Abstract
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA)-type glutamate receptors mediate the majority of excitatory signaling in the CNS, and the functional properties and subcellular fate of these receptors depend on receptor subunit composition. Subunit assembly is thought to occur in the endoplasmic reticulum (ER), although we are just beginning to understand the underlying mechanism. Here we examine the trafficking of Caenorhabditis elegans glutamate receptors through the ER. Our data indicate that neurons require signaling by the unfolded protein response (UPR) to move GLR-1, GLR-2, and GLR-5 subunits out of the ER and through the secretory pathway. In contrast, other neuronal transmembrane proteins do not require UPR signaling for ER exit. The requirement for the UPR pathway is cell type and age dependent: impairment for receptor trafficking increases as animals age and does not occur in all neurons. Expression of XBP-1, a component of the UPR pathway, is elevated in neurons during development. Our results suggest that UPR signaling is a critical step in neural function that is needed for glutamate receptor assembly and secretion.
INTRODUCTION
Ion channels conduct electrochemical signaling in the nervous system, and α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA)-type ionotropic glutamate receptors (AMPAR) in particular mediate the bulk of excitatory transmission in the CNS. AMPAR subunits (up to 4 in mammals, referred to as GluR1-R4) are multi-transmembrane–spanning proteins that can assemble into tetrameric channels of differing subunit composition (Hollmann and Heinemann, 1994; Dingledine et al., 1999). The specific subunit composition of a given AMPAR channel plays a critical role in determining the functional properties of that channel, including its channel opening probability, ion selectivity, and cytosolic binding partners (Hollmann and Heinemann, 1994; Dingledine et al., 1999; Sheng, 2001; Shi et al., 2001; Malinow, 2003). Subunit composition can also control the subcellular localization and regulated cell biological fate of a channel (Beattie et al., 2000; Lin et al., 2000). For example, heteromeric complexes of GluR1-R2 receptors have been shown to be added to hippocampal synapses in an activity-dependent manner, whereas GluR2-R3 complexes appear to cycle into synaptic membranes in a constitutive manner (Passafaro et al., 2001; Shi et al., 2001). To better understand AMPAR function in the nervous system, it is important to determine how individual subunits assemble into specific complexes of channels.
Multisubunit channel assembly in general is tied to the movement of channels through the secretory pathway. Most proteins exit the ER without the need for specialized export signals (Wieland et al., 1987). However, many channels and receptors seem to defy this general rule by requiring channel assembly, sometimes specific export signals, and sometimes the interaction of scaffolding proteins and certain signal transduction events before exiting the ER (Ma and Jan, 2002). One explanation is that subunit assembly acts to mask ER retention signals that would otherwise be exposed in an unassembled subunit. For example, a novel class of RXR endoplasmic reticulum (ER) retention signals has been identified in potassium channels, GABAB (γ-aminobutyric (B)) receptors, and NMDA (N-methyl-d-aspartate)-type glutamate receptors, and it is thought that the correct assembly of these subunits into channels obscures the retention motif and thereby allows these channels to exit the ER (Zerangue et al., 1999; Margeta-Mitrovic et al., 2000; Standley et al., 2000; Scott et al., 2001; Xia et al., 2001). Surprisingly, no such signals have been found in the cytosolically exposed regions of AMPA-type channels; rather, the only major retention signal so far identified has been Arg607 at the GluR2 Q/R editing site in the channel pore (Greger et al., 2002, 2003). Moreover, mutations that block glutamate binding or ion permeation inhibit exit from the ER (Grunwald and Kaplan, 2003). Hence AMPA-type receptors might use a different mechanism for regulating channel assembly, perhaps because of the need for these receptor subunits to form channels of diverse composition.
One mechanism that regulates quality control of protein folding and protein secretion through the ER is the unfolded protein response (UPR), a signaling system that has been previously shown to up-regulate the expression of ER-resident chaperone proteins in response to ER stress (Spear and Ng, 2001; Kaufman et al., 2002). One component of this pathway that is conserved in all known eukaryotes is IRE1, an ER resident membrane-spanning endonuclease that is activated by stress (Nikawa and Yamashita, 1992; Cox et al., 1993; Mori et al., 1993; Tirasophon et al., 1998; Wang et al., 1998). On activation in animals, IRE1 is thought to dimerize and catalyze the splicing of an XBP1 mRNA, thereby allowing for the production of functional XBP1 protein (Yoshida et al., 2001; Calfon et al., 2002). XBP1 is a bZIP transcription factor that can in turn induce the expression of downstream UPR target genes required to respond to the stress event. The role of human XBP1 in the nervous system is of particular interest as a polymorphism in its promoter region that results in reduced XBP1 expression has been identified as a genetic risk factor for bipolar disorder (Kakiuchi et al., 2003). Moreover, a number of mood stabilizing drugs used to treat uni- and bipolar depression have been shown to modulate glutamate signaling, and one in particular, valproate, has been shown to up-regulate the expression of XBP1 (Kakiuchi et al., 2003).
We hypothesized that the UPR pathway might have a critical role in neurons to assure the proper assembly and secretion of synaptic proteins, including AMPARs. To better understand the role of the UPR in neurons, we used the Caenorhabditis elegans model system to examine the requirement of ire-1 and xbp-1, homologues of mammalian IRE1 and XBP1, in neuronal protein trafficking (Shen et al., 2001; Calfon et al., 2002; Urano et al., 2002). In particular, we examined the trafficking of GLR-1, a C. elegans ionotropic glutamate receptor subunit most similar to the AMPA-type receptors (Hart et al., 1995; Maricq et al., 1995). We find that GLR-1 secretion and synaptic transport require components of the UPR signaling pathway. In the absence of either ire-1 or xbp-1, GLR-1 subunits accumulate in the ER, whereas other synaptic and membrane proteins are capable of ER export. Our data suggest that the UPR pathway has a specialized role in neurons for trafficking of AMPA receptors.
MATERIALS AND METHODS
Transgenes and Germline Transformation
Transgenic strains were isolated by microinjecting various plasmids (typically at 50 ng/ml) using either lin-15(+) (J. Mendel) or rol-6dm (C. Mello) as a marker. The following transgenes were integrated into the genome by either gamma irradiation or UV/trimethylpsoralen as previously described (Yandell et al., 1994; Rongo et al., 1998). All transgenes were backcrossed to wild-type strains to remove other mutations introduced during the integration. The glr-1::cfp and glr-2::yfp transgenes were generated by injecting the respective plasmids (V. Maricq) into nematodes and integrating them into the genome to generate odIs25[glr-1::cfp] and odIs18[glr-2::yfp]. The Pglr-1::rfp transgene was generated by subcloning monomeric RFP (R. Tsien) into the glr-1 promoter vector pV6 (V. Maricq; Campbell et al., 2002). The resulting plasmid was injected into nematodes and integrated into the genome to generate odIs6.
The following transgenes were introduced into the germline and followed as extrachromosomal arrays. The Pglr-1::ire-1 transgene was generated by subcloning exons 1–3 and part of 4 from genomic DNA and exons 5–10 plus the remainder of 4 from the ire-1 cDNA from yk8e9 (Y. Kohara) into pV6. The resulting plasmid was injected into ire-1(ok799) nematodes and followed as an extrachromosomal array. The Pglr-1::xbp-1A and Pglr-1::xbp-1B transgenes were generated by subcloning the corresponding open reading frame (ORF) from the xbp-1 cDNA yk878d06 (Y. Kohara) into pV6. The resulting plasmids were injected into xbp-1(zc12) nematodes and followed as an extrachromosomal array. The xbp-1(1–60)::gfp transgene was generated by introducing genomic upstream sequences and the first exon of xbp-1 into the GFP reporter vector pPD95.75 (A. Fire). The XB3 and XB7 transgenes were generated by introducing genomic upstream sequences and the full-length ORFs from xbp-1A and xbp-1B, respectively, into pPD95.75 to create translational fusions. The XB12 transgene was generated by removing sequences for the regulatory intron from the xbp-1 cDNA yk878d06. The ire-1(1–29)::gfp transgene was generated by introducing genomic upstream sequences and the first exon of ire-1 into pPD95.75. The tram::yfp transgene was generated by introducing a genomic sequence containing the tram ORF into pV6 (Rolls et al., 2002). Sequences encoding YFP were then introduced to generate a TRAM::YFP chimera with YFP at the carboxy-terminal end of TRAM. The resulting plasmids were injected into nematodes and followed as extrachromosomal arrays.
Fluorescent Microscopy
Immunohistochemistry of nematodes was performed using a heat fixation method. Nematodes were collected from plates and washed with S Basal buffer. Fixation buffer (70 mM NaCl, 0.03% Triton X-100) was preheated to 80°C and then added to samples for 20 s at 80°C. Samples were then flash-frozen and thawed in liquid N2. Samples were blocked in PBST-A (1× phosphate-buffered saline [PBS], pH 6.8, 0.5% Triton X-100, 1% bovine serum albumin [BSA], 10% normal goat serum) and then incubated with anti-GLR-1 antibodies in PBST-A (Kass et al., 2001). Samples were washed in PBST-B (1× PBS, pH 6.8, 0.5% Triton X-100, 0.1% BSA) and incubated in Cy3-conjugated secondary (Jackson ImmunoResearch, West Grove, PA). We noted that GLR-1 immunofluorescence from fixed nematodes is more distended from the nucleus than GLR-1::GFP from living nematodes, probably from ER membrane distention during fixation, which has been observed previously for the ER of fixed nematode neurons (Rolls et al., 2002).
GFP-, CFP-, YFP-, and RFP-tagged fluorescent proteins were visualized in nematodes by mounting larvae on 2% agarose pads with 10 mM levamisole at room temperature. Fluorescent images were observed using a Zeiss Axioplan II (Thornwood, NY) and 100× 1.4 NA PlanApo objective, and imaged with an ORCA digitally cooled CCD camera (Hamamatsu, Middlesex, NJ) using ImagePro v4.1 and VayTek v6.2 software (Farifield, IA). Exposure times were chosen to fill the 12-bit dynamic range without saturation. Animals were optically sectioned (0.4 μm), and out-of-focus light was removed with a constrained interative deconvolution algorithm (VayTek).
To quantitate the fluorescence levels of GFP-tagged proteins, images of nematodes were captured by CCD as above using a 20× 0.75 NA PlanApo objective and a constant gain and exposure time (filled the 12-bit dynamic range) for all samples. A fluorescent standard (Labsphere, North Sutton, NH) showed <1% drift in signal throughout the imaging session. Images were corrected for coverslip fluorescence by subtracting a background image. Pixel intensity was measured for each animal by quantifying an area of interest that collected the fluorescent neuron cell bodies, nerve ring, and ventral nerve cord for each sample. For each nematode sample, an integrated optical density (IOD) score was obtained by summing the pixel intensity values for all of the fluorescent tissue within the area of interest. Images of nematodes expressing fluorescent proteins from extrachromosomal arrays were only captured if the array was present in all cells previously reported; animals with mosaic nervous systems were not examined.
Behavioral Assays
Nose touch sensory responses were assayed as previously described (Hart et al., 1995). Each animal was tested for reversal of locomotion after a forward collision with a hair. Light body touch sensory responses were assayed by lightly touching the animal with a hair on the anterior half of its body and then scoring for reversal of locomotion (Chalfie et al., 1985). For both assays, each animal was tested 10 times, and 20 or more animals were tested for each genotype.
RNA Isolation and Reverse Transcription–Polymerase Chain Reaction
Total RNA from L1 and young adult worms was prepared using Trizol (Invitrogen, Carlsbad, CA), and mRNA from total RNA using Oligotex mRNA mini Kit (Qiagen, Chatsworth, CA). Cloned AMV First-Strand Synthesis Kit (Invitrogen) was used to perform reverse transcriptions (RTs) according to the manufacturer's instructions. RT reactions were conducted with 4.8 μg of total RNA and 100 ng of mRNA. For quantitative RT–polymerase chain reaction (RT-PCR) using competitor DNA, fragments of genomic DNA for atf-6 and pek-1 were subcloned into a TOPO-TA vector (Invitrogen), and plasmid concentration was determined for a range of competitor amounts. Competitor DNA (1 μl) from the highest concentration (1 ng/μl) to the lowest concentration (0.1 pg/μl) was added into each reverse-transcribed RNA reaction tube, and the other conditions such as primers and template cDNA concentration were kept equal. Each mixture was then amplified using a set of primers that yield different size bands depending on whether cDNA or competitor was used as template. PCR products were run out on 2.5% agarose. Competitive RT-PCR reactions were repeated two times with total RNA and three times with mRNA, with every experiment showing the same result
To determine the presence of the 23-base pair regulatory intron and characterize the products from the XB3 and XB7 transgenes, RNA was isolated as above. The RT-PCR products of oligo R74.3-7 and oligo pPD9575-1 from the transgenic lines were subcloned into a TOPO-TA vector (Invitrogen), generating 48 independent subclones. Each subclone was then PCR amplified using oligos R74.3-5 and R74.3-4 to test for the presence of the regulatory intron. Four of the clones that lacked the regulatory intron were sequenced to determine intron and exon boundaries.
RESULTS
GLR-1 Accumulates in the ER of UPR Mutants
To better understand how GluRs are assembled and trafficked out of the ER, we observed the subcellular localization of channels containing the AMPA-type glutamate receptor subunit GLR-1. Chimeric GLR-1 receptors tagged with the green fluorescent protein (GLR-1::GFP) can be used to visualize glutamate receptors in living animals (Rongo et al., 1998). GLR-1::GFP is localized to synaptic clusters at neuron-neuron synapses within the C. elegans neuropil, and GLR-1::GFP synaptic localization is dependent on Ca2+ signaling and the PDZ protein LIN-10 (Rongo et al., 1998; Rongo and Kaplan, 1999). The synaptic abundance of GLR-1::GFP is regulated by the ubiquitination and subsequent endocytosis of the GLR-1 subunit (Burbea et al., 2002). Interestingly, mutations in the pore domain and the ligand-binding domain have been shown to reduce trafficking of GLR-1 from the ER to synapses, suggesting that there might be a quality control mechanism in the ER that monitors GluR channel assembly and activity (Grunwald and Kaplan, 2003).
In a search for genes that when mutated result in defects in GLR-1::GFP localization, we found that mutations in ire-1 and xbp-1, components of the UPR, result in GLR-1 accumulation in the cell body. We examined the localization of GLR-1::GFP in previously identified molecular null mutants for these genes: ire-1(ok799), in which the three exons containing the kinase and endonuclease domains are deleted, and xbp-1(zc12), in which there is a nonsense mutation at codon 11 (Yoshida et al., 2001; Calfon et al., 2002). Previous studies of GFP-tagged ER resident proteins in C. elegans neurons indicate that ER residents are primarily concentrated in the neuron cell body in a compact ER that is closely associated with the nuclear envelope (Rolls et al., 2002). We examined GLR-1 localization within the neuron cell body either by coexpressing GLR-1::GFP with monomeric DsRed (RFP) to fill the cytosolic compartment (Figure 1, A–D), by detecting GLR-1::GFP costained with 4′,6-amidino-phenylindole · 2HCl (DAPI) to visualize nuclei (Figure 1, E–H) or by detecting endogenous GLR-1 with anti-GLR-1 antibodies (costained with DAPI; Figure 1, I–L). In wild-type animals, both endogenous GLR-1 and GLR-1::GFP can be found throughout the cell body cytosol in punctate structures (Figure 1, A and I) as well as at punctate structures throughout the neurites previously shown to correspond to synaptic inputs (Figure 1B; Rongo et al., 1998; Burbea et al., 2002). Mutant nematodes that lack glr-1 show no fluorescence, indicating the specificity of the antibody (Figure 1L; Hart et al., 1995). To determine whether the punctate structures in the cell body corresponded to Golgi-localized receptor, we coexpressed GLR-1::CFP and mannosidase-YFP (MAN::YFP) and found that they colocalized to punctate structures within the cell bodies (Figure 1, M–O; Rolls et al., 2002).
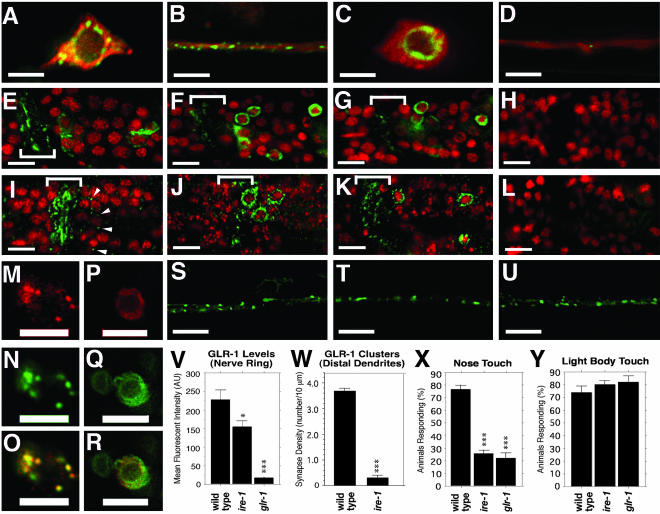
Figure 1.
GLR-1 accumulates in the ER of ire-1 and xbp-1 mutants. (A–D) GFP and RFP fluorescence from nematode neuron cell bodies (A and C) and ventral cord neurites (B and D) expressing GLR-1::GFP and RFP. Percentages of nematodes showing indicated phenotypes for a typical sample are given below, along with the number sampled. (A and B) Wild-type nematodes contain GLR-1::GFP in punctate structures in the cell body and neurites (100%, n = 42). (C and D) ire-1 (100%, n = 36) and (unpublished data) xbp-1 (100%, n = 45) null mutants retain GLR-1::GFP in the ER and contain few clusters in neurites. (E–H) GLR-1::GFP (green) costained with DAPI (red) showing the proximal neurites of the nerve ring (bracket). (I–L) Endogenous GLR-1 detected by immunofluorescence (green) and costained with DAPI (red). (E and I) Wild-type nematodes contain GLR-1 in the proximal neurites of the nerve ring (bracket, 100%, n = 46) and in punctate structures in the cell body (arrowheads; 76%, n = 46). (L) No GLR-1 immunofluorescence is detected in the cell bodies of glr-1 null mutants (100%, n = 30), and (H) no GLR-1::GFP is detected in the cell bodies of nematodes lacking the transgene. GLR-1 accumulates in the ER of (F and J) ire-1 (63%, n = 48) and (G and K) xbp-1 (68%, n = 31) mutants. Nematodes that express GLR-1::CFP (M) and the Golgi marker MANS::YFP (N) show colocalization at puncta in the cell body (O, merge, 100%, n = 22). Nematodes that express GLR-1::CFP (P) and the ER marker EMR::YFP (Q) in xbp-1 mutants show colocalization at ER membranes in the cell body (R, merge, 100%, n = 25). (V) Mean fluorescent pixel intensity (±SEM) was measured for the nerve ring of wild-type, ire-1, and glr-1 mutants stained with anti-GLR-1 antibodies. (W) Density of GLR-1 clusters along the distal dendrites of the ventral nerve cord (±SEM) was measured for wild-type and ire-1. Percent of responses showing a locomotion reversal in response to (X) nose touch (±SEM) or (Y) light body touch per animal was measured for wild-type, ire-1, and glr-1. Scale bar in A–D, S–U, and M–R, 5 μm; in E–L, 10 μm. *p < 0.01 and ***p < 0.0001, compared with wild-type by ANOVA/Bonferoni.
Mutants lacking either ire-1 or xbp-1 accumulate high levels of GLR-1 and GLR-1::GFP in the neuron cell body (Figure 1, C, F, G, J, and K). In particular, GLR-1::GFP is found in close association with the nuclear envelope, suggesting ER retention. To confirm this, we coexpressed GLR-1::CFP and YFP fusions to several ER resident proteins, including TRAM, SP12, and emerin (EMR), using the glr-1 promoter (Figure 1, P–R, and unpublished data). As previously described, we found that these ER resident proteins localized around the nuclear envelope and reticular membranes (Rolls et al., 2002). GLR-1::CFP colocalized with ER markers both in ire-1 and xbp-1 mutants, consistent with the idea that GLR-1 is retained in the ER in the absence of UPR signaling.
GLR-1 is observed in the proximal neurites of the nerve ring in ire-1 and xbp-1 mutants, similar to what is observed for wild-type animals (Figure 1, F, G, J, and K). However, there is a 35% reduction in the proximal neurite levels of GLR-1 (Figure 1V). Furthermore, there is a dramatic reduction in the amount of receptor found in the distal neurites of the ventral nerve cord, such that only a small amount of GLR-1::GFP can be found in punctate structures (Figure 1, D and W). GLR-1 receptors receive and transduce signals from nose-touch mechanosensory neurons, but not from light body touch mechanosensory neurons. To determine whether a reduction in the levels of GLR-1 in neurites results in a reduction in responsiveness to nose-touch, we tested ire-1 and xbp-1 mutants for nose-touch sensitivity. We found that like glr-1 mutants, ire-1 and xbp-1 mutants are significantly impaired for nose-touch mechanosensation (Figure 1X and unpublished data). Moreover, ire-1 and xbp-1 mutants are fully capable of responding to light body touch (Figure 1Y and unpublished data), which although not mediated by GLR-1 nevertheless requires the function of GLR-1–expressing interneurons (Chalfie et al., 1985). Taken together, these results suggest that ire-1 and xbp-1 mutant interneurons, although impaired for GLR-1–and GLR-1–mediated modalities, are capable of receiving and transmitting sensory information in general.
PERK and ATF6 are additional ER-resident proteins that respond to ER stress in parallel to IRE-1/XBP-1 by inhibiting translation and regulating the expression of downstream target genes to cope with folding problems in the ER (Spear and Ng, 2001; Kaufman et al., 2002). We examined GLR-1 localization in pek-1(ok275) and atf-6(ok551) mutants (both are out-of-frame deletions within the genes and are likely nulls) for the C. elegans homologues of these genes (Shen et al., 2001; Calfon et al., 2002; Urano et al., 2002). We also examined sel-1(e1948), a mutant for an ER-associated degradation (ERAD) component, inasmuch as ERAD genes are up-regulated by XBP1 and play an important role in ER stress and the UPR (Urano et al., 2002). We found that GLR-1 in pek-1 mutants, atf-6 mutants, and sel-1 mutants is localized to neurites as in wild-type nematodes (Figure 1, S–U), suggesting that of the three known parallel pathways that respond to ER stress, only the IRE-1/XBP-1 pathway seems to be required for glutamate receptor trafficking.
Age-dependent Requirement for the UPR Pathway
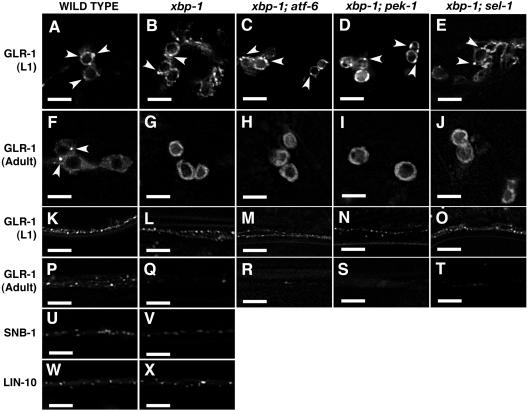
Our initial studies examined GLR-1 localization in UPR mutants of adult animals. To determine whether the UPR pathway is required during the initial synthesis of GLR-1 early in development, we examined wild-type and UPR mutants at different developmental stages. Surprisingly, we found that GLR-1 exits the ER and is localized to the ventral cord structures both in wild-type and UPR mutants as young larvae (Figure 2, A, B, K, and L). However, as UPR mutants age (after stage L2 in development), they accumulate GLR-1 at the ER and fail to localize GLR-1 to the distal neurites (Figure 2, G and Q). One explanation could be that alternate pathways (e.g., atf-6, pek-1, or sel-1) compensate for xbp-1 and ire-1 loss during early development. To test this, we examined GLR-1 localization in double mutant combinations of xbp-1 with either atf-6, pek-1, or sel-1. We found that xbp-1; atf-6 doubles and xbp-1; sel-1 doubles were slow growing but viable and were identical to xbp-1 single mutants with respect to GLR-1 localization (Figure 2, C, E, H, J, M, O, R, and T). We found that xbp-1; pek-1 doubles (>95%) arrested during early larval development; nevertheless, they resembled xbp-1 in phenotype (Figure 2, D, M, and N). The handful that progressed to adulthood failed to export GLR-1 from the ER (Figure 2, I and S). Triple mutants were inviable at too early a stage of development to observe GLR-1.
Figure 2.
Age-dependent requirement for the UPR in GLR-1 ER export. GFP fluorescence from nematodes that express (A–T) GLR-1::GFP, (U and V). SNB-1::GFP, and (W and X) LIN-10::GFP. Percentages of nematodes showing indicated phenotypes for a typical sample are given below, along with the number sampled. (A) L1 stage wild-type nematodes (100%, n = 20), (B) L1 stage xbp-1 nematodes (100%, n = 20), (C) L1 xbp-1; atf-6 (100%, n = 24), (D) L1 xbp-1; pek-1 (100%, n = 18), (E) L1 xbp-1; sel-1 (100%, n = 12), and (F) wild-type adults (100%, n = 20) all contain GLR-1 at punctate structures in their cell bodies (arrowheads). In contrast, (G) xbp-1 adults (100%, n = 20), (H) xbp-1; atf-6 adults (100%, n = 24), (I) xbp-1; pek-1 adults (100%, n = 3), and (J) xbp-1; sel-1 adults (100%, n = 10) accumulate GLR-1 at the ER. L1 stage nematodes that are (K) wild-type, (L) xbp-1, (M) xbp-1; atf-6, (N) xbp-1; pek-1, and (O) xbp-1; sel-1 all contain GLR-1 at punctate structures along distal neurites, as do (P) wild-type adults. In contrast, (Q) xbp-1 adults, (R) xbp-1; atf-6 adults, (S) xbp-1; pek-1 adults, and (T) xbp-1; sel-1 adults lack GLR-1 in distal neurites. Both (U and W) wild-type and (V and X) xbp-1 mutant nematodes localize (U and V) SNB-1::GFP (100%, n = 30 for wild-type, n = 19 for xbp-1) and (W and X) LIN-10::GFP (100%, n = 24 for wild-type, n = 22 for xbp-1) to clusters in neurites. Scale bar, 5 μm.
If atf-6 or pek-1 compensate for xbp-1 loss during early development, then we might expect elevated expression of these genes at that time. We examined whether atf-6 or pek-1 expression is higher in L1 larvae than in young adults by performing quantitative RT-PCR from total and poly(A)+ RNA isolated from wild-type L1 and adults. We find that both genes are expressed throughout development and that the levels of both increase as nematodes reach adulthood (Supplementary Figure 1). Our results suggest that the atf-6 and pek-1 pathways do not compensate for xbp-1 loss during early development. Rather, the UPR pathway is not continually needed for GLR-1 secretion, even during the stages when GLR-1 is initially synthesized. Our results suggest that there is an age-dependent requirement for the UPR pathway in GLR-1 trafficking.
Localization of Synaptic Components in UPR Mutants
A requirement for the UPR pathway in C. elegans has previously been shown by treating nematodes with tunicamycin, which causes ER stress by inhibiting glycosylation (Calfon et al., 2002). By contrast, we have found that the UPR pathway is required for GLR-1 exit from the ER even in the absence of ER stress-inducing agents such as tunicamycin, suggesting that neurons might have a basal-level requirement for UPR signaling in order to export GluRs from the ER. GLR-1 is coexpressed in an overlapping set of neurons with other glutamate receptor subunits, one of which (GLR-2) is thought to partner with GLR-1 (Brockie et al., 2001; Mellem et al., 2002). To determine whether other glutamate receptor subunits require the UPR to exit the ER, we examined the localization of GLR-2::YFP and GLR-5::GFP in wild-type and UPR mutant nematodes (Brockie et al., 2001). Like GLR-1, GLR-2::YFP and GLR-5::GFP accumulate in the ER of UPR mutant neurons (Figure 3, D and F). Interestingly, some neurons that express GLR-5 can export it from the ER in UPR mutants, suggesting that the UPR pathway is required in specific cell types, at least for GLR-5.
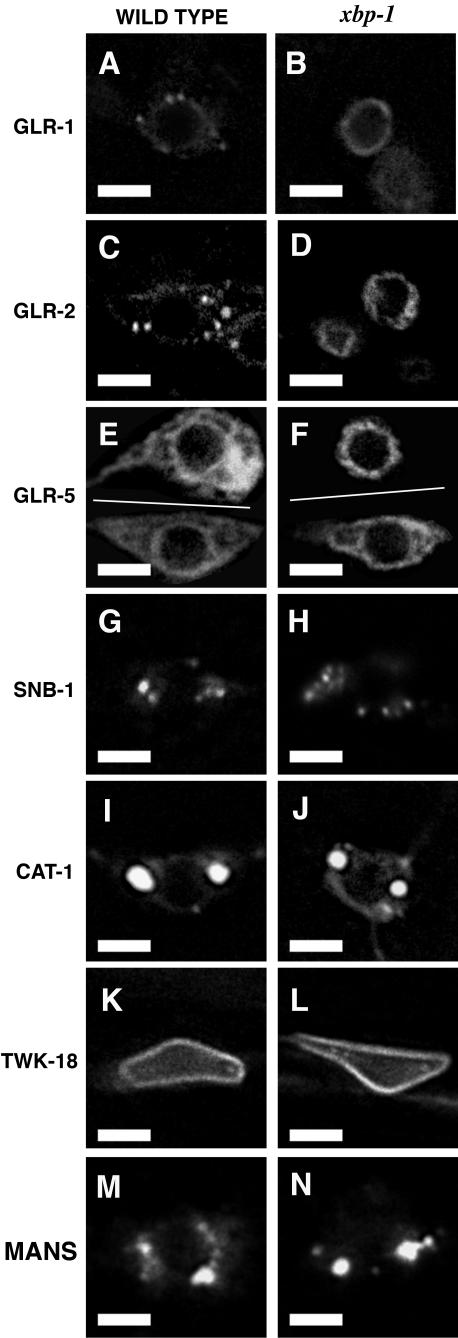
Figure 3.
The requirement for the UPR is biased toward glutamate receptors. Fluorescence from either (A, C, E, G, I, K, and M) wild-type or (B, D, F, H, J, L, and N) xbp-1 mutants expressing (A and B) one copy of the GLR-1::GFP transgene (n = 18 for wild-type, n = 20 for xbp-1), (C and D) GLR-2::YFP (n = 16 for wild-type, n = 20 for xbp-1), (E and F) GLR-5::GFP (n = 17 for wild-type, n = 17 for xbp-1), (G and H) SNB-1::GFP (n = 20 for wild-type, n = 20 for xbp-1), (I and J) CAT-1::GFP (n = 18 for wild-type, n = 14 for xbp-1), (K and L) TWK-18::GFP (n = 16 for wild-type, n = 15 for xbp-1), and (M and N) MAN::YFP (n = 18 for wild-type, n = 14 for xbp-1). Phenotypes shown represent what was observed in 100% of nematodes sampled. In E and F, two different cells are shown in composite: AVB (top) and DVA (bottom). In xbp-1 mutants, GLR-5::GFP accumulates in the ER of 100% of neurons like AVB (neurons in which other GluR subunits are expressed). In contrast, GLR-5::GFP is exported from the ER of DVA, a neuron that only expresses GLR-5, in 100% of the same nematodes. Scale bar, 5 μm.
We reasoned that different membrane-spanning proteins in neurons might have distinct requirements for UPR signaling, even in the absence of ER stress-inducing agents. We examined the subcellular localization of several different membrane-spanning proteins in wild-type and mutant neurons by expressing GFP- or YFP-tagged proteins. To monitor the exit of membrane proteins from the ER, we used the glr-1 promoter to express and examine the localization of mannosidase-YFP (MAN::YFP, Golgi-resident protein) and TWK-18::GFP (TWK-type K+ channel that is uniformly distributed to the membrane; Kunkel et al., 2000; Rolls et al., 2002). We found that MAN::YFP is exported from the ER and accumulates in punctate structures in the cell body in UPR mutants, similar to those found in wild-type neurons (Figure 3, M and N). TWK-18::GFP is also exported from the ER of both mutant and wild-type neurons, accumulating at cell body membranes (Figure 3, K and L). We also examined the trafficking of presynaptic proteins, including a synaptobrevin-GFP chimera (SNB-1::GFP, a single spanning membrane protein found in synaptic vesicles) and a vesicular monoamine transporter-GFP chimera (CAT-1::GFP, a multiple membrane-spanning protein that forms homomeric transporters in synaptic vesicles; Nonet et al., 1998; Duerr et al., 1999). In wild-type animals, CAT-1::GFP and SNB-1::GFP exit the ER and are incorporated into synaptic vesicles that are transported to synapses. Unlike GLR-1::GFP, which is trapped in the ER in UPR mutants, we found that CAT-1::GFP and SNB-1::GFP exit the ER and translocate to neurites in UPR mutants (Figures 2, U and V, and 3, G–J). We also examined the localization of ODR-10::GFP, a transmembrane odorant receptor that is localized to sensory endings, and found no difference between wild-type and UPR mutant nematodes (unpublished data; Dwyer et al., 2001).
One explanation for the differential requirement of the UPR could be the difference in expression levels from the different trafficking proteins. To test this, we measured the levels of different GFP-tagged chimeric proteins by quantitative epifluorescence. We measured the relative levels of GLR-1 from nematodes that carried either one or two copies of the GLR-1::GFP transgene and found that we could detect a roughly twofold difference in expression levels (Table 1). We also examined the expression levels of two proteins expressed under the same glr-1 promoter (SNB-1::GFP and TWK-18::GFP) and two proteins expressed under their own promoters (GLR-5::GFP and CAT-1::GFP, both of which are expressed in about the same number of neurons per animal as GLR-1). We found that nematodes that carry one copy of the GLR-1 transgene express approximately twofold lower levels of GFP chimeric protein relative to nematodes that carry two copies of the GLR-1 transgene or the other GFP chimeric proteins tested (Table 1 and unpublished data). Nevertheless, GLR-1 accumulates in the ER in nematodes carrying only one copy of glr-1::gfp if they lack UPR signaling, suggesting that the absolute expression level of a GFP-chimeric secretory protein is not sufficient to determine whether it requires UPR signaling (Figure 3, A and B). Interestingly, we found that the levels of SNB-1::GFP were reduced in UPR mutants relative to wild-type nematodes, suggesting that although the UPR signaling pathway is not required for these synaptic vesicles proteins to exit the ER, either UPR signaling or the presence of synaptic glutamate receptors are nevertheless needed to maintain wild-type levels of at least some synaptic components.
Table 1.
Relative expression levels of GFP-tagged proteins
| Transgene | Genotype | IOD (×103) | Normalized | N |
|---|---|---|---|---|
| 1 copy glr-1::gfp | wild type | 294 ± 50a | 1.00 | 17 |
| 2 copies glr-1::gfp | wild type | 704 ± 52 | 2.39 | 13 |
| 1 copy glr-1::gfp | xbp-1 | 249 ± 37a | 0.85 | 17 |
| 2 copies glr-1::gfp | xbp-1 | 739 ± 42 | 2.51 | 22 |
| 2 copies snb-1::gfp | wild type | 817 ± 80 | 2.78 | 21 |
| 2 copies snb-1::gfp | xbp-1 | 590 ± 52b | 2.01 | 21 |
| Ex twk-18::gfp | wild type | 686 ± 83 | 2.33 | 12 |
| Ex twk-18::gfp | xbp-1 | 519 ± 51 | 1.77 | 10 |
| Ex glr-5::gfp | wild type | 570 ± 67 | 1.94 | 14 |
| Ex glr-5::gfp | xbp-1 | 533 ± 40 | 1.81 | 13 |
Relative expression levels are shown based on measurements of GFP fluorescence from different transgenes. Images were collected from the cell bodies and neurites of individual nematodes, and pixel values were summed for each nematode to give an Integrated Optical Density (IOD). The average IOD per nematode is shown, including the s.e.m. For each integrated transgene, the number of genomic copies of the transgene is indicated. Transgenes that were examined as extrachromosomal arrays are indicated by “Ex.” For the extrachromosomal arrays, animals with mosaic nervous systems were excluded.
p < 0.0001 compared with 2 glr-1::gfp copies (ANOVA with Bonferroni correction)
p < 0.01 compared with snb-1::gfp in wild type
One possible explanation for the severe ER retention of GLR-1 in UPR mutants is that GLR-1 ER retention is a secondary consequence of the reduction in synaptic signaling observed in these mutants. We previously observed GLR-1 localization in mutants that are defective for synaptic signaling either because they have severe deficits in synaptic vesicle release (e.g., unc-18 and unc-13) or because they fail to transport synaptic vesicles from the cell body to the synapse (e.g., unc-104; Rongo and Kaplan, 1999). These mutants have a severe uncoordinated locomotion defect; nevertheless, they are indistinguishable from wild-type nematodes in their ability to traffic GLR-1, suggesting that the defects in GLR-1 trafficking that we observe in UPR mutants are not simply due to a more general defect in synaptic signaling (Rongo and Kaplan, 1999).
We also examined the localization of a LIN-10::GFP chimera expressed under the glr-1 promoter (LIN-10::GFP) as LIN-10 is a cytosolic PDZ protein that colocalizes with GLR-1 at Golgi and at synapses and is required for GLR-1 localization (Rongo et al., 1998; Whitfield et al., 1999). Interestingly, the LIN-10::GFP localization to perinuclear Golgi structures and to clusters in the ventral nerve cord of UPR mutants is indistinguishable from the LIN-10::GFP localization pattern in wild-type nematodes (Figure 2, W and X, and unpublished data). This result demonstrates that although LIN-10 colocalizes with GLR-1 and is required for GLR-1 synaptic localization, LIN-10 does not require GLR-1 transport in order to form clusters in the distal neurites of the ventral cord.
UPR Genes Are Expressed in C. elegans Neurons
The differential requirement for the UPR pathway in different tissues and for different proteins might in part reflect a tissue-specific difference in expression of the pathway in metazoans. To address this issue, we generated GFP transcriptional and translational reporter constructs to examine where in nematodes IRE-1 and XBP-1 are expressed. In the presence of IRE1 signaling in most eukaryotes examined, an additional intron in XBP1 mRNA, which is not processed by the nuclear splicing apparatus and contains a reading frame shift, is removed by IRE1 endonuclease activity (Yoshida et al., 2001; Calfon et al., 2002). This results in an in-frame mature message and hence stable accumulation of the full-length XBP1 protein. In the absence of IRE1 signaling in mammals, this conserved intron is not removed from the XBP1 mRNA, resulting in a frame shift that prematurely terminates translation of the protein. The C. elegans xbp-1 gene consists of three exons and two introns comprising a 1807-nucleotide pre-mRNA (Shen et al., 2001). Moreover, xbp-1 contains a 23-base pair intron sequence in the second exon (nucleotides 953–976) that is conserved with the regulatory sequence found in other eukaryotes (Calfon et al., 2002). We have designated the protein produced from mRNA containing this intron XBP-1A, whereas protein produced from the spliced mRNA and containing the correctly frame shifted carboxy-terminus we have designated XBP-1B (Figure 4A). To determine the expression and localization of XBP-1, we generated a transgene, xbp-1(1–60)::gfp, that contains 1.8 kb of upstream sequence and the first exon of XBP-1 (nucleotides 1–180, amino acids 1–60) fused in frame to GFP. We introduced this transgene into nematodes and found that XBP-1(1–60)::GFP is expressed in all cells of embryos and adult larvae (unpublished data). XBP-1(1–60)::GFP contains a potential basic bipartite nuclear localization signal but lacks the regulatory intron sequence (nucleotides 954–976). We found that XBP-1(1–60)::GFP protein was produced even in the absence of ER stress and was localized to the nucleus, consistent with its role as a transcription factor thought to regulate the transcription of heat shock proteins and chaperones. We also found that XBP-1(1–60)::GFP expression was higher in neurons than in nonneural cells (unpublished data). To determine which forms of full-length XBP-1 are produced in nematodes, we generated transgenes that contained the 1.8-kb upstream promoter and either sequences encoding the XBP-1A (transgene XB3) or XBP-1B (transgene XB7) ORF fused in frame to GFP (Figure 4A). We introduced each transgene separately into nematodes and used RT-PCR to examine the mRNA they produced. Using a pair of primers that anneal to the xbp-1 5′end and the gfp 5′ end, we were able to detect a product of the correct size for xbp-1a::gfp (850 base pairs) by RT-PCR analysis of RNA from nematodes carrying XB3 (Supplementary Figure 2). We subcloned 24 independent RT-PCR products and found that three of them had spliced out the 23 nucleotide regulatory intron (Supplementary Figure 3). We were also able to detect a product of the correct size (1060 base pairs) for xbp-1b::gfp using the same primers on RNA from nematodes carrying XB7 (Supplementary Figure 2). We subcloned 24 independent RT-PCR products and found that two of them had spliced out the 23 nucleotide regulatory intron (Supplementary Figure 3). We have also sequenced the RT-PCR products, which confirms the intron-exon boundaries (Supplementary Figure 4). Our results suggest that a basal level of spliced XBP-1 is made even in the absence of ER stress.
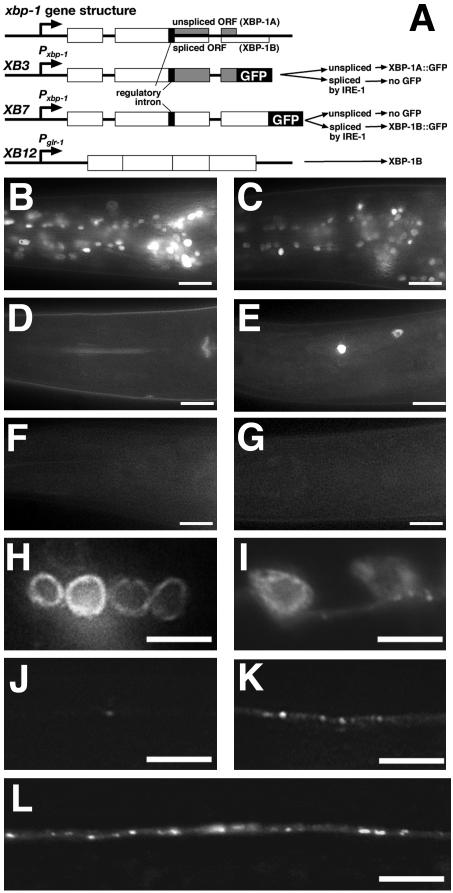
Figure 4.
UPR signaling in neurons. (A) Schematics of the xbp-1 genomic organization and the XBP-1 expression constructs (XB3, XB7, and XB12). White boxes indicate common exons, and gray boxes indicate exons present in the spliced product. The GFP translation fusion is indicated by a black box. The thin black box indicates the 23-base pair regulatory intron. The predicted products from each transgene are indicated, assuming either splicing or not of the regulatory intron. Nematodes either carry (B and C) the XB3 transgene, or (D–G) the XB7 transgene. (B) XBP-1A::GFP is expressed in all cells, whereas (D) XBP-1B::GFP is not detectable. (C) After ER stress induction, XBP-1A::GFP expression is decreased, whereas (E) XBP-1B::GFP expression can now be detected in some neurons. XBP-1B::GFP expression requires IRE-1 as expression is not detected in ire-1 mutants either (F) before or (G) after ER stress induction. (H–L) ire-1 mutants expressing GLR-1::GFP. GLR-1::GFP is exported out of the ER in (I) neuron cell bodies and localized to punctate structures in (K) distal neurites of ire-1 mutants that also express an ire-1 minigene under the glr-1 promoter. Mutants that lack the rescuing transgene (H) accumulate GLR-1::GFP in the ER and (J) show little GLR-1 in ventral cord neurites. Mutants that express a “prespliced” xbp-1 message from the XB12 transgene (L) export GLR-1 from the ER and localize it to punctate structures in the ventral cord. Scale bar in B–G, 10 μm; in H–L, 5 μm.
We examined these transgenic nematodes for the expression of their respective GFP fusion proteins. We found that XBP-1A::GFP protein, which is produced from mRNA that has not spliced out the 23 nucleotide intron, is expressed in most cells carrying the XB3 transgene (Figure 4B). However, we found that XBP-1B::GFP, which is produced from the ORF that results from the removal of the 23 nucleotide intron, is below the level of detection in neurons carrying XB7 (Figure 4D) The low level of protein produced by the spliced ORF confirms our RT-PCR results, which indicate that the spliced mRNA represents only ∼10% of the total. We induced ER stress in the three reporter transgenic strains by either a 3-h heat shock incubation at 30°C or treatment with tunicamycin. XBP-1(1–60)::GFP expression did not increase after heat shock, and XBP-1A::GFP expression decreased after ER stress, consistent with a switch in splicing to the XBP-1B ORF (Figure 4C and unpublished data). After ER stress induction, XBP-1B::GFP expression could now be detected in some cells, including neurons (Figure 4E and unpublished data). To determine whether IRE-1 is required for XBP-1B expression, we introduced the XB7 transgene into ire-1 mutants. We did not detect XBP-1B expression in the presence or absence of ER stress induction, demonstrating that IRE-1 is required for XBP-1B up-regulation (Figure 4, F and G, and unpublished data).
We also examined the expression of IRE-1 by generating a GFP reporter construct. We generated a transgene, ire-1::gfp, that contained 3 kb of upstream sequence and the first exon of IRE-1 (nucleotides 1–87, amino acids 1–29) fused in frame to GFP. We introduced this transgene into nematodes and found that IRE-1(1–29)::GFP is expressed in all cells of embryos and adult larvae (unpublished data). Unlike XBP-1 expression, we did not detect an enrichment for IRE-1 in neurons.
UPR Gene Activity Is Required Autonomously in C. elegans Neurons for GLR-1 Trafficking
The GLR-1 trafficking defects observed in UPR mutants could reflect a specific requirement for UPR signaling in the GLR-1–expressing neurons or a more general requirement for the pathway in other tissues of the animal. To test this hypothesis, we used the glr-1 promoter to express either an ire-1, xbp-1A, or xbp-1B cDNA in GLR-1–expressing cells of the corresponding mutants. We found that a transgene expressing IRE-1 under the glr-1 promoter was sufficient to restore GLR-1 trafficking and exit from the ER in ire-1 mutants to levels observed in wild-type nematodes (Figure 4, I and K). We also found that a transgene expressing XBP-1B but not XBP-1A was sufficient to restore GLR-1 trafficking in xbp-1 mutants (unpublished data). These results indicate that the UPR pathway is required in the same cells as GLR-1 for proper trafficking of GLR-1. Moreover, the XBP-1B reading frame that results from IRE-1–mediated splicing is required for GLR-1 trafficking.
We hypothesized that if IRE-1 catalyzed the splicing of xbp-1A mRNA into xbp-1B mRNA by removing the regulatory intron, then we should be able to preclude the requirement for IRE-1 by expressing “prespliced” xbp-1B transcripts, which already have the regulatory intron removed, in ire-1 mutants. We generated XB12, a transgene containing the glr-1 promoter and xbp-1B, which lacks all three introns including the regulatory intron (Figure 4A). We introduced XB12 into ire-1 mutants and found that it was sufficient to restore GLR-1 trafficking and exit from the ER in ire-1 mutants to wild-type levels (Figure 4L). These results demonstrate that the removal of the regulatory intron can allow xbp-1 to bypass the requirement for ire-1, suggesting that the role of IRE-1 protein in regulating GLR-1 trafficking is strictly for the regulated splicing of the xbp-1 message.
DISCUSSION
AMPA receptor subunit composition has been shown to play an important role in the functional and physiological properties of these channels as well as in their cell biological fate. The ability to form channels of different subunit composition within the same neuron might represent a unique challenge for AMPA receptor assembly and secretion through the ER. To begin to explore this idea, we examined the subcellular localization of GLR-1, a C. elegans receptor subunit similar to mammalian AMPA-type receptors, in mutant nematodes that lack ire-1 or xbp-1 activity. We showed that although GLR-1, GLR-2, and GLR-5 receptor subunits exit the ER of wild-type nematodes, they accumulate in the ER of UPR mutant nematodes even in the absence of ER stress events like heat shock or tunicamycin treatment. We examined other secreted proteins for their reliance on the UPR pathway under nonstressful conditions and found that all of the proteins that we tested, including several synaptic proteins, have little if any requirement for basal level signaling by the UPR pathway. Our data indicates that glutamate receptors have a special requirement for the UPR pathway either for their assembly into a channel or for the exit of receptor-containing channels that are assembled but retained in the ER for other reasons. Moreover, this requirement is age dependent and cell type specific.
XBP-1, a transcription factor that up-regulates targets of the UPR pathway, is expressed in most cells and enriched in neurons during larval stages. Vertebrate XBP1 has previously been shown to facilitate B-cell differentiation into plasma cells. B-cells are stimulated to differentiate by antigen and cytokine receptor signaling, and the cells increase their expression of immunoglobulins (Ig), presumably triggering signaling by IRE1 and causing the subsequent splicing of XBP1. This, in turn, leads to the activation of the UPR target genes needed to handle the Ig synthesis and secretion that occurs during plasma cell differentiation (Iwakoshi et al., 2003). In the absence of XBP1, B cells fail to activate the UPR, leading to the accumulation of Igs and other secreted proteins in the ER, which precludes their differentiation into plasma cells.
Does an analogous situation occur for GLR-1 trafficking? Late stage embryos and L1 larvae synthesize GLR-1, which is able to exit the ER and be delivered to neurites in both wild-type and UPR mutant nematodes. However, as development proceeds from larval stages into adulthood, GLR-1 translocation becomes dependent on the UPR pathway, suggesting that there might be an additional step in this process during the adult stages that requires XBP-1. One simple change that might be occurring during this step in differentiation is an increase in glutamate receptor expression, analogous to the change in the Ig production rate during plasma cell differentiation. We think that this is unlikely for several reasons. First, there is not a dramatic difference in the levels of GLR-1 in adults vs. L1 larvae. Second, when we tested other membrane proteins (e.g., SNB-1, TWK-18, mannosidase) by expressing them under the glr-1 promoter, they accumulated to levels similar to that of GLR-1 (also expressed under the identical glr-1 promoter) throughout all stages of development. Nevertheless, these similar levels of secreted proteins did not require the UPR pathway as did GLR-1. We suggest that activation of the UPR pathway is an important step in making older neurons competent to export receptor from the ER; however, unlike the requirement of XBP-1 for B-cell differentiation, we do not think that activation of the pathway is caused by a simple increase in GLR-1 levels during development.
Another explanation for why AMPA receptors specifically require the UPR could be their requirement for forming heteromeric channels of different subunit composition. For example, the receptors might require UPR signaling (and the subsequent chaperone response) in neurons where they need time to partner with other subunits, whereas they might not require such signaling in neurons where they would normally form a homomeric channel. We found this to be the case for GLR-5, which unlike GLR-1 and GLR-2 was not retained in the ER of all neurons in UPR mutant animals. GLR-5 is coexpressed with GLR-1 and GLR-2 in many of the interneurons that govern locomotion, and GLR-5 was retained in the ER of these cells in UPR mutants (Brockie et al., 2001). In contrast, GLR-5 is also expressed in neurons like DVA in which no other glutamate receptor subunit is expressed, suggesting that it forms homomeric channels in these neurons. GLR-5 is exported from the ER of DVA in both wild-type and UPR mutant nematodes, consistent with the idea that glutamate receptor subunits require the UPR for ER exit when they are faced with a choice of forming channels with other subunits.
Recent work indicates that edited GluR2 subunits, which contain Arg in their P-loop editing site, accumulate to high levels as monomers in the ER because of the poor ability of this subunit to form homomeric channels (Greger et al., 2002, 2003). The entry of limiting amounts of GluR1 and/or GluR3 (both have Gln in the editing site) allows GluR2 subunits to heteromultimerize and exit the ER. Presumably the excess of ER resident GluR2 coupled with its poor homotypic affinity results in most AMPA channels exiting the ER with only one or two GluR2 subunits. It is not clear why such an excess of monomeric GluR2 is retained in the ER and is not removed by ERAD (Ellgaard and Helenius, 2003). Interestingly, knockout mice that lack GluR2 subunits are forced to form GluR1/GluR3 heteromers, GluR1 homomers, and GluR3 homomers, but these channels are poorly translocated to synapses (Sans et al., 2003). These results suggest that GluR2 plays a critical role in channel assembly and trafficking and that there are mechanisms in place to assure the assembly of channels with only the appropriate combination of subunits. We speculate that the UPR pathway is one such mechanism, required to elevate the level of chaperones and other ER resident proteins that facilitate channel assembly between appropriate subunits and hence export from the ER.
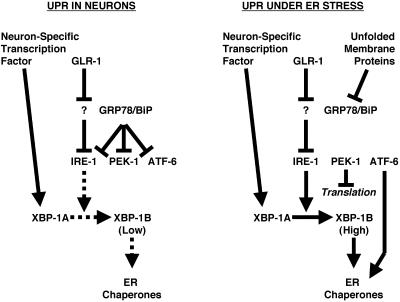
Does glutamate receptor trafficking require the other two known ER stress response pathways that function in parallel to IRE-1/XBP-1? We examined GLR-1 localization in pek-1 and atf-6 mutants, C. elegans homologues of the PERK and ATF6 proteins that signal in response to ER stress in parallel with IRE-1 (Shen et al., 2001; Calfon et al., 2002; Urano et al., 2002). We also examined sel-1 mutants, which lack an ERAD component (Urano et al., 2002). We found that GLR-1 translocates through the ER of these mutants and is localized as in wild-type nematodes, suggesting that of the three known parallel pathways that respond to ER stress, only IRE-1/XBP-1 seems to be required for glutamate receptor trafficking. All three pathways are thought to monitor BiP, which becomes sequestered by unfolded proteins in the ER. That GLR-1 trafficking requires IRE-1 but not PERK or ATF6 raises the interesting possibility that GLR-1 entry into the ER might activate IRE-1 by a mechanism other than BiP sequestration, which should activate all three pathways (Figure 5).
Figure 5.
Model for IRE-1 and XBP-1 signaling. In the absence of ER stress, glutamate receptors are able to trigger the partial activation of IRE-1/XBP-1 signaling, perhaps through a yet to be identified ER resident protein (indicated by the question mark). On stimulation by an ER stress event, the presence of unfolded proteins inhibits GRP78/BiP, thereby resulting in the activation of IRE-1, PEK-1, and ATF-6. Arrows indicate positive genetics interactions. Bars indicate negative genetics interactions. Dashed lines indicate weak interactions, whereas solid lines indicate strong interactions.
Glutamate receptors have been implicated in a number of CNS disorders and psychiatric conditions, raising the question of whether defects in glutamate receptor folding, assembly, and trafficking could be an underlying cause of these disorders (Lee et al., 2002; Tsai and Coyle, 2002; Barnes and Slevin, 2003). Recently, promoter mutations in human XBP1 that impair XBP1 expression have been shown to be a genetic risk factor for bipolar disorder. Given the particular predisposition of glutamate receptors to depend on UPR signaling for their proper ER export, it is tempting to speculate that glutamate receptor assembly and/or trafficking might be impaired in the bipolar disorder subpopulation affected with this XBP1 allele. Lower levels of XBP1 could produce lower levels of synaptic receptors, resulting in depressed glutamatergic signaling in affected individuals. Consistent with this idea, drugs that promote glutamate receptor function have been found to enhance biogenic amine-based therapies used to treat depression, and valproate, which can stimulate the UPR, is an effective treatment for bipolar disorder (Kakiuchi et al., 2003; Li et al., 2003).
One common link in many neuropathologies is the production of misfolded and/or aggregated proteins, which accumulate and eventually cause neuronal loss (Paschen and Frandsen, 2001; Kudo et al., 2002; Soto, 2003). In many of the diseases in which misfolded proteins are detected, impaired neural function can be observed before neurodegeneration. As many of the resulting misfolded proteins share the ER with glutamate receptors, it is possible that these misfolded proteins are overtaxing the UPR, thereby inhibiting glutamate receptor assembly and trafficking, which in turn might underlie some of the earliest symptoms of affected individuals. Small molecules that potentiate the UPR pathway or AMPA receptor function might be effective therapeutics for treating such disorders.
Supplementary Material
Acknowledgments
We thank A. Fire, A. Fraser (Sanger Institute), R. Herman (CGC), J. Kaplan, Y. Kohara, M. Kunkel, V. Maricq, J. Mendel, C. Mello, M. Nonet, S. Nurrish, T. Rapoport, M. Rolls, L. Salkoff, T. Stiernagle (CGC), and R. Tsien for reagents and strains. We also thank Bonnie Firestein and Ruth Steward for critical comments on the manuscript. We were assisted with the transgene integrations by A. Caravello, C. H. Chang, and L. Monahan. T.U. was supported by a Japan Society for the Promotion of Science Fellowship. C.R. is a Pew Scholar in the Biomedical Sciences. Funding was provided by the National Institutes of Health (R01 NS42023) and a Grant-In-Aid from the American Heart Association.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04–02–0108. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–02–0108.
The online version of this article contains supplementary material accessible through http://www.molbiolcell.org.
References
- Barnes, G.N., and Slevin, J.T. (2003). Ionotropic glutamate receptor biology: effect on synaptic connectivity and function in neurological disease. Curr. Med. Chem. 10, 2059-2072. [DOI] [PubMed] [Google Scholar]
- Beattie, E.C., Carroll, R.C., Yu, X., Morishita, W., Yasuda, H., von Zastrow, M., and Malenka, R.C. (2000). Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat. Neurosci. 3, 1291-1300. [DOI] [PubMed] [Google Scholar]
- Brockie, P.J., Madsen, D.M., Zheng, Y., Mellem, J., and Maricq, A.V. (2001). Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J. Neurosci. 21, 1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbea, M., Dreier, L., Dittman, J.S., Grunwald, M.E., and Kaplan, J.M. (2002). Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35, 107-120. [DOI] [PubMed] [Google Scholar]
- Calfon, M., Zeng, H., Urano, F., Till, J.H., Hubbard, S.R., Harding, H.P., Clark, S.G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92-96. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie, M., Sulston, J.E., White, J.G., Southgate, E., Thomson, J.N., and Brenner, S. (1985). The neural circuit for touch sensitivity in C. elegans. J. Neurosci. 5, 956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J.S., Shamu, C.E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197-1206. [DOI] [PubMed] [Google Scholar]
- Dingledine, R., Borges, K., Bowie, D., and Traynelis, S.F. (1999). The glutamate receptor ion channels. Pharmacol. Rev. 51, 7-61. [PubMed] [Google Scholar]
- Duerr, J.S., Frisby, D.L., Gaskin, J., Duke, A., Asermely, K., Huddleston, D., Eiden, L.E., and Rand, J.B. (1999). The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J. Neurosci. 19, 72-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, N.D., Adler, C.E., Crump, J.G., L'Etoile, N.D., and Bargmann, C.I. (2001). Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron 31, 277-287. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell. Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- Greger, I.H., Khatri, L., Kong, X., and Ziff, E.B. (2003). AMPA receptor tetramerization is mediated by q/r editing. Neuron 40, 763-774. [DOI] [PubMed] [Google Scholar]
- Greger, I.H., Khatri, L., and Ziff, E.B. (2002). RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron 34, 759-772. [DOI] [PubMed] [Google Scholar]
- Grunwald, M.E., and Kaplan, J.M. (2003). Mutations in the ligand-binding and pore domains control exit of glutamate receptors from the endoplasmic reticulum in C. elegans. Neuropharmacology 45, 768-776. [DOI] [PubMed] [Google Scholar]
- Hart, A.C., Sims, S., and Kaplan, J.M. (1995). Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378, 82-84. [DOI] [PubMed] [Google Scholar]
- Hollmann, M., and Heinemann, S. (1994). Cloned glutamate receptors. Annu. Rev. Neurosci. 17, 31-108. [DOI] [PubMed] [Google Scholar]
- Iwakoshi, N.N., Lee, A.H., and Glimcher, L.H. (2003). The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194, 29-38. [DOI] [PubMed] [Google Scholar]
- Kakiuchi, C. et al. (2003). Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet. 35, 171-175. [DOI] [PubMed] [Google Scholar]
- Kass, J., Jacob, T.C., Kim, P., and Kaplan, J.M. (2001). The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J. Neurosci. 21, 9265-9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R.J., Scheuner, D., Schroder, M., Shen, X., Lee, K., Liu, C.Y., and Arnold, S.M. (2002). The unfolded protein response in nutrient sensing and differentiation. Nat. Rev. Mol. Cell. Biol. 3, 411-421. [DOI] [PubMed] [Google Scholar]
- Kudo, T., Katayama, T., Imaizumi, K., Yasuda, Y., Yatera, M., Okochi, M., Tohyama, M., and Takeda, M. (2002). The unfolded protein response is involved in the pathology of Alzheimer's disease. Ann. NY Acad. Sci. 977, 349-355. [DOI] [PubMed] [Google Scholar]
- Kunkel, M.T., Johnstone, D.B., Thomas, J.H., and Salkoff, L. (2000). Mutants of a temperature-sensitive two-P domain potassium channel. J. Neurosci. 20, 7517-7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.G., Zhu, X., Ghanbari, H.A., Ogawa, O., Raina, A.K., O'Neill, M.J., Perry, G., and Smith, M.A. (2002). Differential regulation of glutamate receptors in Alzheimer's disease. Neurosignals 11, 282-292. [DOI] [PubMed] [Google Scholar]
- Li, X., Witkin, J.M., Need, A.B., and Skolnick, P. (2003). Enhancement of antidepressant potency by a potentiator of AMPA receptors. Cell Mol. Neurobiol. 23, 419-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.W., Ju, W., Foster, K., Lee, S.H., Ahmadian, G., Wyszynski, M., Wang, Y.T., and Sheng, M. (2000). Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat. Neurosci. 3, 1282-1290. [DOI] [PubMed] [Google Scholar]
- Ma, D., and Jan, L.Y. (2002). ER transport signals and trafficking of potassium channels and receptors. Curr. Opin. Neurobiol. 12, 287-292. [DOI] [PubMed] [Google Scholar]
- Malinow, R. (2003). AMPA receptor trafficking and long-term potentiation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358, 707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta-Mitrovic, M., Jan, Y.N., and Jan, L.Y. (2000). A trafficking checkpoint controls GABA(B) receptor heterodimerization. Neuron 27, 97-106. [DOI] [PubMed] [Google Scholar]
- Maricq, A.V., Peckol, E., Driscoll, M., and Bargmann, C.I. (1995). Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378, 78-81. [DOI] [PubMed] [Google Scholar]
- Mellem, J.E., Brockie, P.J., Zheng, Y., Madsen, D.M., and Maricq, A.V. (2002). Decoding of polymodal sensory stimuli by postsynaptic glutamate receptors in C. elegans. Neuron 36, 933-944. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ma, W., Gething, M.J., and Sambrook, J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743-756. [DOI] [PubMed] [Google Scholar]
- Nikawa, J., and Yamashita, S. (1992). IRE1 encodes a putative protein kinase containing a membrane-spanning domain and is required for inositol phototrophy in Saccharomyces cerevisiae. Mol. Microbiol. 6, 1441-1446. [DOI] [PubMed] [Google Scholar]
- Nonet, M.L., Saifee, O., Zhao, H., Rand, J.B., and Wei, L. (1998). Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J. Neurosci. 18, 70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen, W., and Frandsen, A. (2001). Endoplasmic reticulum dysfunction—a common denominator for cell injury in acute and degenerative diseases of the brain? J. Neurochem. 79, 719-725. [DOI] [PubMed] [Google Scholar]
- Passafaro, M., Piech, V., and Sheng, M. (2001). Subunit-specific temporal and spatial patterns of AMPA receptor exocytosis in hippocampal neurons. Nat. Neurosci. 4, 917-926. [DOI] [PubMed] [Google Scholar]
- Rolls, M.M., Hall, D.H., Victor, M., Stelzer, E.H., and Rapoport, T.A. (2002). Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell 13, 1778-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo, C., and Kaplan, J.K. (1999). CaMKII regulates the density of central glutamatergic synapses in vivo. Nature 402, 195-199. [DOI] [PubMed] [Google Scholar]
- Rongo, C., Whitfield, C.W., Rodal, A., Kim, S.K., and Kaplan, J.M. (1998). LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94, 751-759. [DOI] [PubMed] [Google Scholar]
- Sans, N. et al. (2003). Aberrant formation of glutamate receptor complexes in hippocampal neurons of mice lacking the GluR2 AMPA receptor subunit. J. Neurosci. 23, 9367-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D.B., Blanpied, T.A., Swanson, G.T., Zhang, C., and Ehlers, M.D. (2001). An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21, 3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893-903. [DOI] [PubMed] [Google Scholar]
- Sheng, M. (2001). Molecular organization of the postsynaptic specialization. Proc. Natl. Acad. Sci. USA 98, 7058-7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, S., Hayashi, Y., Esteban, J.A., and Malinow, R. (2001). Subunit-specific rules governing AMPA receptor trafficking to synapses in hippocampal pyramidal neurons. Cell 105, 331-343. [DOI] [PubMed] [Google Scholar]
- Soto, C. (2003). Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 4, 49-60. [DOI] [PubMed] [Google Scholar]
- Spear, E., and Ng, D.T. (2001). The unfolded protein response: no longer just a special teams player. Traffic 2, 515-523. [DOI] [PubMed] [Google Scholar]
- Standley, S., Roche, K.W., McCallum, J., Sans, N., and Wenthold, R.J. (2000). PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron 28, 887-898. [DOI] [PubMed] [Google Scholar]
- Tirasophon, W., Welihinda, A.A., and Kaufman, R.J. (1998). A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12, 1812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, G., and Coyle, J.T. (2002). Glutamatergic mechanisms in schizophrenia. Annu. Rev. Pharmacol. Toxicol. 42, 165-179. [DOI] [PubMed] [Google Scholar]
- Urano, F., Calfon, M., Yoneda, T., Yun, C., Kiraly, M., Clark, S.G., and Ron, D. (2002). A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J. Cell Biol. 158, 639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Z., Harding, H.P., Zhang, Y., Jolicoeur, E.M., Kuroda, M., and Ron, D. (1998). Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17, 5708-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, C.W., Benard, C., Barnes, T., Hekimi, S., and Kim, S.K. (1999). Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol. Biol. Cell 10, 2087-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland, F.T., Gleason, M.L., Serafini, T.A., and Rothman, J.E. (1987). The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell 50, 289-300. [DOI] [PubMed] [Google Scholar]
- Xia, H., Hornby, Z.D., and Malenka, R.C. (2001). An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology 41, 714-723. [DOI] [PubMed] [Google Scholar]
- Yandell, M.D., Edgar, L.G., and Wood, W.B. (1994). Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 91, 1381-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881-891. [DOI] [PubMed] [Google Scholar]
- Zerangue, N., Schwappach, B., Jan, Y.N., and Jan, L.Y. (1999). A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron 22, 537-548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.