy et al., 2002) in PIN1 localization. In addition, ADL1A function may be required for the polar localization of other recently identified, asymmetrically distributed, PM-associated proteins and cell wall polysaccharides (Freshour et al., 1996; Schindelman et al., 2001; Friml and Palme, 2002). Further genetic and biochemical characterization of ADL1A and interacting proteins should advance our understanding of the involvement of ADL1 in membrane dynamics during division and plant cell expansion.
METHODS
General Reagents
Enzymes for the manipulation of nucleic acids were purchased from New England Biolabs (Beverly, MA) or Pharmacia Biotech (Piscataway, NJ), unless indicated otherwise. All other reagents, unless specified, were from Sigma Chemical (St. Louis, MO).
Oligonucleotides Used in This Study
Oligonucleotide sequences shown in Table 1 were synthesized by Integrated DNA Technologies (Coralville, IA). Lowercase letters in the oligonucleotide sequences indicate added restriction sites used for cloning.
Table 1.
Oligonucleotides Used in This Study
| Name | Sequence |
|---|---|
| SB46 | 5′-tcactgcagCCGATTCCAACCGAATGG-3′ |
| SB47 | 5′-tgaggatccCTTGTTAACCAGAGAGATCAG-3′ |
| SB48 | 5′-tgaggatccCTCCAGGACTGAAGACTTCCC-3′ |
| SB64 | 5′-CAGTTATTTTGTTGTTTAGTGTCATCCTT-3′ |
| SB91 | 5′-CTTGGAGTCTGTTATCAGGACCCGTATTC-3′ |
| SB92 | 5′-GTTCTCTAAAACCATCACCAGAAACACAC-3′ |
| SB106 | 5′-AAGTACAGAAACCCCTCTCCACAACAATC-3′ |
| SB245 | 5′-cgggatccTCATAGCTGCGGGACACCATCTT-3′ |
| SB246 | 5′-cgggatccTTTCATCGTCGCCATGTTTCCTGG-3′ |
| SB248 | 5′-tcactgcagATCTGTGGCGTGGTAATGAATCT-3′ |
| SB249 | 5′-cgggatccTAACCCAATCAAACTCTCCATA-3′ |
| SB258 | 5′-GCACGTCCGCCATGGGTAAAGGAGAACTT-3′ |
| SB259 | 5′-ggaattcAGGTCACTGGATTTTGGTTTTAG-3′ |
| JL202 | 5′-CATTTTATAATAACGCTGCGGACATCTACA-3′ |
Plant Transformation Vector Construction
To generate ADL1 promoter–β-glucuronidase (GUS) fusion vectors, the bacterial UIDA (GUS) gene (Jefferson et al., 1987) and the nopaline synthase terminator from pBI121 (Clontech, Palo Alto, CA) were subcloned into the EcoRI-HindIII sites of pPZP211 (Hajdukiewicz et al., 1994) and designated pBK08. The ADL1A (2019 bp upstream of the ADL1A start codon) and ADL1E (1955 bp upstream of the ADL1E start codon) promoter regions were amplified by PCR from BACs MJC20 (Kotani et al., 1997) and T209 (Mozo et al., 1998) using primer pairs SB46-SB47 and SB248-SB249 and subcloned into pBK08 to generate pBK16 and pBK20, respectively. For the ADL1A-mGFP5 fusion construct, the coding sequence of mGFP5 (Siemering et al., 1996) was amplified by PCR from pAVA393 (kindly provided by A. von Arnim, University of Tennessee, Knoxville) with primer pair SB258-SB259 and cloned into pPZP211 (Hajdukiewicz et al., 1994) using EcoRI-SacI sites (pBK22). The ADL1A promoter (1554 bp upstream of the ADL1A start codon) and cDNA fusion construct were subcloned into pBK22 as a HindIII-XhoI fragment to generate the C-terminal ADL1A-mGFP5 translational fusion expression vector pBK32. For molecular complementation of adl1A/adl1A adl1E/adl1E embryo-defective seeds, a 9.7-kb EcoRI fragment of BAC T209 containing ADL1E and ∼4 kb of the 5′ putative promoter and the 3′ untranslated DNA was cloned into the modified pPZP221 (Hajdukiewicz et al., 1994) binary transformation vector pPZP221-B (Kang et al., 2001) to generate pBK04.
Plant Material and Growth Conditions
Wild-type and syngenic adl1E-1 Arabidopsis thaliana ecotype Wassilewskija, isolated from the University of Wisconsin–Madison T-DNA knockout collection (http://www.biotech.wisc.edu/Arabidopsis/) of independent T-DNA–tagged mutant lines (kanamycin resistant), were grown as described by Kang et al. (2001). ADL1A/adl1A; adl1E/adl1E plants were isolated from the F2 progeny of a cross between homozygous adl1E-1 and heterozygous adl1A-2 plants. Wild-type Wassilewskija and mutants were transformed using the Agrobacterium tumefaciens–mediated floral-dip method (Clough and Bent, 1998). Transgenic plants transformed with pBK16, pBK20, and pBK32 were selected on solidified (0.6% phytoagar) Murashige and Skoog (1962) medium (Gibco BRL, Rockville, MD) containing 40 μg/mL kanamycin. Wild-type transgenic lines containing pBK32 were crossed to homozygous adl1A-2 mutants to generate the homozygous adl1A-2 line expressing ADL1A-mGFP5. To test for molecular complementation of adl1E-1, pBK04 was transformed into ADL1A/adl1A; adl1E/adl1E plants and transgenic lines were selected for growth on Murashige and Skoog medium containing 20 μg/mL glufosinate (Crescent Chemicals, Hauppauge, NY). Total protein isolation and immunoblot analysis were performed as described previously (Kang et al., 2001).
Immunoblot Analysis
For preparation of total protein extract from wild-type, adl1A-2, adl1E-1 plants and transgenic lines expressing ADL1A-mGFP5, ∼50 mm2 of rosette leaf tissue was homogenized in 100 μL of SDS-PAGE sample buffer (Laemmli, 1970) and incubated at 95°C for 5 min. The samples were cleared of insoluble debris by centrifugation at 16,100g for 5 min at room temperature, and 10 μL of the supernatant was resolved on a 7.5% (w/v) SDS-polyacrylamide minigel and analyzed by immunoblotting as described (Kang et al., 2001) with anti-ADL1 GTPase domain–specific (α-GTPase) and α-ADL1A antibodies. To analyze the expression and intactness of ADL1A-mGFP5, total protein extracts were resolved on a 12.5% (w/v) SDS-polyacrylamide minigel and immunoblotted with α-GFP antibodies. Before immunoblotting, the membrane was analyzed by Ponceau S staining to confirm equal protein loading. GFP antiserum was a generous gift from R. Vierstra (University of Wisconsin–Madison). Affinity-purified polyclonal α-AtCDC48 (Rancour et al., 2002), α-ADL1A, and α-GTPase (Kang et al., 2001) antibodies were generated and used as described. Donkey anti-rabbit IgG horseradish peroxidase was purchased from Amersham Life Sciences (Arlington Heights, IL). Detection by the enhanced chemiluminescence protein gel blot detection system (Amersham Pharmacia Biotech) was performed according to the manufacturer's instructions.
Histochemical Localization of GUS Expression
Tissues from T2 transgenic embryos, seedlings, and plants containing either pBK16 or pBK20 were rinsed with 50 mM sodium phosphate buffer, pH 7.0, and vacuum infiltrated in the presence of the staining solution (1.0 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 2.5 mM K3[Fe(CN)6], 2.5 mM K4[Fe(CN)6], 0.1% [v/v] Triton X-100, 50 mM sodium phosphate, pH 7.0, and 0.1% [v/v] β-mercaptoethanol) for 15 min at room temperature. After incubation for 6 to 12 h at 37°C, samples were cleared by incubation in 70% (v/v) ethanol for 2 h at room temperature to remove chlorophyll. Embryos from the transgenic lines were removed from the seed coat and stained. Photomicrographs were taken with a Leica MZ6 (Leica Microsystems, Bannockburn, IL) stereomicroscope equipped with a SPOT digital imaging system (Diagnostic Instruments, Sterling Heights, MI) or an Axioskop (Carl Zeiss, Thornwood, NY) microscope equipped with a MicroMax (Princeton Instruments, Trenton, NJ) digital camera using IPLab Spectrum (Signal Analytics, Vienna, VA).
Bright-Field Microscopy and Transmission Electron Microscopic Analysis
Transmission electron microscopic analysis of developing stigmatic papillae cells was performed on Wassilewskija wild-type and homozygous adl1A stage-11 to -16 (Smyth et al., 1990) flower buds that were fixed in 5.0% (w/v) glutaraldehyde and processed and analyzed as described (Kang et al., 2001). For analysis of developing embryos, siliques were fixed in 2% (w/v) glutaraldehyde (Ted Pella, Redding, CA), 2% (w/v) paraformaldehyde (Ted Pella), 100 mM Hepes, pH 7.2, and 0.05% (v/v) Tween 20 for 2 h under vacuum at room temperature. The samples were rinsed thoroughly with 100 mM Hepes, pH 7.2, postfixed with 2% (w/v) OsO4 for 2 h at room temperature, and embedded in Spurr's resin as described (Kang et al., 2001). Thin sections (60 to 80 nm) were cut with a Reichert-Jung Ultracut model E microtome (Vienna, Austria), and images were made using a Philips CM120 scanning transmission electron microscope (Eindhoven, The Netherlands). For bright-field microscopy, semithin sections (∼1.5 μm) of double mutant embryos were cut and stained with toluidine blue O as described previously (Kang et al., 2001). For scanning electron microscopy, flowers and trichomes were fixed, attached to stubs, dissected as needed, and coated as described (Kang et al., 2001).
Epifluorescence Imaging
For indirect immunofluorescence microscopy, 5-day-old seedlings were washed once with 50 mM PIPES-KOH, 5 mM EGTA, and 5 mM MgSO4, pH 7.0 (MTSB), and incubated in MTSB containing 4% (w/v) formaldehyde (Ted Pella) for 1 h under vacuum at room temperature. Fixed seedlings were washed three times with MTSB and incubated in 2% (w/v) driselase in phosphate buffer saline, pH 7.4, to digest cell walls. Root tissues were dissected from seedlings and mounted on glass slides. Root tissues were incubated in permeabilization buffer (MTSB containing 10% [v/v] DMSO and 3% [v/v] Nonidet P-40) for 1 h at room temperature and transferred to blocking buffer (3% [w/v] BSA in MTSB [BSA/MTSB]). After blocking for 1 h at room temperature, root tissues were incubated in affinity-purified rabbit α-GTPase or rabbit α-ADL1A antiserum diluted with blocking buffer for 2 h at 37°C. The primary antibodies were detected with fluorescein isothiocyanate–conjugated or Cy3-conjugated secondary goat anti-rabbit antibodies (Jackson ImmunoResearch, West Grove, PA) diluted to 6.25 or 4.5 μg/mL, respectively, in BSA/MTSB for 1 h at 37°C. Samples were counterstained with 10 mM propidium iodide for 10 min at room temperature to visualize nuclei. Live cell imaging of ADL1A-mGFP5 in roots was performed on 3-day-old seedlings grown on solid Murashige and Skoog medium supplemented with 1% (w/v) sucrose. Confocal imaging was performed with a Zeiss Axiovert 100M inverted microscope equipped with a Bio-Rad MRC1024 laser scanning unit and a ×63 (numerical aperture, 1.4) PlanAPOChroma oil-immersion objective lens.
For callose detection, developing siliques were fixed and washed as described above (without OsO4 postfixation) and embedded in LR White resin (Ted Pella) according to the manufacturer's instructions. Three-micrometer samples were affixed to poly-Lys–coated slide glass with heat and stained with aniline blue solution (100 μg/mL in 0.1 M K3PO4, pH 12.0) for 20 min at room temperature. Callose–aniline blue complexes were visualized by UV light epifluorescence microscopy (excitation filter, 355 nm; dichroic mirror, 400 nm; emission filter, 420 nm) using a ×20 Plan Neofluor (Carl Zeiss) dry objective lens.
All images were processed using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA) and/or ImageJ 1.28v (National Institutes of Health; http://rsb.info.nih.gov/ij/) imaging software on MacIntosh computers (Apple Computer, Cupertino, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We are grateful to R. Amasino, D. Fernandez, L.A. Staehelin, D. Rancour, and members of the laboratory for discussion and critical reading of the manuscript. We also thank J. Kimble for the generous use of her confocal microscope and the Electron Microscope Facility at the University of Wisconsin Medical School for help with transmission electron microscopy analysis. This research was supported partially by the College of Agriculture and Life Sciences at the University of Wisconsin–Madison, by a Department of Biochemistry predoctoral fellowship to B.-H.K., by the U.S. Department of Agriculture Cooperative State Research Education and Extension Service Project (Grant WIS04430), and by an award from the Milwaukee Foundation to S.Y.B.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009670.
Footnotes
Online version contains Web-only data.
References
- Arimura, S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99, 5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, F.F., Mayer, U., Wanner, G., and Jürgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253, 267–277. [DOI] [PubMed] [Google Scholar]
- Battey, N.H., James, N.C., Greenland, A.J., and Brownlee, C. (1999). Exocytosis and endocytosis. Plant Cell 11, 643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek, S.Y., and Falbel, T.G. (2002). Membrane trafficking during plant cytokinesis. Traffic 3, 621–629. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler, S., and Ehrhardt, D. (2002). Polarized cytokinesis in vacuolate cells of Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 2812–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Freshour, G., Clay, R.P., Fuller, M.S., Albersheim, P., Darvill, A.G., and Hahn, M.G. (1996). Developmental and tissue-specific structural alterations of the cell-wall polysaccharides of Arabidopsis thaliana roots. Plant Physiol. 110, 1413–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., and Palme, K. (2002). Polar auxin transport: Old questions and new concepts? Plant Mol. Biol. 49, 273–284. [PubMed] [Google Scholar]
- Geldner, N., Frimi, J., Stierhof, Y.-D., Jürgens, G., and Palme, K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Goldberg, R.B., de Paiva, G., and Yadegari, R. (1994). Plant embryogenesis: Zygote to seed. Science 266, 605–614. [DOI] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P. (1996). Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15, 695–704. [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and Verma, D.P.S. (1997). Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cell division. Plant Cell 9, 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- Hinshaw, J.E., and Schmid, S.L. (1995). Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H.F., and Hettema, E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein, S.E. (2002). Clathrin and plant endocytosis. Traffic 3, 614–620. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Delauney, A.J., and Verma, D.P. (2001). A cell plate–specific callose synthase and its interaction with phragmoplastin. Plant Cell 13, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp, M., Misra, S., and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J.B., Kim, Y.A., Kim, S.J., Lee, S.H., Kim, D.H., Cheong, G.W., and Hwang, I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13, 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.-H., Busse, J.S., Dickey, C., Rancour, D.M., and Bednarek, S.Y. (2001). The Arabidopsis cell plate-associated dynamin-like protein, ADL1a, is required for multiple stages of plant growth and development. Plant Physiol. 126, 47–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.-H., Rancour, D.M., and Bednarek, S.Y. (2003). The dynamin-like protein ADL1C is essential for plasma membrane maintenance during pollen maturation. Plant J., in press. [DOI] [PubMed]
- Kang Shin, G., Jin Jing, B., Piao Hai, L., Pih Kyeong, T., Jang Hyun, J., Lim Jeong, H., and Hwang, I. (1998). Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38, 437–447. [DOI] [PubMed] [Google Scholar]
- Kim, Y.W., Park, D.S., Park, S.C., Kim, S.H., Cheong, G.W., and Hwang, I. (2001). Arabidopsis dynamin-like 2 that binds specifically to phosphatidylinositol 4-phosphate assembles into a high-molecular weight complex in vivo and in vitro. Plant Physiol. 127, 1243–1255. [PMC free article] [PubMed] [Google Scholar]
- Klockow, B., Tichelaar, W., Madden, D.R., Niemann, H.H., Akiba, T., Hirose, K., and Manstein, D.J. (2002). The dynamin A ring complex: Molecular organization and nucleotide-dependent conformational changes. EMBO J. 21, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, H., Sato, S., Fukami, M., Hosouchi, T., Nakazaki, N., Okumura, S., Wada, T., Liu, Y.G., Shibata, D., and Tabata, S. (1997). A fine physical map of Arabidopsis thaliana chromosome 5: Construction of a sequence-ready contig map. DNA Res. 4, 371–378. [DOI] [PubMed] [Google Scholar]
- Kropf, D.L., Bisgrove, S.R., and Hable, W.E. (1998). Cytoskeletal control of polar growth in plant cells. Curr. Opin. Cell Biol. 10, 117–122. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., and De Camilli, P. (2002). Dynamin at actin tails. Proc. Natl. Acad. Sci. USA 99, 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84, 61–71. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98, 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1990). Endosperm cellularization in Arabidopsis thaliana L. Arabidopsis Inf. Serv. 27, 65–72. [Google Scholar]
- Mansfield, S.G., and Briarty, L.G. (1991). Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69, 461–476. [Google Scholar]
- McNiven, M.A., Cao, H., Pitts, K.R., and Yoon, Y. (2000). The dynamin family of mechanoenzymes: Pinching in new places. Trends Biochem. Sci. 25, 115–120. [DOI] [PubMed] [Google Scholar]
- Mozo, T., Fischer, S., Shizuya, H., and Altmann, T. (1998). Construction and characterization of the IGF Arabidopsis BAC library. Mol. Gen. Genet. 258, 562–570. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Murphy, A., Hoogner, K., Peer, W.A., and Taiz, L. (2002). Identification, purification and molecular cloning of N-1-napthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128, 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry, P., Mayer, U., and Jürgens, G. (2000). Genetic dissection of cytokinesis. Plant Mol. Biol. 43, 719–733. [DOI] [PubMed] [Google Scholar]
- Nakayama, M., Yazaki, K., Kusano, A., Nagata, K., Hanai, N., and Ishihama, A. (1993). Structure of mouse Mx1 protein: Molecular assembly and GTP-dependent conformational change. J. Biol. Chem. 268, 15033–15038. [PubMed] [Google Scholar]
- Nickle, T.C., and Meinke, D.W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15, 321–332. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa, G.C., Slepnev, V.I., Neff, L., Ringstad, N., Takei, K., Daniell, L., Kim, W., Cao, H., McNiven, M., Baron, R., and De Camilli, P. (2000). A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150, 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Pollock, M.A., Vacik, J., Szymanski, D.B., Ericson, B., Feldmann, K., and Marks, M.D. (1997). Essential role of a kinesin-like protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, 6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth, J.D., Krueger, E.W., Cao, H., and McNiven, M.A. (2002). The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc. Natl. Acad. Sci. USA 99, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M., and Staehelin, L.A. (2000). Syncytial-type cell plates: A novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12, 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Mastronarde, D.N., Kang, B.-H., Bednarek, S.Y., and Staehelin, L.A. (2001). Three-dimensional analysis of syncytial-type cell plates during endosperm cellularization visualized by high resolution electron tomography. Plant Cell 13, 2033–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., Keegan, B.R., Brisch, E., Thatcher, J.W., Hermann, G.J., Bleazard, W., and Shaw, J.M. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancour, D.M., Dickey, C.E., Park, S., and Bednarek, S.Y. (2002). Characterization of AtCDC48: Evidence for multiple membrane mechanisms at the plane of cell division in plants. Plant Physiol. 130, 1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, A.L., and Bisalputra, T. (1990). Endocytosis in elongating root cells of Lobelia erinus. J. Cell Sci. 97, 157–166. [Google Scholar]
- Samuels, A.L., Giddings, T.H., and Staehelin, L.A. (1995). Cytokinesis in tobacco BY-2 and root tip cells: A new model of cell plate formation in higher plants. J. Cell Biol. 130, 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni, V., Kieffer, S., Desclaux, D., Masson, F., and Rabilloud, T. (2000). Membrane proteomics: Use of additive main effects with multiplicative interaction model to classify plasma membrane proteins according to their solubility and electrophoretic properties. Electrophoresis 21, 3329–3344. [DOI] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemering, K.R., Golbik, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663. [DOI] [PubMed] [Google Scholar]
- Smirnova, E., Shurland, D.L., Ryazantsev, S.N., and van der Bliek, A.M. (1998). A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 143, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner, R., Glässer, G., Wanner, G., Somerville, C.R., Jürgens, G., and Assaad, F.F. (2002). Cytokinesis-defective mutants of Arabidopsis. Plant Physiol. 129, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness, M.K., Kandasamy, M.K., Nasrallah, M.E., and Nasrallah, J.B. (1993). Genetic ablation of floral cell in Arabidopsis. Plant Cell 5, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek, A.M. (1999). Functional diversity in the dynamin family. Trends Cell Biol. 9, 96–102. [DOI] [PubMed] [Google Scholar]
- Vaughn, K.C., Hoffman, J.C., Hahn, M.G., and Staehelin, L.A. (1996). The herbicide dichlobenil disrupts cell plate formation: Immunogold characterization. Protoplasma 194, 117–132. [Google Scholar]
- Verma, D.P. (2001). Cytokinesis and building of the cell plate in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 751–784. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Hong, Z., and Verma, D.P. (2000). Phragmoplastin polymerizes into spiral coiled structures via intermolecular interaction of two self-assembly domains. J. Biol. Chem. 275, 8779–8784. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Nishizawa, N., Wu, Y., Kost, B., and Chua, N.H. (2000). KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.