ABSTRACT
Meiotic recombination initiates following the formation of DNA double-strand breaks (DSBs) by the Spo11 endonuclease early in prophase I, at discrete regions in the genome coined “hot spots.” In mammals, meiotic DSB site selection is directed in part by sequence-specific binding of PRDM9, a polymorphic histone H3 (H3K4Me3) methyltransferase. However, other chromatin features needed for meiotic hot spot specification are largely unknown. Here we show that the recombinogenic cores of active hot spots in mice harbor several histone H3 and H4 acetylation and methylation marks that are typical of open, active chromatin. Further, deposition of these open chromatin-associated histone marks is dynamic and is manifest at spermatogonia and/or pre-leptotene-stage cells, which facilitates PRDM9 binding and access for Spo11 to direct the formation of DSBs, which are initiated at the leptotene stage. Importantly, manipulating histone acetylase and deacetylase activities established that histone acetylation marks are necessary for both hot spot activity and crossover resolution. We conclude that there are functional roles for histone acetylation marks at mammalian meiotic recombination hot spots.
KEYWORDS: crossover, double-strand break initiation, histone acetylation, meiotic recombination, mouse meiosis
INTRODUCTION
Meiotic recombination commences only following the double-stranded cleavage of DNA by the meiosis-specific Spo11 endonuclease in leptotene to early zygotene meiosis I cells (1–4), at discrete regions of the genome coined meiotic recombination “hot spots” (5, 6). Meiotic DNA double-strand breaks (DSBs) are then repaired by homologous recombination (HR) pathways, mediated by the meiosis-specific Rad51 and DMC1 repair proteins in mid- and late-zygotene-stage cells (4, 7, 8). The chromatin features, regulatory factors, and histone modifications that contribute to the control of mammalian meiotic DSB site selection by Spo11 and to the initiation, crossover (CO) activity, and resolution of meiotic recombination events are generally poorly understood.
In mice and humans, one key regulator of DSB site selection is PRDM9 (5, 9–13), a polymorphic histone H3 methyltransferase that binds in a sequence-specific fashion to polymorphic sequences that are present in nearly 90% of hot spot cores (14) and that generates the trimethylated histone H3 lysine 4 (H3K4Me3) mark that is a hallmark of open chromatin (15, 16). However, in addition to the known role for the H3K4Me3 mark at hot spot cores, an emerging body of evidence suggests that several histone modifications associated with open chromatin, including acetylation, methylation, and ubiquitination, are involved in regulating meiotic DSBs (17–24). For example, in Saccharomyces cerevisiae, histone H2B ubiquitylation by the Rad6-Bre1 ubiquitin ligase complex is necessary for the proper recruitment and/or stabilization of the meiotic DSB-forming machinery that contains Spo11 (24). Further, ubiquitylated histone H2B is known to promote methylation of open chromatin-associated marks on histone H3, at K4 and K79, suggesting that histone H2B ubiquitylation influences DSB formation through methylation of histone H3 (24–26). Moreover, the Schizosaccharomyces pombe histone acetyltransferase (HAT) SpGcn5 is required for optimal recombination activity of the ade6-M26 meiotic hot spot (21), and X nondisjunction factor 1 (xnd-1) of Caenorhabditis elegans controls CO activity and DSB formation at least in part by modulating acetylation of H2A lysine 5 (19). Finally, loss of the Sir2 histone deacetylase (HDAC) alters the frequency of meiosis-specific DSBs in up to 12% of all yeast genes (27).
Roles for histone acetylation and methylation have also been established for modulation of DNA DSB repair pathways. First, the human histone acetyltransferase Tip60 induces acetylation of histones surrounding DNA damage sites, resulting in chromatin relaxation and loading of HR repair proteins (28). Second, both yeast and human histone acetyltransferase 1 proteins are required for acetylating histones H3 and H4 at DSB sites, which facilitates Rad51 recruitment to promote efficient homologous recombination (29, 30). Third, the SET domain histone methyltransferase (HMT) Metnase, which methylates histone H3 K4 and K36, creating marks of open chromatin, appears to be important for recruiting repair factors and for facilitating the joining of DNA ends during nonhomologous end-joining repair (31). Fourth, H3K4Me2 and H3K9Ac marks are associated with the mouse Psmb9 hot spot core, and histone H4 hyperacetylation is a feature of the mouse Hlx1 hot spot core during meiotic DSB repair (32). Finally, in yeast the histone acetyltransferases Gcn5 and Esa1 and the histone deacetylases Rpd3, Sir2, and Hst1 are dynamically recruited to the HO lesion during homologous recombination repair in Saccharomyces cerevisiae (33).
At present, it is unclear if there are dynamic and select changes in histone modifications at mouse recombination hot spots throughout meiosis and if such marks play roles in the control of recombination. Using highly purified fractions of cells from all stages of meiosis I and native chromatin immunoprecipitation (nChIP)/real-time PCR profiling, we show that there are dynamic changes in acetylated and methylated histone marks found in open chromatin at recombinogenically active meiotic hot spot cores and that, specifically, histone acetylation plays profound functional and necessary roles in laying down histone methylation marks and in hot spot crossover activity.
RESULTS
Expression analysis of genes encoding histone modification enzymes during meiosis I.
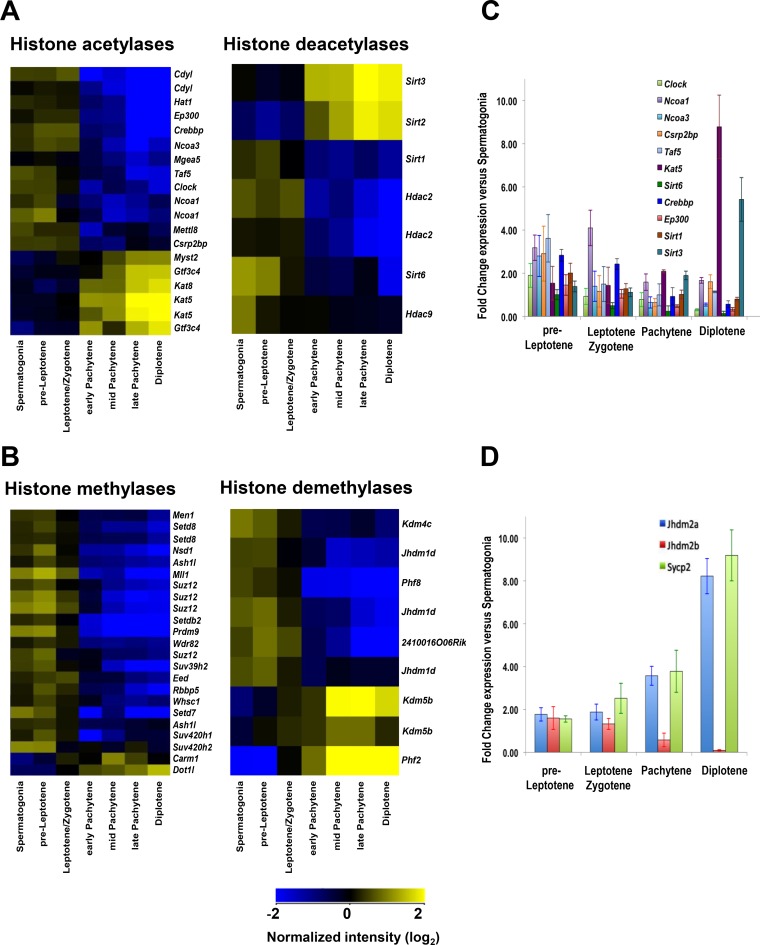
To initially assess the possible dynamics of histone modifications in meiosis I, gene expression analysis was performed on highly fluorescence-activated cell sorter (FACS)-purified populations of meiotic cells (34–36). Expression profiling and quantitative reverse transcription-PCR (qRT-PCR) analyses established differential gene expression for 11 histone acetyltransferases (HATs), 16 histone methyltransferases (HMTs), and 4 histone deacetylases and demethylases (HDACs and HDMTs) in early meiotic spermatogonium, preleptotene, and leptotene-zygotene cells versus pachytene and later meiotic-stage cells (Fig. 1; see Data Set S1 and Table S1 in the supplemental material). Further, several of the HATs (e.g., those encoded by Hat1, Ep300, Crebbp, and Taf5) and HMTs (e.g., those encoded by Ash1l, Mll1, Prdm9, and Suz12) expressed at high levels during early meiosis I (Fig. 1A; Table S1) target lysine residues of histones H3 and H4 and are essential for the transcription of meiotic genes, for germ cell and embryonic development, and/or for the DNA damage response (30, 37–42). Finally, some of the HAT-, HDAC-, and HMT-encoding genes that are upregulated at later stages of meiosis I (e.g., Kat5, Sirt3, and Dot1l) (Fig. 1A and B; Data Set S1) have functions in DSB repair (42–44). Notably, the yeast homolog of Sirt3, Sir2, functions as a meiotic checkpoint and controls meiotic recombination (27, 45, 46).
FIG 1.
Expression of histone-modifying enzymes during meiosis. (A) Profiling of differentially expressed genes encoding histone acetylases and deacetylases in all meiosis I cell stages from wild-type B6/DBA mouse testes. (B) Profiling of differentially expressed genes encoding histone methylases and demethylases in all meiosis I cell stages from wild-type B6/DBA mouse testes. (A and B) Meiotic cells were isolated using FACS sorting as described in Materials and Methods. For all heat maps, an absolute fold change filter of 1.5-fold for the spermatogonium/average pachytene expression ratio was applied to obtain differentially expressed genes, followed by hierarchical clustering. Only significantly changed probe sets with P values (corrected) of <0.05 are shown. Microarray data were normalized across the median for all samples, and microarray analyses are described in Materials and Methods. Details of statistical analysis are provided in Data Set S1 in the supplemental material, and the findings were confirmed by qRT-PCR. (C) qRT-PCR analyses of genes encoding HATs and HDACs were performed for all stages of meiosis I. (D) qRT-PCR analyses of specific meiotic stage marker genes that are selectively expressed at different stages of meiosis I (90). (C and D) The y axis shows the normalized expression ratio for the indicated meiotic stage versus expression in spermatogonia. Results are the means and standard errors of the means (SEM) for technical replicates (n = 3). qRT-PCR analysis details are described in Materials and Methods. Real-time PCR primer pairs used for panel C are shown in Table S1.
Dynamic regulation of histone H3 and H4 acetylation and methylation marks at recombination hot spot cores during meiosis I.
The differential regulation of histone-modifying enzymes suggested that histone marks might be controlled dynamically at hot spot cores prior to and/or following the formation of DSBs. To test this, we performed native chromatin immunoprecipitation (nChIP) followed by real-time PCR analyses of several histone H3 and H4 acetylation and methylation marks throughout meiosis, from spermatogonia to diplotene-stage cells, at four well-characterized meiotic recombination hot spots, HS22, HS59.4, HS59.5, and HS61.1, which are present on mouse chromosome 19 (Fig. 2 to 4; Fig. S1) (34, 47–49). These four hot spots have crossover (CO) rates ranging from 0.05 × 10−4 to 6.0 × 10−4 crossover per meiosis, which are comparable to the CO rates for the majority of mouse meiotic hot spots, which have recombination frequencies of 10−5 to 10−3 (6, 49, 50).
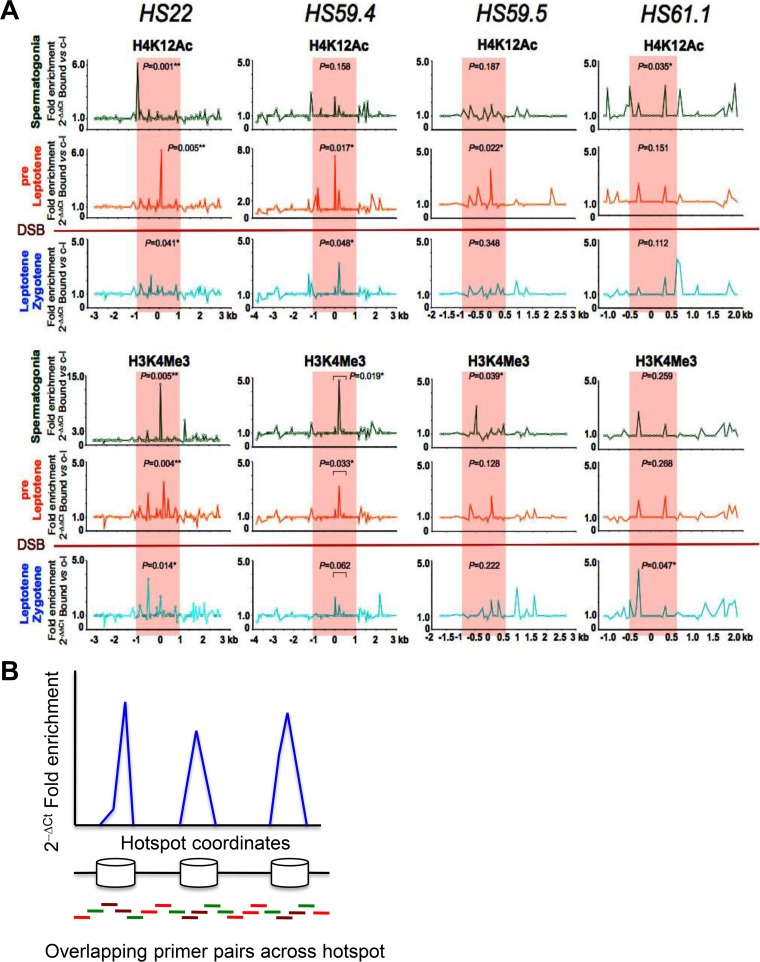
FIG 2.
Profiles of active histone marks at mouse meiotic recombination hot spots in early meiotic prophase. (A) Representative normalized native ChIP profiles of the active H4K12Ac and H3K4Me3 histone marks are shown for the HS22, HS59.4, HS59.5, and HS61.1 hot spots in spermatogonium, preleptotene, and leptotene-zygotene meiotic cells. Normalized native ChIP profiles were obtained by real-time PCR data analysis for validated primer pairs that overlap these hot spots (see Materials and Methods and panel B). The y axis indicates the normalized absolute ratio of bound native ChIP fractions for a given histone mark versus the no-antibody control, using the 2−ΔΔCT formula. The x axis represents the location across the analyzed HS locus. Initial native ChIP profiles were obtained by calculating the absolute fold enrichment between bound and input ChIP DNA fractions, using the 2−ΔCT formula (see panel B). Each of the ChIP profiles shown is the average for three independent, normalized ChIP experiments. The Mann-Whitney test was performed to determine the statistical significance of the difference in normalized bound fractions for a given histone mark and the no-antibody control within either the entire hot spot core region or a specific subregion of the hot spot core, indicated with a bar, as described in Materials and Methods. Corresponding P values are depicted for each of the histone H4K12Ac and H3K4Me3 mark ChIPs at the HS22, HS59.4, HS59.5, and HS61.1 hot spot cores for three meiotic cell stages. *, P < 0.05; **, P < 0.01. Note that statistically significant P values were obtained for active histone mark enrichments within hot spot cores for at least one of the meiotic stages, and usually for at least one of or both the early spermatogonium and preleptotene meiotic stages, which precede the formation of DSBs. In the H3K4Me3 mark profile at the HS61.1 hot spot core, the most significant enrichment was observed for the leptotene-zygotene meiotic population. Red shading indicates the hot spot cores. The red line between the preleptotene and leptotene-zygotene meiotic stages indicates the stage when Spo11 generates DSBs. (B) Schematic of the real-time PCR technique used to obtain native ChIP profiles of histone marks and nucleosome occupancy profiles for mouse meiotic hot spots on chromosome 19. The native ChIP method used to generate profiles of histone modification marks across meiotic hot spots involved isolation of immunoprecipitation-enriched mononucleosomal, histone mark-associated DNAs from FACS-purified meiotic cell fractions, which were then further analyzed by real-time PCR using validated overlapping primer pairs across a given hot spot locus (96, 94, 48, and 41 primer pairs were used to tile the HS22, HS59.4, HS59.5, and HS61.1 hot spots, respectively) (34, 47) (also see Table S2). For nucleosome occupancy profiles (Fig. S2), mononucleosomal DNA was purified and used as a template for quantitative PCR, using the same overlapping primer pairs across several hot spot regions. In brief, nucleosomal DNA regions (blue peaks) were amplified with brown (robust PCR signal) and green (intermediate PCR signal) primer pairs, while DNA gaps between the nucleosomes were amplified with green and red primer pairs (trace PCR signal). Finally, native ChIP profiles and nucleosome occupancy profiles were obtained using the 2−ΔCT formula for bound and input ChIP DNA fractions and for the mononucleosome MNase-digested DNA fraction and undigested genomic DNA, respectively.
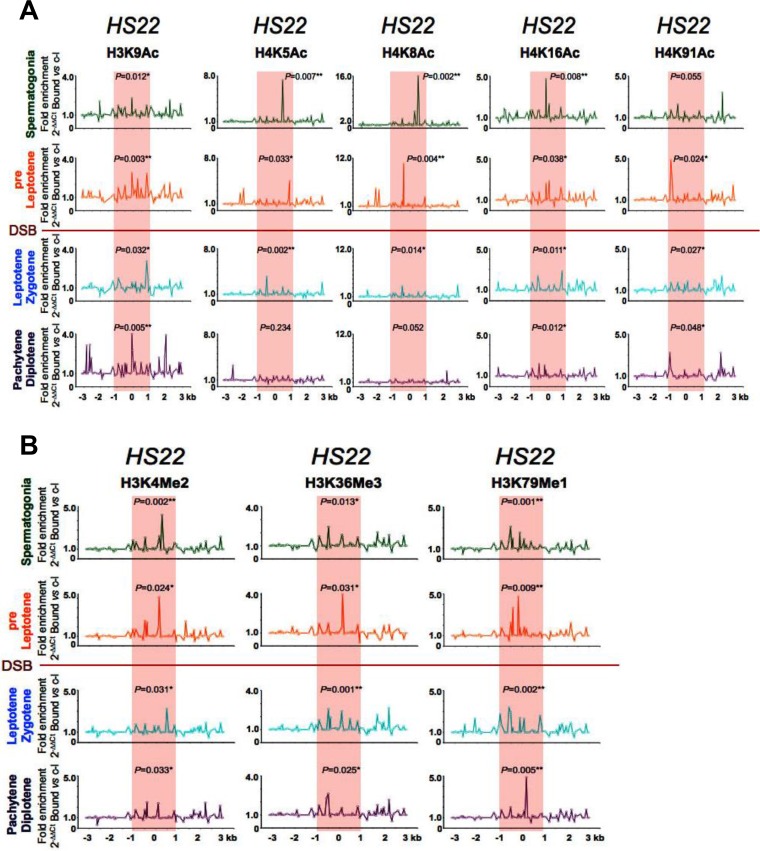
FIG 3.
Profiles of active acetylated and methylated histone marks at the HS22 hot spot in early meiotic prophase. (A) Representative normalized native ChIP profiles of the active H3K9Ac, H4K5Ac, H4K8Ac, H4K16Ac, and H4K91Ac acetylated histone marks are shown for spermatogonium, preleptotene, leptotene-zygotene, and pachytene-diplotene meiotic cells for the HS22 hot spot. (B) Representative normalized native ChIP profiles of the active H3K4Me2, H3K36Me3, and H3K79Me1 methylated histone marks are shown for spermatogonium, preleptotene, leptotene-zygotene, and pachytene-diplotene meiotic cells for the HS22 hot spot. Normalized histone modification profiles in panels A and B were obtained as described in the legend to Fig. 2. ChIP profiles shown are the averages for three independent, normalized ChIP experiments. The statistical significance of the histone mark enrichments within the HS22 hot spot core and P values were defined using the Mann-Whitney test as described in the legend to Fig. 2. *, P < 0.05; **, P < 0.01. Red shading, hot spot core; red line, stage where Spo11 generates DSBs.
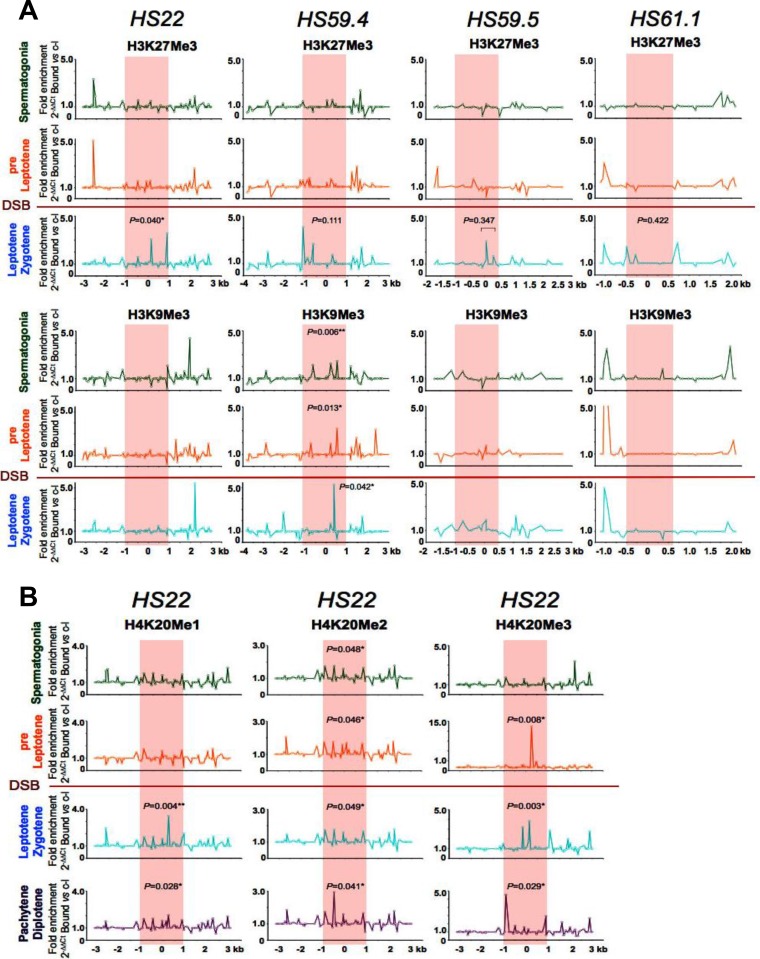
FIG 4.
Profiles of repressive methylated histone marks at mouse meiotic recombination hot spots in early meiotic prophase. (A) Representative normalized native ChIP profiles of the repressive H3K27Me3 and H3K9Me3 methylated histone marks are shown at the HS22, HS59.4, HS59.5, and HS61.1 hot spots in spermatogonium, preleptotene, and leptotene-zygotene meiotic cells. (B) Additional representative normalized native ChIP profiles of the repressive H4K20Me1, H4K20Me2, and H4K20Me3 methylated histone marks are shown at the HS22 hot spot for spermatogonium, preleptotene, leptotene-zygotene, and pachytene-diplotene meiotic cells. Normalized histone modification profiles in both panels were obtained as described in the legend to Fig. 2. ChIP profiles shown are averages for three independent, normalized ChIP experiments. The statistical significance of the histone mark enrichments within the HS22 hot spot core (if any) and P values were defined using the Mann-Whitney test as described in the legend to Fig. 2. *, P < 0.05; **, P < 0.01. Red shading, hot spot cores; red line, stage where Spo11 generates DSBs.
Notably, nChIP analyses using validated antibodies specific for histone marks (data are presented as profiles normalized to no-antibody controls) revealed that active histone marks that are typical of open chromatin are a hallmark of hot spot cores in spermatogonia and/or preleptotene cells, the stages of meiosis I that immediately precede the formation of DSBs (Fig. 2 and 3), a scenario that should facilitate access of these sites by the Spo11 endonuclease that directs DSBs at the leptotene stage (1, 3, 5). These active histone marks include, for example, H4K12Ac and H3K4Me3, which have known roles in marking actively transcribed open chromatin regions (Fig. 2A) (15, 16). Moreover, the HS22 hot spot cores of spermatogonia and pre-leptotene-stage cells harbored several other histone modifications that are associated with open chromatin, including the H3K9Ac, H4K5Ac, H4K8Ac, H4K16Ac, and H4K91Ac acetylated marks (Fig. 3A) as well as the active methylated marks H3K4Me2, H3K36Me3, and H3K79Me1 (Fig. 3B) (16, 51, 52). Notably, the active histone marks in spermatogonia and/or preleptotene meiotic cells were mostly observed within the cores of the meiotic HS22, HS59.4, HS59.5, and HS61.1 hot spots, which are the most active recombinogenic regions in hot spot loci (34, 47–49). However, in some instances, comparable active histone mark levels were also seen for zones adjacent to the core regions of the hot spots, which likely represent recombinogenic sites outside the hot spot cores. These included, for example, the H4K12Ac mark at the HS59.4 and HS61.1 hot spots and the H3K9Ac mark at the HS22 hot spot in spermatogonia, the H4K12Ac mark at the HS61.1 hot spot, and the H4K5Ac and H4K8Ac marks at the HS22 hot spot in preleptotene cells (Fig. 2A and 3A). Importantly, there was concordance in the overlap of the histone mark ChIP profiles with nucleosome occupancy maps, which were determined by real-time PCR and whole-genome sequencing (WGS) analyses of micrococcal nuclease (MNase) resistance regions across these four hot spots (Fig. S2). Finally, in some instances, the active histone marks were reduced to background levels at hot spot cores in leptotene-zygotene cells and/or during later stages of meiosis (e.g., pachytene-diplotene cells) (Fig. 2 and 3; Fig. S1A).
To assess if active histone marks were manifest outside recombinogenic hot spot regions, we also performed nChIP analyses of two of the active histone marks, H4K12Ac and H3K4Me3, at 1-kb intervals at flanking sites surrounding the HS22 hot spot core, i.e., at kb −25, −10, +10, and +25 (Fig. S3, top and middle panels). Importantly, DSB profiling of the flanking sites of HS22 using whole-genome sequencing data for meiotic DSBs in mice (10) showed that HS22-flanking regions were indeed recombinogenically inactive, with only trace DSB levels observed at these loci, while recombination sites were highly enriched at the HS22 hot spot core (Fig. S3, bottom panels). Notably, the signal intensities for both the H4K12Ac and H3K4Me3 active histone marks were comparable to background levels at upstream and downstream regions flanking the HS22 hot spot, in contrast to their marked enrichment at the HS22 hot spot core in spermatogonium, pre-leptotene-, and leptotene-zygotene-stage cells. Finally, and importantly, the nChIP-qPCR profile of the active histone H3K4Me3 mark at the HS22 meiotic hot spot core and flanking regions highly overlapped the respective WGS mapping of this mark obtained from genome-wide H3K4Me3 ChIP-DNA sequencing (ChIP-Seq) analyses of mouse meiotic cells (Fig. S3, middle and bottom panels) (10).
Repressive histone methylation marks that are usually associated with heterochromatic regions (H3K27Me3, H3K9Me3, H4K20Me1, H4K20Me2, and H4K20Me3) (15, 16, 52, 53) were generally absent in hot spot cores in spermatogonia and pre-leptotene-stage cells (Fig. 4). Some repressive histone marks were evident at these cores during DSB formation at the leptotene-zygotene stage and at later stages of meiosis (Fig. 4; Fig. S1B). However, not all repressive histone marks followed this pattern. For example, H3K9Me3 and H4K20Me3 marks were observed at the HS59.4 and HS22 cores, respectively, in preleptotene cells (Fig. 4). This may reflect the dual nature of these two histone marks, which have been observed in both active and silenced genes (52, 54). Alternatively, the presence of such repressive marks might correlate with the reduced recombination rates of some meiotic hot spots, such as HS59.4 (34).
Note that the generally modest enrichment of histone modification marks detected by nChIP at meiotic hot spot cores reflects the fact that the data are averages for the entire meiotic population and represent only about 10% of the total of such marks, as only 10% of meiotic DSBs are successfully repaired via CO events in mice (55, 56). These findings are also consistent with a genome-wide analysis of H3K4Me3 marks at mouse meiotic recombination hot spots (14), which showed that the H3K4Me3 signals at the sites of DSB hot spots are generally much weaker than those that are found at transcription start sites.
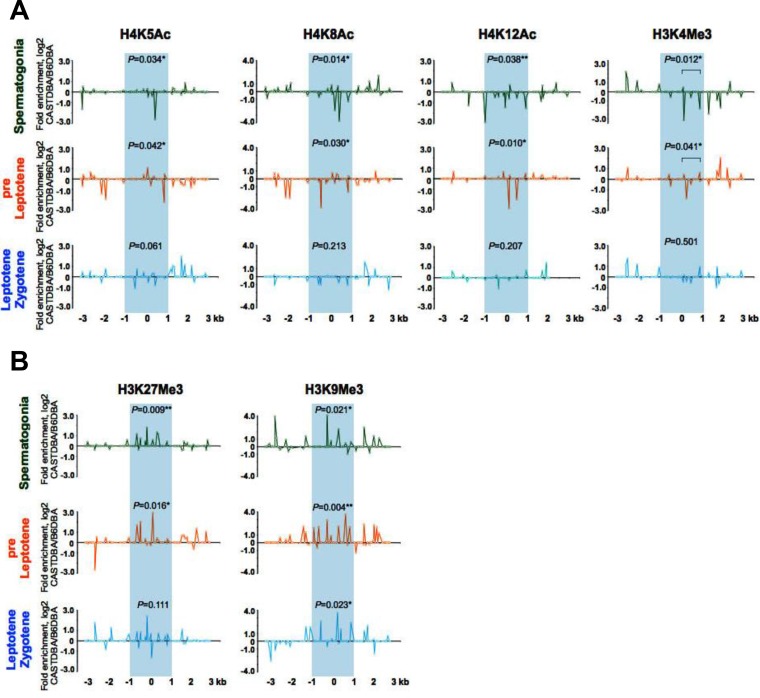
Repressive histone H3 and H4 marks predominate at hot spot cores in inactive mouse backgrounds.
To assess if changes in histone marks at a given hot spot core correlate with the recombination activity of the hot spot, we compared the dynamics of the histone marks at the HS22 hot spot during early meiosis I in the CAST/DBA strain, in which this hot spot is inactive (47), and the B6/DBA background, in which HS22 is active (48) (Fig. 5; Fig. S4). Notably, the active acetylated and methylated histone marks H4K5Ac, H4K8Ac, H4K12Ac, and H3K4Me3 were reduced to background levels at the HS22 core in the inactive CAST/DBA strain, whereas they were enriched in B6/DBA spermatogonium, pre-leptotene-, and leptotene-zygotene-stage cells (Fig. 5A; Fig. S4). In contrast, the repressive H3K9Me3 and H3K27Me3 histone modifications were enriched in early-stage meiotic cells at the HS22 hot spot core in the inactive CAST/DBA strain versus the active B6/DBA strain (Fig. 5B; Fig. S4). Thus, histone marks typical of active, open chromatin at hot spot cores are indeed associated with the activity of meiotic recombination hot spots.
FIG 5.
Profiles of active and repressive acetylated and methylated histone H3 and H4 marks at the HS22 hot spot in inactive versus active mouse strain backgrounds. (A) Normalized log2(ΔΔCT) ratios of bound native ChIP fractions of active H4K5Ac, H4K8Ac, and H4K12Ac acetylated and H3K4Me3 methylated histone marks at the HS22 hot spot core in the inactive CAST/DBA strain versus the active B6/DBA mouse strain for spermatogonium, preleptotene, and leptotene-zygotene meiotic cells. (B) Normalized log2(ΔΔCT) ratios of bound native ChIP fractions of repressive H3K9Me3 and H3K27Me3 methylated histone marks at the HS22 hot spot core in the inactive CAST/DBA strain versus the active B6/DBA strain for spermatogonia and preleptotene and leptotene-zygotene meiotic cells. Inactive HS22 hot spot cores are indicated by blue shading in both panels. Normalized histone modification profiles were obtained as described in Materials and Methods. Data shown are averages for three independent, normalized ChIP experiments. The Mann-Whitney test was performed to determine the statistical significance (if any) of the difference in normalized bound fractions for a given histone mark within the HS22 hot spot core region or a specific subregion of the HS22 hot spot core, indicated with a bar, in the CAST/DBA and B6/DBA mouse strains, as described in Materials and Methods. Corresponding P values are depicted for each of the histone H4K5Ac, H4K8Ac, and H3K4Me3 mark ChIPs (A) and histone H3K27Me3 and H3K9Me3 mark ChIPs (B) at the HS22 hot spot core for three meiotic cell stages. *, P < 0.05; **, P < 0.01. Corresponding nonnormalized histone modification profiles at the HS22 core for three meiotic cell stages in the CAST/DBA and B6/DBA mouse strains are shown in Fig. S4.
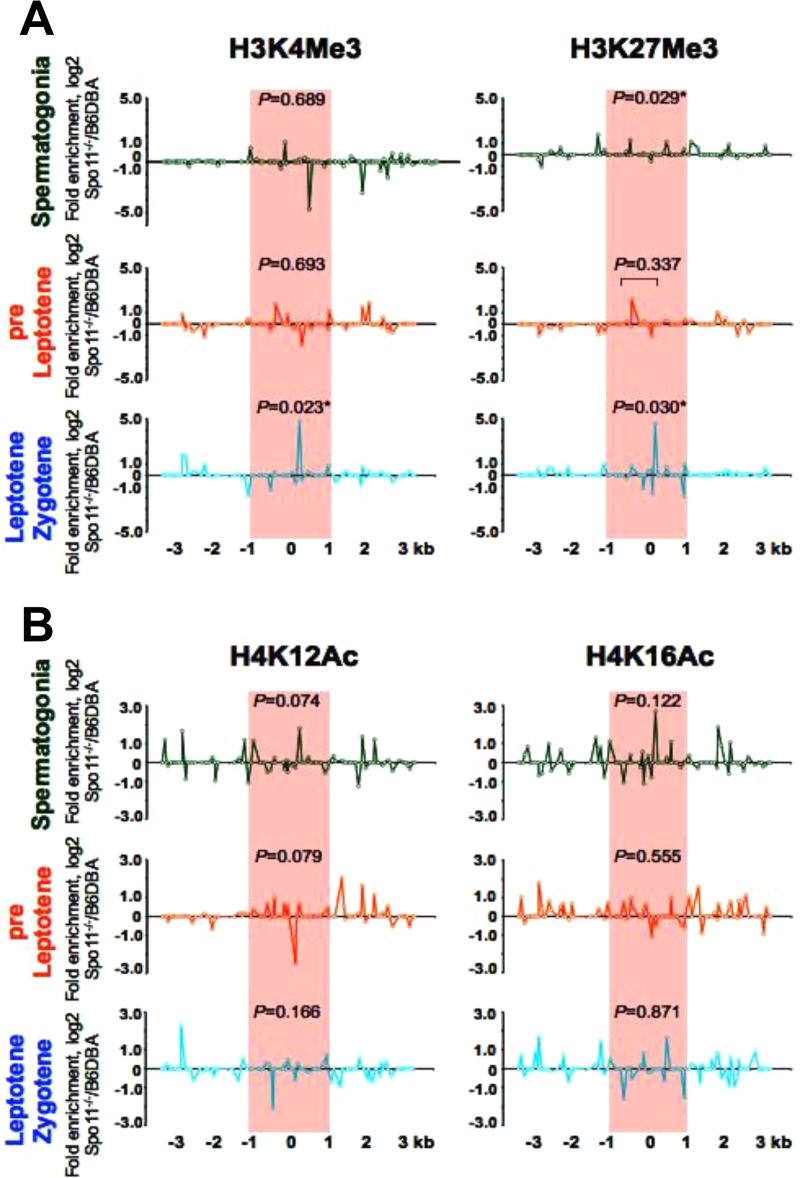
Chaotic histone marks are manifest at meiotic hot spots in Spo11−/− meiotic cells.
In mouse Spo11−/− spermatocytes, DSBs are not generated, the chromosomes fail to synapse, and meiosis is arrested in prophase I, at the zygotene stage (2, 4, 57, 58). Native ChIP analyses demonstrated that the pattern of histone modifications at hot spot cores in early meiotic Spo11−/− germ cells is rather chaotic, with both active and repressive marks being present (Fig. 6; Fig. S5). For example, both active and repressive H3K4Me3 and H3K27Me3 methylated histone marks are present at the HS22 core in early meiotic Spo11−/− cells, and these accumulate to very high levels, with 23- to 27-fold increases in Spo11−/− leptotene-zygotene-like cells compared to the levels of these histone marks in wild-type (WT) early meiotic cells (Fig. 6A; Fig. S5). Further, levels of the active H4K12Ac mark are reduced in early meiotic Spo11−/− cells, whereas the levels of the active H4K16Ac mark either are not significantly changed or are enriched at the HS22 hot spot core in Spo11−/− versus wild-type spermatogonia (Fig. 6B; Fig. S5).
FIG 6.
Profiles of active and repressive histone H3 and H4 marks at the HS22 hot spot of Spo11−/− versus wild-type early meiotic cells. (A) Normalized log2(ΔΔCT) ratios of bound native ChIP fractions of active and repressive methylated H3K4Me3 and H3K27Me3 histone marks at the HS22 hot spot were determined for spermatogonia and preleptotene and leptotene-zygotene-like meiotic cells from Spo11−/− versus wild-type B6/DBA mice. (B) Normalized log2(ΔΔCT) ratios of bound native ChIP fractions of active acetylated H4K12Ac and H4K16Ac histone marks at the HS22 hot spot were determined for spermatogonium, preleptotene, and leptotene-zygotene-like meiotic cells from Spo11−/− versus wild-type B6/DBA mice. The hot spot core found at HS22 in B6/DBA mice is indicated by red shading in both panels. Normalized histone modification profiles were obtained as described in Materials and Methods. Data shown are averages for at least two independent, normalized ChIP experiments. The Mann-Whitney test was performed to determine the statistical significance (if any) of the difference in normalized bound fractions for a given histone mark within the HS22 hot spot core region or a specific subregion of the HS22 hot spot core, indicated with a bar, in Spo11−/− and wild-type B6/DBA mice, as described in Materials and Methods. Corresponding P values are depicted for each of the histone H3K4Me3 and H3K27Me3 mark ChIPs (A) and histone H4K12Ac and H4K16Ac mark ChIPs (B) at the HS22 hot spot core for three meiotic cell stages. *, P < 0.05. Corresponding nonnormalized histone modification profiles at the HS22 hot spot core for three meiotic cell stages in the Spo11−/− and wild-type B6/DBA mouse strains are provided in Fig. S5.
The observed changes in histone methylation and acetylation mark levels at hot spot cores at different Spo11−/− meiotic cell stages are in accord with changes in the expression patterns of genes encoding HMTs, HDMTs, HATs, and HDACs in Spo11−/− meiotic cells (Fig. S6 and S7 and Data Sets S1 and S2). The histone-modifying genes analyzed were defined as those that were differentially expressed in wild-type meiotic cells (Fig. 1; Data Set S1). In general, histone-modifying genes that were upregulated in wild-type early meiotic cells were also mostly upregulated in Spo11−/− meiotic cells, yet genes that were upregulated in wild-type pachytene and diplotene meiotic cells had reduced expression levels in Spo11−/− mouse meiotic cells (Fig. 1A and B; Fig. S6 and S7). These included genes encoding several HMTs (Carm1 and Dot1l), HATs (Kat5, Kat8, Gtf3c4, and Myst2), HDMTs (Kdm5b and Phf2), and HDACs (Sirt2 and Sirt3). However, there were a few examples of HDMT and HDAC genes (e.g., Phf2 and Sirt2) that were upregulated in Spo11−/− versus wild-type preleptotene cells and spermatogonia/preleptotene cells, respectively (Fig. S6C and D and S7C and D).
Among histone-modifying genes that were highly expressed in wild-type early meiotic cells, the most profound differences in Spo11−/− versus wild-type gene expression ratios were observed for histone methyltransferase gene clusters, which were upregulated in Spo11−/− spermatogonia and leptotene-zygotene-like cells (Fig. S6A and B and Data Set S2). Upregulated genes in these clusters included Prdm9, Suz12, Setdb2, Wdr82, and Suv420h1, which encode histone methyltransferases that deposit both active and repressive histone H3 marks. In contrast, some of the histone methyltransferase genes that were enriched in wild-type early meiotic cells and that direct deposition of repressive methylated histone H3 marks, including Setd8, Whsc1, Eed, Suv39h2, Suv420h1, and Suv420h2, had reduced expression in Spo11−/− pre-leptotene-stage meiotic cells (Fig. S6A and B and Data Set S2).
Among genes that encode histone acetylases and that are highly expressed in wild-type early meiotic cells, only a few HAT genes were upregulated in Spo11−/− spermatogonia (Ncoa1, Ncoa3, and Gtf3c4) and leptotene-zygotene-stage cells (Cdyl and Clock) (Fig. S7A and B and Data Set S2). However, there were no significant changes in the expression of these HAT genes in Spo11−/− preleptotene cells (Fig. S7A and B and Data Set S2).
The expression patterns of genes encoding HDMTs and HDACs in Spo11−/− mouse germ cells were in agreement with the Spo11−/− HMT and HAT gene expression patterns in these cells (Fig. S6 and S7 and Data Set S2). For example, upregulated HDMTs in Spo11−/− preleptotene cells (encoded by Kdm4c, Phf2, Phf8, and Jhdm1d) mostly target repressive methylated histone H3 marks, while downregulated HDMTs (encoded by Kdm5b and 2410061O06Rik) target active methylated histone H3 marks (Fig. S6C and D and Data Set S2). Interestingly, the histone deacetylase Sirt6, which is downregulated in Spo11−/− preleptotene cells (Fig. S7C and D and Data Set S2), is known to control acetylation of histone H3 in telomeric chromatin and DNA DSB repair and is required for genomic stability (59).
Histone acetylation is necessary for activating histone methylation marks at hot spot cores and for meiotic recombination hot spot activity.
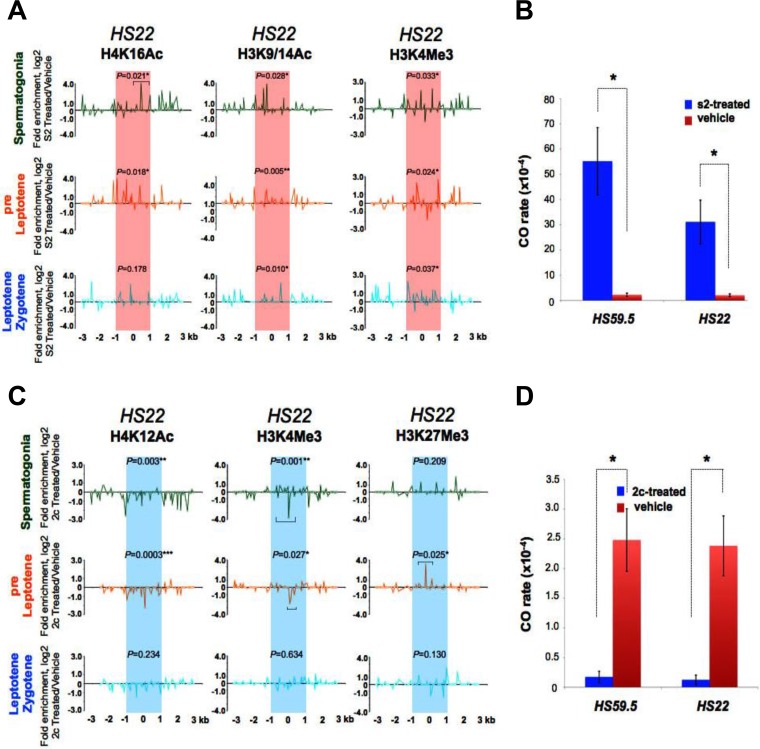
To test if histone acetylation plays functional roles in the deposition of other histone marks at hot spots and/or in hot spot site selection, activity, or CO resolution, we assessed the effects of in vivo treatments with validated small-molecule HDAC and HAT inhibitors (Fig. 7; Fig. S8 to S11). As an HDAC inhibitor, we used the mercaptoacetamide compound s2 (Fig. S8A, left panel), a broad-spectrum HDAC inhibitor (50% inhibitory concentration [IC50] of 0.2 μM) (60) that was originally designed as an anticancer drug, has prolonged activity, and markedly increases levels of total acetylated histone H4 in vivo (61, 62). Treatment with s2 did not affect meiosis, as reflected by FACS profiles (Fig. S8B, left panel), and also led to expected increases in the total levels of acetylated histone H3 in meiotic cells (Fig. S8C). Notably, s2 treatment significantly increased the levels of H4K16Ac and H3K9/14Ac marks (up to 17- and 14-fold, respectively) at the hot spot cores tested (HS22 and HS59.5) in spermatogonium, preleptotene, and leptotene-zygotene meiotic cells (Fig. 7A; Fig. S9 and S11A). Further, s2 treatment also significantly increased (up to 6-fold) the levels of the active histone H3K4Me3 mark at the HS22 and HS59.5 hot spot cores in these cells (Fig. 7A; Fig. S9 and S11A). Thus, increasing the levels of histone acetylation marks also augments the deposition of the activating histone H3K4 methylation mark at hot spot cores. Most importantly, increased histone acetylation marks at hot spot cores correlated with marked increases in CO recombination rates at these hot spots of up to 24- and 15-fold for the HS59.5 and HS22 hot spots, respectively (Fig. 7B; Table S4). Finally, s2 treatment also led to an increase of H4K12 acetylation at the inactive HS22 core in early meiosis I cells from CAST/DBA mice (Fig. S11B), but this did not activate hot spot core activity in this strain (data not shown). Thus, deposition of histone acetylation marks is not sufficient to initiate DSBs at meiotic hot spot cores in inactive strain backgrounds.
FIG 7.
Histone acetylation controls the activity of mouse meiotic recombination hot spots. (A) Normalized log2(ΔΔCT) native ChIP analyses of active H4K16Ac and H3K9/14Ac acetylated histone and H3K4Me3 methylated histone marks were performed at the HS22 hot spot for spermatogonium, preleptotene, and leptotene-zygotene meiotic cells from 10-week-old B6/DBA mice treated i.p. (1 mg/kg) with the HDAC inhibitor s2 (Fig. S8A and B, left panel) versus vehicle. Red shading indicates the s2-activated HS22 hot spot core. (B) CO rates at the HS59.5 and HS22 hot spots for s2-treated (blue bars) versus vehicle-treated (red bars) sperm samples. (C) Normalized log2(ΔΔCT) native ChIP analyses of active H4K12Ac acetylated and H3K4Me3 methylated histone marks and of the inactive H3K27Me3 methylated histone mark were performed at the HS22 hot spot for spermatogonium, preleptotene, and leptotene-zygotene meiotic cells from 10-week-old B6/DBA mice treated i.p. (50 mg/kg) with the liposomal HAT inhibitor 2c (Fig. S8A and B, right panel) versus vehicle. Blue shading indicates the 2c-inactivated HS22 hot spot core. (D) CO rates at the HS59.5 and HS22 hot spots for 2c-treated (blue bars) versus vehicle-treated (red bars) sperm samples. The normalization method and inhibitor treatment details for panels A and C are described in Materials and Methods. Data shown are averages for at least two independent, normalized ChIP experiments. The Mann-Whitney test was performed to determine the statistical significance (if any) of the difference in normalized bound fractions for a given histone mark within the HS22 hot spot core region or a specific HS22 core subregion, indicated with a bar, in the s2- and vehicle-treated (A) or 2c- and vehicle-treated (C) B6/DBA mice, as described in Materials and Methods. In panels A and C, corresponding P values are depicted for each of the histone marks at the HS22 hot spot core for three meiotic cell stages. *, P < 0.05; **, P < 0.01. Corresponding nonnormalized histone modification profiles at the HS22 hot spot core for three meiotic cell stages in s2- or vehicle-treated and 2c- or vehicle-treated B6/DBA mice are provided in Fig. S9 and S10. In panels B and D, results are means and SEM for biological replicates (n = 3). *, P < 0.05 by Student's t test.
To test if histone acetylation is necessary for hot spot activity, mice were treated with the cinnamoyl small-molecule HAT inhibitor compound 2c (Fig. S8A, right panel), which inhibits p300 HAT activity (IC50 of 5 μM) and blocks histone acetylase activity in HeLa cells (63). For in vivo treatments, we prepared a liposome-encapsulated 2c drug that was soluble in saline, based on a protocol developed for curcumin (64). Treatment with liposomal 2c had no toxicity for treated mice and had essentially no effect on meiotic progression as judged by FACS analyses (Fig. S8B, right panel), yet it led, as expected, to marked reductions in the levels of total acetylated histones H3 and H4 in meiotic cells (Fig. S8D). Notably, 2c treatment essentially abolished histone H4K12Ac marks at the HS22 and HS59.5 recombination cores in spermatogonium, preleptotene, and leptotene-zygotene meiotic cells (Fig. 7C; Fig. S10 and S11C). Moreover, HAT inhibition also led to marked decreases in the levels of the activating histone H3K4Me3 mark and to increases in the repressive histone H3K27Me3 mark at the HS22 and HS59.5 cores (up to 10-fold) in early meiotic cells (Fig. 7C; Fig. S10 and S11C). Thus, histone acetylation controls the deposition of chromatin-activating and -repressing histone methylation marks at hot spot cores. Most importantly, hypoacetylation of histones H3 and H4 at hot spot cores was associated with marked reductions in CO rates, as recombination rates dropped 15- to 20-fold at the HS59.5 and HS22 cores, respectively (Fig. 7D; Table S4).
DISCUSSION
Roles of histone acetylation and methylation marks during mouse meiosis I.
Several of the histone-modifying enzymes that are upregulated in mouse early meiotic cells (Fig. 1; see Data Set S1 in the supplemental material) are essential for development, as their deletion in the mouse leads to prenatal or postnatal lethality, defects in spermatogenesis, chromosomal aberrations, and/or infertility (e.g., the deletion of Hat1, Suv39h1/2, Suv4-20h1/2, Mll1, Prdm9, Sirt1, or Sirt6) (39–41, 65–70). Further, some of the histone acetylation and methylation marks that are deposited by these HATs and HMTs have defined roles in mammalian germ cell development and meiosis. For example, mammalian germ cells, similar to embryonic stem cells, form bivalent (poised) chromatin domains that bear both activating H3K4Me3 and repressive H3K27Me3 histone marks at the promoters of a large set of developmental genes; this architecture appears to be important for maintenance of germ cell identity and for poised expression of regulators of somatic cell fate (38, 71, 72). In addition, H3K9 and H3K4 methylation is required for higher-order hetero- and euchromatin structures that are needed for homologous chromosome synapsis and sex body formation (39, 41, 65). Finally, the H3K4Me3 mark is a known marker of meiotic DSBs in yeast and mammals and also has functions in activating the transcription of meiotic genes (5, 41).
Dynamic control of select histone acetylation and methylation marks at recombination hot spot cores during meiosis I.
The data reported herein establish that, unlike nucleosome occupancy (34, 47), histone marks at hot spot cores are dynamic. First, histone acetylation and methylation marks that are typical of open chromatin are present at hot spot cores at meiotic stages that precede the formation of DSBs by Spo11 (i.e., in spermatogonia and preleptotene cells), and such marks would be predicted to open chromatin and facilitate Spo11 binding and cleavage. Accordingly, at these early stages of meiosis there is a dearth of repressive histone methylation marks at hot spot cores. Later in meiosis, in leptotene-diplotene cells, several of the active histone marks are reduced and repressive marks often appear. Second, activating histone marks are not found at hot spot cores in strains where these hot spots are inactive, which are rather decorated by repressive histone marks. Collectively, these findings suggest that an open chromatin environment is necessary for meiotic DSBs. However, as shown in our in vivo HDAC inhibitor studies, the deposition of activating histone acetylation marks is not sufficient to confer activity on an otherwise inactive hot spot core.
Profiling of a cast of histone marks revealed that select modifications have distinct dynamics at hot spot cores. For example, histone H4 acetylated at K5, K8, K12, and K16 is enriched at hot spot cores in spermatogonia and pre-leptotene-stage cells, prior to the formation of DSBs, and these marks then decrease in leptotene-zygotene cells and are reduced to background levels following DSB repair in pachytene-diplotene cells (Fig. 2 and 3A; Fig. S1A). In contrast, the histone H3K9Ac mark increases from spermatogonia to diplotene cells, and the histone H4K91Ac is enriched in both preleptotene and pachytene-diplotene cells but is reduced in leptotene-zygotene cells at the HS22 hot spot core (Fig. 3A). In part, these differences may reflect specific functions of such modified histones during the formation or repair of meiotic DSBs. Indeed, H4K91 acetylation plays essential functions in chromatin assembly and DNA damage repair (73). Further, the changes in histone acetylation marks at mouse hot spots noted herein have corollaries in yeast, as during HO endonuclease-mediated recombination, which resembles Spo11-induced events (74), histone H4 acetylation marks accumulate at the sites flanking the DSB, followed by deacetylation of these marks during DSB repair (33). Notably, several histone acetyltransferases (Hat1p, Gcn5, and Esa1) and histone deacetylases (Rpd3, Sir2, and Hst1) are recruited to DSBs during homologous recombination repair in Saccharomyces cerevisiae (30, 33).
The H3K4Me3 mark is a known hallmark of mouse and human meiotic hot spots that is deposited by the meiosis-specific histone methyltransferase PRDM9 (5, 11, 13). In the present study, H3K4Me3 was also shown to mark meiotic hot spot cores in spermatogonia and preleptotene meiotic cells, which are stages that precede DSB formation. Thus, at these stages, the deposition of the H3K4Me3 mark at hot spot cores may facilitate opening of adjacent chromatin regions for the subsequent DSB event (Fig. 2). H3K4Me3 mark levels were also either enriched (HS22 and HS61.1) (P < 0.05) or slightly reduced (HS59.4 and HS59.5) in leptotene-zygotene cells during the DSB formation and repair stages, in agreement with the reduced levels of histone acetylation marks observed at meiotic hot spot cores in these cells (Fig. 2A and 3A). Profiling also revealed that the active H3K4Me2, H3K36Me3, and H3K79Me1 marks, as well as the repressive H4K20Me3 mark, were also enriched at the HS22 hot spot in preleptotene meiotic cells, just prior to the formation of DSBs (Fig. 3B and 4B), and that the H3K79Me1 and H4K20Me3 marks were also present at the HS22 hot spot core in leptotene-zygotene meiotic cells and were augmented in pachytene-diplotene cells (Fig. 3B and 4B). Notably, these H3K79 and H4K20 marks have been linked to the recruitment of the DNA repair factor 53BP1/CRB2 to sites of DNA damage in mammalian cells and yeast (75–79), and thus it is feasible that they might play similar roles in DNA repair in meiosis.
Generally, active acetylated and methylated histone marks are reduced and repressive methylated marks are increased at hot spot cores in leptotene-zygotene and/or pachytene-diplotene cells, right after the formation of DSBs (Fig. 2 to 4; Fig. S1). This scenario suggests a resetting of the chromatin conformation after the DSB is formed and then repaired. In accord with this notion, both H3K4Me3 and H3K27Me3 marks, which are hallmarks of bivalent chromatin regions, are enriched at meiotic recombination hot spots in human sperm (80).
Spo11 is necessary for proper chromatin structure at mouse recombination hot spots.
In the absence of DSBs in Spo11−/− mice, the data suggest that a bivalent-like chromatin structure that has both active (H3K4Me3) and repressive (H3K27Me3) histone methylation marks is formed at hot spot cores at all early meiotic stages and that this is especially manifest in leptotene-zygotene-like cells (Fig. 6A; Fig. S5). As noted above, bivalent chromatin domains are associated with promoters of developmental genes having roles in embryonic development and in lineage specification in undifferentiated embryonic stem (ES) cells and mammalian germ cells (38, 72, 81). For example, bivalent regions of chromatin have been proposed to silence developmental genes in ES cells and primordial germ cells, as well as in meiotic and postmeiotic germ cells, while keeping them poised for activation (71, 72, 81, 82). Notably, some histone acetylation marks are reduced (H4K12Ac, at all three meiotic stages) or enriched (H4K16Ac, mostly in spermatogonia) at recombination hot spot cores in Spo11−/− mice (Fig. 6B; Fig. S5).
HAT/HDAC and HMT/HDMT genes that are differentially expressed in wild-type early and late meiotic stages (Fig. 1) have similar expression patterns in Spo11−/− germ cells, where similar histone-modifying genes are upregulated in both early meiotic Spo11−/− and early meiotic wild-type cells (Fig. 1; Fig. S6 and S7). The most significant changes in the gene expression values for Spo11−/− meiotic cells were in clusters of upregulated HMT genes in spermatogonia and leptotene-zygotene-like cells that direct deposition of both active and repressive histone H3 methylation marks (Fig. S6A and B and Data Set S2). Changes in expression levels of these histone methylases and demethylases in Spo11−/− germ cells may contribute to the formation of the chaotic bivalent-domain chromatin structure that is observed at hot spot cores in Spo11−/− meiotic cells and that may also be present in genes that are essential for meiotic progression. Such a scenario would keep these genes silent in a Spo11-deficient context and would cause early meiotic arrest in Spo11−/− mice. Further, such a bivalent chromatin structure may also contribute to keeping recombination hot spots in Spo11−/− meiotic cells in an inactive state. These findings also establish that Spo11, alone or in complex with chromatin binding factors, contributes to the organization of hot spot cores and is required to initiate and/or sustain specific histone marks at DSBs.
Histone acetylation is necessary but not sufficient for mouse meiotic hot spot activity.
Notably, our in vivo HAT and HDAC inhibitor studies revealed functional roles for histone acetylation in controlling meiotic hot spots. First, inhibition of HDACs augmented histone acetylation marks at hot spot cores in spermatogonium, preleptotene, and leptotene-zygotene meiotic cells and, surprisingly, also led to increases in activating histone methylation marks, for example, H3K4Me3, at these sites (Fig. 7A; Fig. S9 and S11A). Conversely, inhibition of HATs abolished histone acetylation and histone methylation marks typical of open chromatin and rather led to the accumulation of repressive histone methylation marks (e.g., H3K27Me3) at hot spot cores in early meiotic cells (Fig. 7C; Fig. S10 and S11C). These data suggest that histone acetylation is necessary to initiate or sustain an open chromatin structure at hot spots and is also required for subsequent histone methylation and hot spot activity. Alternatively, this may reflect hierarchical histone modifications at recombination hot spot cores. We favor the former alternative, whereby histone acetylation at hot spot cores facilitates the deposition of activating histone methylation marks (e.g., via PRDM9) that then fully open chromatin to allow access of the protein recombination machinery and cleavage by Spo11. In support of this notion, HDAC inhibitor treatment augments levels of the H4K16Ac mark at meiotic hot spot cores (Fig. 7A; Fig. S9 and S11A), and H4K16Ac has known roles in disrupting the compaction of higher-order chromatin domains (83). In addition, HDAC inhibitor treatment also augments acetylation of H3K9 and H3K14 at meiotic hot spot cores in early meiotic cells (Fig. 7A; Fig. S9 and S11A), which promotes access of the repair machinery to damaged DNA (42). The doubly acetylated H3K9/14Ac mark has also been shown to be a hallmark of loose/open chromatin regions (84).
Notably, our studies revealed that manipulating histone acetylation levels at hot spot cores leads to concordant, profound changes in hot spot activity and that this may reflect effects on H3K4Me3 levels at these sites (Fig. 7B and D; Table S4). We propose that the opening of chromatin at meiotic hot spots by histone acetylation at stages that precede the DSB increases access of the core to the protein recombination machinery and stimulate first the DSB event (e.g., PRDM9 binding and H3K4 methylation, followed by the Spo11-induced DSB) and eventually CO formation at these sites (Fig. 7B; Table S4). In contrast, compaction of chromatin at recombination cores by blocking of histone acetylation restricts access of PRDM9 and Spo11 to meiotic hot spots, leading to a decrease in the frequency of DSBs and to marked reductions in hot spot activity (Fig. 7D; Table S4). Our findings strongly support the notion that histone acetylation is necessary for meiotic recombination, and they are consistent with analyses of hot spot activity in epigenetic mutant strains of yeasts and plants (21, 85–87). For example, deletion of the S. pombe HAT gene Gcn5, which directs hyperacetylation of the ade6-M26 hot spot, leads to reductions in DSBs and CO rates at this hot spot (21). Conversely, deletion of the S. cerevisiae HDAC gene Rpd3 augments HIS4 hot spot activity (86). However, our HDAC inhibitor studies with the CAST/DBA mouse strain, in which the HS22 hot spot is inactive, have also shown that augmenting histone acetylation (Fig. S11B) is not sufficient to confer recombinogenic activity. Thus, other levels of control in addition to histone acetylation marks must also be manifest in licensing hot spot cores for DSB cleavage and recombination.
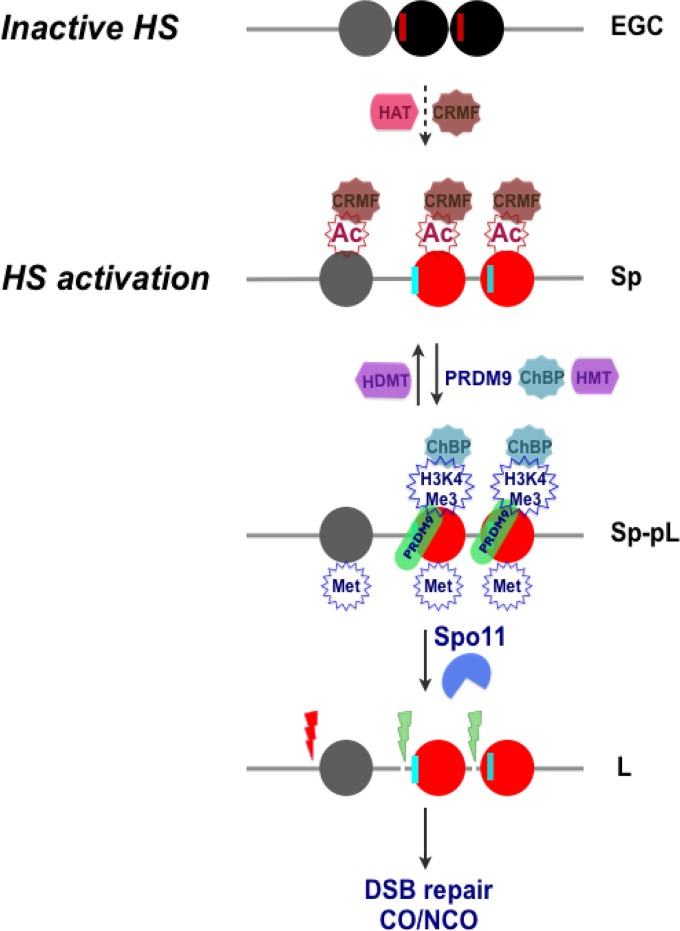
Collectively, the data establish a dynamic role for histone modifications in controlling the activity and resolution of mammalian recombination hot spots. Specifically, these studies reveal functional roles for acetylated histones at mouse recombination hot spot cores, with acetylated histones revealed as being necessary but not sufficient for imbuing hot spot activity. The data support a model (Fig. 8; Fig. S12) in which initially inactive condensed chromatin that is present at hot spot cores in early germ cells, i.e., primordial germ cells (37, 88, 89), shifts to an open conformation at the onset of meiosis in spermatogonia and pre-leptotene-stage cells, and they suggest that open chromatin is driven, at least in part, by the deposition of acetylated marks on histones H3 (K9) and H4 (K5, K8, K12, K16, and K91) by histone acetyltransferases expressed in these cells (Fig. 1A and 8; Fig. S12B). In turn, acetylation of histone H3 and H4 tails opens up chromatin at the hot spot core, making them more accessible for PRDM9 binding, followed by PRDM9-driven deposition of H3K4Me3 marks at these sites, and the deposition of additional active methylated histone H3 marks (H3K4Me2, H3K36Me3, and H3K79Me1) by HMTs expressed in early meiotic cells (Fig. 1B and 8; Fig. S12B). Collectively, this creates a chromatin environment conducive to Spo11 binding and the initiation of meiotic recombination and affects the crossover frequency at a given hot spot locus (Fig. 8). Thus, histone acetylation, along with PRDM9, controls mammalian recombination hot spot site selection and activity.
FIG 8.
Model for meiotic recombination hot spot (HS) activation by histone acetylation. (Top) A condensed chromatin structure is proposed to be manifest at the HS22 hot spot core in early germ cells (EGC), as represented by three central, closely juxtaposed nucleosomes at the core (black and gray circles). Potential PRDM9 binding sites (red vertical lines) are positioned next to “black” nucleosomes (Fig. S12A and Data Set S3). (Second panel) Licensing of hot spot cores for activation is posited to occur in spermatogonia (Sp) and preleptotene (pL) meiotic cells and requires deposition of acetylated marks on histones H3 and H4 that are directed by HATs expressed in these cells (Fig. 1A; Fig. S12B). This leads to the formation of open chromatin, which is likely facilitated by binding of chromatin remodeling factors (CRMFs) to acetylated histone tails (18, 38, 99). (Third panel) Acetylated open chromatin (red nucleosomes) then allows PRDM9 binding at its cognate binding sequences (vertical cyan lines), followed by its deposition of H3K4Me3 marks, as well as the deposition of other active methylated histone H3 marks, by HMTs expressed in these cells (Fig. 1B; Fig. S12B). (Bottom) DNA regions juxtaposed to active (red) nucleosome sites then become accessible for DSB cleavage by Spo11 at the leptotene (L) stage, which appears to occur via the recruitment of chromatin binding proteins (ChBP) to H3K4Me3 tails (11, 38, 100). DSBs are then repaired by the meiotic recombination machinery, leading to a CO or to gene conversion without a CO (NCO).
MATERIALS AND METHODS
Mouse strains.
Mice were bred and maintained at the Animal Resource Center of The Scripps Research Institute-Scripps Florida under the Institutional Animal Care and Use Committee (IACUC) guidelines and an IACUC-approved protocol. Mouse strains C57BL/6J, DBA/2J, CAST/EiJ, and C57BL/6J × DBA/2J F1 (males; B6D2F1/J) were purchased from the Jackson Laboratory (Bar Harbor, ME). Spo11+/− breeding pairs were generously provided by R. Daniel Camerini-Otero (NIDDK/NIH, Bethesda, MD).
Dissociation of testis cells and FACS sorting.
Spermatogonium, preleptotene, leptotene-zygotene, and pachytene-diplotene cell fractions were purified as described previously (34, 35). Briefly, C57BL/6J × DBA/2J F1 or CAST/EiJ × DBA/2J F1 meiotic cell fractions (here called B6 for C57BL/6J, DBA for DBA/2J, and CAST for CAST/EiJ) were purified using a FACS-based method (34, 35). Sorting was performed on a Becton Dickinson Aria cell sorter as described previously (35). Sorts typically took 3 to 6 h to collect 0.2 × 106 to 1.0 × 106 cells of each population, which were more than 95% pure for preleptotene, leptotene-zygotene, and pachytene-diplotene cells (34). Usually two to four testes were processed per sort.
Sample collection, RNA extraction, and array hybridization of Spo11−/− meiotic cells.
Spermatogonia, preleptotene, and leptotene-zygotene sorted meiotic fractions were collected from the testes of 10- to 16-week-old Spo11−/− mice. RNA was extracted from sorted cells as described previously (36). Each independent RNA pool included cells from two sorts of meiotic cells, one from four and one from three Spo11−/− mouse testes, which were combined for microarray hybridizations. RNA labeling and microarray hybridizations using MOE430v2.0 arrays (Affymetrix) were performed as described previously (36).
Microarray analysis of histone-modifying enzymes during meiosis in wild-type and Spo11−/− mice.
The initial analysis of C57BL/6J × DBA/2J F1 mouse meiotic samples was described previously (36), and the data are found under GEO accession number GSE21447. In the present study, 3 Spo11−/− meiotic cell samples (spermatogonium, preleptotene, and leptotene-zygotene cells) and 8 wild-type B6/DBA meiotic cell samples (spermatogonium, preleptotene, leptotene-zygotene, early pachytene, mid-pachytene, late pachytene, diplotene, and round spermatid cells), or a total of 23 Affymetrix Mouse430v2.0 CEL files, were analyzed using GeneSpring GX v12.1. The summarization algorithm was set to GCRMA. Quantile normalization was applied, and the baseline transformation was set to the median for all samples. Replicate samples were grouped for each cell type. The GeneSpring GX GO analysis function was used to identify genes (probe sets) encoding proteins that have histone acetylase, histone deacetylase, histone methylase, and histone demethylase activities. Probe sets with signals higher than the chip median for one cell type or more were analyzed.
To generate the heat map for each gene list (Fig. 1; see Fig. S6A and C and S7A and C in the supplemental material), the GeneSpring GX hierarchical clustering algorithm was utilized. For all heat maps, an absolute fold change filter of 1.5-fold for the wild-type spermatogonium/average pachytene expression ratio was applied to obtain the differentially expressed gene list, followed by hierarchical clustering. The gene expression data for the secondary spermatid stage were omitted. We identified 19 (of a total of 48), 7 (of a total of 23), 23 (of a total of 82), and 9 (of a total of 30) differentially expressed gene probes for HAT-, HDAC-, HMT-, and HDMT-encoding genes, respectively, that were represented across spermatogonium, preleptotene, leptotene-zygotene, early-, mid-, and late-pachytene, and diplotene meiotic cells from wild-type mice for the heat maps shown in Fig. 1. The same differentially expressed gene probes were analyzed across 3 meiotic stages (spermatogonium, preleptotene, and leptotene-zygotene cells) in Spo11−/− versus wild-type mouse samples for the heat maps presented in Fig. S6A and C and S7A and C. Only significantly changed probe sets, with P values (corrected) of <0.05, are shown.
Microarray data were normalized across the median for all 11 Spo11−/− and wild-type (WT) meiotic stage cell samples. Details regarding statistical analyses, as well as the complete and differentially expressed original probe listings for genes encoding histone-modifying enzymes, are provided in Data Set S1. Spo11−/−/WT expression ratios were also plotted, and the respective graphs are provided in Fig. S6B and D and S7B and D. The log2 fold change values for Spo11−/−/WT expression ratios are also shown in Data Set S2.
qRT-PCR.
Total RNAs were isolated from the sorted spermatogonium, preleptotene, leptotene-zygotene, and pachytene-diplotene meiotic cell fractions as described previously (36). cDNAs were generated using an iScript cDNA synthesis kit (Bio-Rad) and were quantified with a Quant-iT OliGreen ssDNA assay kit (Invitrogen). Real-time PCR primers for qRT-PCR analyses of genes encoding HATs and HDACs (Fig. 1C) were designed using Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) to yield short (200 to 250 bp) PCR fragments with a melting temperature of about 60°C (Table S1). Primer pairs that were used for qRT-PCR analyses of specific meiotic stage marker genes (Fig. 1D) were described previously (90). For each reaction, 0.9 ng of cDNA was mixed with 2.5 pmol of each primer in 5 μl (final volume) of SYBR green master mix (Quanta Biosciences) and ROX dye (Invitrogen), as a reference. All amplifications were run in triplicate.
Histone mark profiling across meiotic recombination hot spots on mouse chromosome 19.
Histone acetylation and methylation mark profiling across meiotic recombination hot spots HS22, HS59.4, HS59.5, and HS61.1 on mouse chromosome 19 was performed using native chromatin immunoprecipitation (nChIP) followed by real-time PCR analysis using the procedures described below.
Preparation of mononucleosomes by MNase digestion of native chromatin.
Mononucleosomes were isolated from native chromatin of B6 × DBA F1, CAST × DBA F1, or Spo11−/− mouse meiotic cells as described previously (34), with minor modifications. In addition to a protease inhibitor cocktail (Roche), all cell processing solutions were supplemented with 10 mM sodium butyrate for performing nChIP analyses of acetylated histone marks. After micrococcal nuclease (MNase) digestion and the first round of nucleosome extraction to an S1 supernatant, the remaining chromatin pellet was resuspended in 200 μl of lysis buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.4% NP-40, 1 mM CaCl2, 0.25 mM EDTA), kept on ice, and slightly sonicated in a water bath at 4°C. Sonication was performed on a Misonix 3000 sonicator at power level 5 for 1 min, followed by a 1-min pause and then sonication for 1 min, for a 2-min total sonication time. The suspension was centrifuged for 10 min at 10,000 rpm at 4°C, and the supernatant was retained (supernatant S2) and processed as described previously (34).
nChIP analysis of histone modification marks.
Following the preparation of mononucleosomes from native chromatin, immunoprecipitations were performed using protein G agarose (Millipore) as described for native histone H3 ChIP (34). Antibodies against the following marks were used for ChIP analyses: H4K12Ac (ab1761 and ab46983 [Abcam]; 39927 [Active Motive]), H4K5Ac (ab1758; Abcam), H4K8Ac (ab15823; Abcam), H4K16Ac (06-762 [Millipore]; ab61240 [Abcam]), H4K91Ac (ab4627; Abcam), H3K9Ac (ab12179; Abcam), H3K4Me3 (07-473 [Millipore]; ab8580 [Abcam]), H3K4Me2 (07-030; Millipore), H3K79Me1 (ab2886; Abcam), H3K36Me3 (ab9050; Abcam), H4K20Me3 (ab9053; Abcam), H4K20Me1 (ab9051; Abcam), H4K20Me2 (07-367; Millipore), H3K9Me3 (ab8898; Abcam), H3K27Me3 (9756; Cell Signaling), and H3K9/K14Ac (PA5-16194; Pierce).
Briefly, 0.2 × 106 to 1.0 × 106 B6 × DBA F1, CAST × DBA F1, or Spo11−/− meiotic cells were isolated by FACS. Typically, 2 to 4 μg of an antibody was then incubated with chromatin. As a control, a no-antibody ChIP was performed. DNAs from input and bound ChIP fractions were isolated as described previously (34), quantified using a PicoGreen kit (Invitrogen), and used for real-time PCR analysis.
Real-time PCR and data analysis.
qPCR-ChIP analyses were performed using enriched mononucleosomal histone mark-associated DNA and previously validated quantitative oligo-tiling PCR assays as described previously (34). In brief, to define histone modification profiles across the four mouse recombination hot spots studied, we used primer pair tiling across the selected loci, using 70- to 140-bp amplicons located, on average, every 30 to 75 bp, which selectively amplified only those DNA regions within the hot spot loci that were initially protected by a nucleosome and were therefore undigested by MNase (34) (Fig. 2B). Validated primer pairs used to tile the HS22, HS59.4, and HS61.1 hot spots on chromosome 19 and the 1-kb flanking regions surrounding the HS22 hot spot core, at kb −25, −10, +10, and +25, were described previously (34). The validated HS59.5 real-time PCR primers (47) are detailed in Table S2. All real-time PCR quantifications were performed using a Mastercycler RealPlex4S machine (Eppendorf) with SYBR green fluorescence (Quanta Biosciences) and ROX dye (Invitrogen), as a reference, and the real-time PCR conditions were described previously (34). The position at the center of each amplicon was used as the abscissa value for points plotted across the hot spot histone modification maps.
All initial nonnormalized histone modification profiles (Fig. 2 to 6 and 7A and C; Fig. S1, S2A, S3 to S5, and S9 to S11) were calculated as absolute fold enrichment values by using the 2−ΔCT formula, where ΔCT = CTIP(Bound) − CT(Input). We used arbitrary CT cutoff values of 31 and 29 for input and IP (bound) DNAs, respectively. For 2−ΔCT enrichment calculations, a value above 0.15 was considered significant. A profile for every histone mark was obtained after averaging enrichment curves from at least two independent ChIP experiments. No-antibody ChIP profiles were averaged through all the meiotic stages. The normalized nChIP profiles were obtained by performing either absolute 2−ΔΔCT calculations (Fig. 2 to 4; Fig. S1 and S3) or log2(ΔΔCT) normalization (Fig. 5, 6, and 7A and C; Fig. S11), where ΔΔCT = ΔCT ChIP1 (histone mark ChIP; CAST/DBA, Spo11−/−, or s2- or 2c-treated mice) − ΔCT ChIP2 (no-antibody control ChIP; B6/DBA or vehicle-treated mice).
Statistical analyses of ChIP qPCR data.
The Mann-Whitney test (91) was used to test the null hypothesis, namely, that the values for normalized bound ChIP fractions obtained for real-time PCR oligonucleotide pairs covering the HS22, HS59.4, HS59.5, and HS61.1 hot spot core regions for the native immunoprecipitation experiments (Fig. 2 to 7; Fig. S1 and S11) were not different between the histone mark and no-antibody control ChIPs, between two mouse strains (CAST/DBA and B6/DBA for the HS22 hot spot core), between Spo11−/− and B6/DBA (wild-type) mice for the HS22 hot spot core, or between inhibitor-treated and vehicle control ChIPs for the HS22 and HS59.5 hot spot cores.
The Mann-Whitney test was performed using Prism 6 software, and two-tailed P values were defined using ΔCT (ChIP1) versus ΔCT (ChIP2) value comparisons. The Mann-Whitney test was applied either to a whole hot spot core region, if ChIP enrichments were distributed across the hot spot core, or to a specific region within a hot spot core where there were narrow localized enrichment distribution patterns. Corresponding P values are indicated in the ChIP profiles shown in Fig. 2 to 7 and Fig. S1 and S11. P values of <0.05 were defined as being statistically significant.
Illumina genome-wide sequencing of mononucleosomal DNA.
Mononucleosomal DNA was isolated from total meiotic populations of 10-week-old B6 male testes (1.3 × 106 cells) by MNase digestion of native chromatin as described previously (34) (Fig. S2B). Final mononucleosomal DNA was purified using phenol-chloroform extraction followed by ethanol precipitation (34), run in a 2% agarose gel, and isolated using a Qiagen gel extraction kit (Fig. S2C). For whole-genome sequencing, nucleosomal DNA was further ligated to adapters, amplified using Illumina primers by following the TruSeq Nano DNA protocol (Illumina), and sequenced with an Illumina GAIIx genome analyzer. Fastq files from 5 sample runs were merged using the Unix cat command and then mapped to the mm9 mouse genome by using Bowtie, version 0.12.7. A combined sam file was converted to a bam file, sorted, and indexed using samtools, version 0.1.9. The sorted bam file and the bai index file were imported into Integrative Genomic Viewer (IGV 2.3) tools to create a tdf file by using the count function. The tdf file was visualized using the IGV browser. We obtained approximately 140 million reads of 75 nucleotides (nt), with about 73 million reads that were uniquely mapped to the mouse genome (mm9; University of California, Santa Cruz [UCSC], genome database).
Identification and crossover analysis of the HS59.5 recombination hot spot on mouse chromosome 19.
The HS22, HS59.4, and HS61.1 hot spots have been described previously (34, 48, 49). The HS59.5 hot spot was identified using a similar allele-specific PCR strategy (48, 49). In brief, batches of sperm DNA from C57BL/6J × DBA/2J F1 mice with multiple heterozygous single nucleotide polymorphisms (SNPs) were amplified across recombinogenically active regions by use of two PCR rounds with nested allele-specific oligonucleotide (ASO) pairs. Recombinant sperm DNA molecules were then selectively amplified, and the locations of crossover breakpoints were identified by mapping of internal SNPs between B6 and DBA sequences. All PCRs were performed with the previously described reaction buffer (48, 92). Amplifiable molecules were determined using Poisson analysis (48).
For the HS59.5 hot spot, DNA was digested with the XhoI restriction enzyme to cleave outside the tested intervals. Both B6-to-DBA and DBA-to-B6 CO orientations were analyzed. SNPs are indicated in bold in the sequences below. For the B6 × DBA F1 sperm DNA, HS59.5 allele-specific 5′ primers were 59.5DBAF1.3 (5′-GGC AAC TGA AAT CAA ATA CAC-3′) and 59.5DBAF2.3 (5′-GAC TGG AAA ACC ATT CCA TTC CAT-3′) for DBA and 59.5B6F1.3 (5′-GGT AAC TGA AAT CAA ATA CAT G-3′) and 59.5B6F2.3 (5′-CGG AAT GGA AAA CCA TTC TAC-3′) for B6. The 3′ primers were 59.5DBAR1.3 (5′-TTG GAA TTC AAG AAC AAA TAC-3′) and 59.5DBAR2.3 (5′-CTG CAC AGT AAG TCC AGG T-3′) for DBA and 59.5B6R1.3 (5′-TTG GAA TTC AAG AAC AAA CAG-3′) and 59.5B6R2.3 (5′-GTG CAC AGT AAG TCC AGT G-3′) for B6. The first round was performed using the 59.5 F1.3 and 59.5 R1.3 primer pair (6.9 kb), and the second round was performed with the 59.5 F2.3 and 59.5 R2.3 primer pair (6.5 kb). The first round of PCRs was performed at 94°C for 1 min followed by 28 cycles at 94°C for 30 s, 54°C for 45 s, and 63°C for 6 min. The second round of PCRs was performed at 94°C for 1 min followed by 30 cycles at 94°C for 30 s, 54°C for 45 s, and 63°C for 6 min.
All primary PCR products were digested with S1 nuclease to remove single-stranded DNA as described previously (48, 92). Secondary PCR mixtures were seeded with 100-fold-diluted S1 nuclease-treated DNA. Secondary PCR products (70% of the total reaction volume) were run in 0.8% agarose gels in 0.5× Tris-borate-EDTA (TBE) and visualized by staining with ethidium bromide and use of UV light. DNA inputs per small-pool PCR consisted of ∼1,000 amplifiable molecules. This was equivalent to 0.2 to 0.4 recombinant molecule per pool. We typically obtained 10% to 30% positive pools per experiment to limit reactions with two recombinant molecules. All recombinant molecules were sequenced with an ABI 3130xl Prism genetic analyzer and analyzed using Sequencher software (v4.9; GeneCodes). The HS59.5 sequencing primers were HS59.5F11 (5′-ACC GAC TGT GTG TGT GTG TGT-3′), HS59.5F12 (5′-GCT TCT ACA CCT GCC ACA ACT-3′), HS59.5F13 (5′-GGA GCA CAT CCA CAC TTC TGT-3′), HS59.5R11 (5′-AGA CCC ACA AAC AGC ACT GAG-3′), HS59.5R12 (5′-GAA GGA TGT CAC CAT GTC ACC-3′), and HS59.5R13 (5′-AAT CAG AAC ATG GCC TCC TG-3′). Recombination rates and 95% confidence intervals were calculated as described previously (48, 49) (Table S3). The CO profile for the HS59.5 hot spot has been described previously (47).
Synthesis of HDAC and HAT inhibitors.
The HDAC inhibitor 4-dimethylamino-N-[5-(2-mercaptoacetylamino)-pentyl]-benzamide (s2) (Fig. S8A, left panel) and the HAT inhibitor 2,6-bis(3-bromo-4-hydroxybenzylidene)cyclohexanone (2c) (Fig. S8A, right panel) were synthesized as described previously (60, 63).
Preparation of liposomal 2c compound.
The 2c drug has poor solubility in water and was toxic to mice when used in a dimethyl sulfoxide (DMSO) solution. Therefore, based on the similarity of the structures of 2c and curcumin, we adapted a protocol developed for liposome-encapsulated curcumin (64, 93) to prepare a liposomal 2c drug that was soluble in saline. In brief, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phospho-rac-(1-glycerol) (DMPG) lipids (Avanti Polar Lipids) at a 9:1 ratio were dissolved in tert-butanol at a concentration of 10 mg/ml. Sterile water (1/20 volume) and 1 part of the 2c compound were added for a final lipid/2c ratio of 10:1. The solution was sterile filtered, frozen in dry ice and ethanol, and lyophilized overnight. The modified drug was soluble in saline at up to 10 mg/ml and had no toxicity in treated male mice.
In vivo HDAC and HAT inhibitor studies.
The HDAC and HAT inhibitor treatment strategy used takes advantage of the linear process of meiosis I, where an acute drug treatment (typically 5 to 10 days) will affect early mouse germ cells at a set time, and the consequences can then be assessed via analyses of sperm DNA. Inhibitors were administered to 10-week-old mice daily for 5 (2c [HAT inhibitor]) or 8 (s2 [HDAC inhibitor]) days by intraperitoneal (i.p.) injection; vehicle alone was injected into a control cohort of male mice. Doses for s2 inhibitor treatment were described previously (61). The dosage of the 2c HAT inhibitor used in vivo was validated in this study. A group of 8 mice were treated. The s2 compound was resuspended in DMSO (0.25%)-sterile 0.9% sodium chloride vehicle and injected i.p. daily (1 mg/kg of body weight) for 8 days. The liposomal 2c compound was solubilized to a final concentration of 8 to 10 mg/ml in sterile 0.9% sodium chloride and injected i.p. daily (50 mg/kg of body weight) for 5 days. Immediately following treatment (i.e., 5 or 8 days of drug injection), FACS profiles of cells isolated from mouse testes were determined to test if drug administration had deleterious effects on meiotic progression, and native ChIP analyses of H4K12Ac, H4K16Ac, and H3K9/14Ac histone acetylation marks and H3K4Me3 and H3K27Me3 histone methylation marks were performed to assess the effects of treatment on histone mark profiles at the HS22 and HS59.5 meiotic hot spots (Fig. 7; Fig. S8A and B and S9 to S11).
To verify the effects of treatment on the total levels of acetylated H3 and acetylated H4, Western blotting was performed on lysates of meiotic cells from the testes of treated animals (Fig. S8C and D). Briefly, meiotic cells isolated as described previously (34, 35) were washed once in phosphate-buffered saline (PBS) supplemented with protease inhibitors (protease inhibitor cocktail [Roche], 1 mM phenylmethylsulfonyl fluoride [PMSF]) and 10 mM sodium butyrate, resuspended in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween 20) containing protease inhibitors and 10 mM sodium butyrate, and lysed by sonication (Sonic Dismembrator model 100; Fisher Scientific) for a few seconds at power level 2. Protein was quantified using a Micro BCA protein assay kit (Thermo Scientific) and immunoblotted with a 1:15,000 dilution of anti-acetyl-histone H3 (06-599; Millipore), a 1:2,000 dilution of anti-acetyl-histone H3 (39139; Active Motif), a 1:1,000 dilution of anti-acetyl-histone H4 (39925; Active Motif), a 1:2,000 dilution of anti-histone H3 (ab1791; Abcam), a 1:2,000 dilution of anti-histone H4 (ab10158; Abcam), or a 1:7,000 dilution of anti-β-actin (NB600-501; Novus Biologicals) antibodies. Immunoreactive bands were detected by enhanced chemiluminescence (Amersham Biosciences) or by use of an Odyssey infrared imaging system (Li-Cor Biosciences).
CO analysis after HDAC and HAT inhibitor treatment.
CO rates for the HS59.5 and HS22 hot spots in treated sperm DNA were determined 6 weeks after the last day of drug injection. The 6-week time point for sperm collection was chosen, as suggested previously (94), to assess the effects of treatment in early spermatogonium premeiotic cells, which eventually develop into mature sperm, after completion of spermatogenesis, which typically takes up to 40 days in mice, followed by sperm transit through the epididymis for 5 to 7 days (95–98). Sperm DNA was isolated from the epididymides of treated mice as described previously (48). As a control, sperm DNA was isolated from vehicle-treated mice.
CO analysis was performed following allele-specific PCR (48, 49). CO assays for the HS59.5 and HS22 hot spots in treated sperm DNA were run for the DBA-to-B6 CO orientation by using DNA samples from 3 independent treatments. The statistical significance of differences in CO rates between treated and control samples was determined using Student's t test. HS59.5 ASOs were the same as those described in the HS59.5 hot spot identification section provided above. HS22 allele-specific 5′ primers were 22DBA-F1.1-1 (5′-ATG ACC CTC AAG GTC CTA CC-3′) and 22DBA-F3.1-2 (5′-ATG GCC AGA CAC TGT AGT-3′) for DBA and 22B6-F1.1-1 (5′-ATG ACC CTC AAG GTC CTA CG-3′) and 22B6-F3.1-2 (5′-ATG GCC AGA CAC TGT AGC-3′) for B6. The 3′ primers were 22DBA-R7.1-1 (5′-TCG CCG ACT GAT GAC-3′) and 22DBA-R6-2 (5′-GGC CGG CAT TTT AAT CTT CAT AC-3′) for DBA and 22B6-R7.1-1 (5′-GCT CGC CGA CTG ATG AT-3′) and 22B6-R6-2 (5′-GGC CGG CAT TTT AAT CTT CAT AG-3′) for B6 (SNPs are indicated in bold). The first round of PCRs was performed using the 22F1.1-1 and 22R7.1-1 primer pair (5.0 kb), and the second round was performed with the 22F3.1-2 and 22R6-2 primer pair (4.6 kb). The first round of PCRs was performed at 94°C for 1 min followed by 33 cycles of 94°C for 45 s, 60°C for 45 s, and 65°C for 5 min 30 s. The second round of PCRs was performed at 94°C for 1 min followed by 33 cycles at 94°C for 45 s and 62°C for 6 min. All primary PCR products were digested with S1 nuclease to remove single-stranded DNA as described previously (48, 92). Secondary PCR mixtures were seeded with 100-fold-diluted S1 nuclease-treated DNA. Secondary PCR products (70% of the total reaction volume) were run in 0.8% agarose gels in 0.5× TBE and visualized with ethidium bromide and UV light. All the bands after the second round of CO PCR were sequenced with an ABI 3130xl Prism genetic analyzer and analyzed using Sequencher software (v4.9; GeneCodes), and recombinant molecules were identified. HS22 sequencing primers were HS22F1 (5′-TCT ATT GGC CTC GTA CCT GTG-3′), HS22F2 (5′-AAC GGT GCC TTT ACC AAC AG-3′), HS22F3 (5′-GCT CTC ACA CAC CAC CAC TTT-3′), HS22R0 (5′-GAT GAG TGG GAC TGG GAT ACA-3′), HS22R2 (5′-TCA GCT CAG TGA GAA CCT AGT G-3′), and HS22R3 (5′-GAG AGC ATG ATG GGA ACA GAC-3′). Recombination rates and 95% confidence intervals were calculated as described previously (48, 49) (Fig. 7B and D; Table S4).
Identification of PRDM9 binding motifs within the HS22 hot spot core.
To define putative binding sites of PRDM9 within the HS22 hot spot core, we scanned the C57BL/6J HS22 sequence (48) for the presence of the short, 15-mer consensus 9R motif specific for the PRDM9 9R (C57BL/6J) allele (10) (Fig. S12A), using a 9R motif position weight matrix (PWM), which was kindly provided by Pavel Khil, and MEME-ChIP software (http://meme-suite.org/tools/fimo). Statistical details and a list of the 9R motif sites present in the HS22 hot spot core (9Rc1 to 9Rc7) are provided in Data Set S3.
Accession number(s).
The combined microarray data set and the whole-genome nucleosome sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under accession numbers GSE57197 and GSE87057, respectively, and are linked under GEO accession number GSE87081.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Pavel Khil (NIDDK/NIH), Matthew Pipkin (Scripps Florida), and David Shibata (University of Tennessee Health Science Center) for reviewing the manuscript, to Howard Petrie, Bivian Torres, and Kimberley Lowe (Scripps Florida Flow Cytometry Core) as well as Brandon Young and Bradley Long (Scripps Florida Genomics Core) for their technical advice and assistance, and to numerous colleagues for discussions and insights. We also thank R. Daniel Camerini-Otero (NIDDK/NIH) for providing Spo11−/− mice.
We have no relevant conflicts of interest to declare.
This project was supported in part by funding from the State of Florida to Scripps Florida and to the H. Lee Moffitt Cancer Center & Research Institute, by monies from the Marie Mayer and PGA Women's Cancer Awareness Days Postdoctoral Fellowships in Cancer Biology, by NIH grant GM085079, and by Cancer Center support grant P30 CA076292 to the Moffitt Cancer Center.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00942-15.
REFERENCES
- 1.Keeney S. 2008. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn Stab 2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. 2005. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J Cell Sci 118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 3.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 4.Romanienko PJ, Camerini-Otero RD. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6:975–987. [DOI] [PubMed] [Google Scholar]
- 5.Baudat F, Imai Y, de Massy B. 2013. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 6.Paigen K, Petkov P. 2010. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet 11:221–233. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. 2008. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180:673–679. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet 4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res 24:724–732. doi: 10.1101/gr.170167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature 485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grey C, Barthes P, Chauveau-Le Friec G, Langa F, Baudat F, De Massy B. 2011. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 9:e1001176. doi: 10.1371/journal.pbio.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shones DE, Zhao K. 2008. Genome-wide approaches to studying chromatin modifications. Nat Rev Genet 9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, Zhao K. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youds J, Boulton SJ. 2011. The choice in meiosis—defining the factors that influence crossover or non-crossover formation. J Cell Sci 124:501–513. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 18.Wahls WP, Davidson MK. 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet 26:202–208. doi: 10.1016/j.tig.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner CR, Kuervers L, Baillie DL, Yanowitz JL. 2010. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467:839–843. doi: 10.1038/nature09429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirota K, Mizuno K, Shibata T, Ohta K. 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol Biol Cell 19:1162–1173. doi: 10.1091/mbc.E07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. 2004. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J 23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, Paigen K. 2016. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet 12:e1006146. doi: 10.1371/journal.pgen.1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Geli V, de La Roche Saint-Andre C. 2004. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J 23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita K, Shinohara M, Shinohara A. 2004. Rad6-Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A 101:11380–11385. doi: 10.1073/pnas.0400078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng HH, Xu RM, Zhang Y, Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 26.Sun ZW, Allis D. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 27.Mieczkowski PA, Dominska M, Buck MJ, Lieb JD, Petes TD. 2007. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104:3955–3960. doi: 10.1073/pnas.0700412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. 2006. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Li L, Liang J, Shi L, Yang J, Yi X, Zhang D, Han X, Yu N, Shang Y. 2013. Histone acetyltransferase 1 promotes homologous recombination in DNA repair by facilitating histone turnover. J Biol Chem 288:18271–18282. doi: 10.1074/jbc.M113.473199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin S, Parthun MR. 2006. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol Cell Biol 26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, Ramsey H, Kwan L, Nickoloff JA, Hromas R. 2005. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci U S A 102:18075–18080. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buard J, Barthes P, Grey C, de Massy B. 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamburini BA, Tyler JK. 2005. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol 25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Getun IV, Wu ZK, Khalil AM, Bois PR. 2010. Nucleosome occupancy landscape and dynamics at mouse recombination hotspots. EMBO Rep 11:555–560. doi: 10.1038/embor.2010.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Getun IV, Torres B, Bois PR. 2011. Flow cytometry purification of mouse meiotic cells. J Vis Exp 50:2602. doi: 10.3791/2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fallahi M, Getun IV, Wu ZK, Bois PRJ. 2010. A global expression switch marks pachytene initiation during mouse male meiosis. Genes 1:469–483. doi: 10.3390/genes1030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kota SK, Feil R. 2010. Epigenetic transitions in germ cell development and meiosis. Dev Cell 19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Chen T, Dent SY. 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Godmann M, Lambort R, Kimmins S. 2009. The dynamic epigenetic program in male germ cells: its role in spermatogenesis, testis cancer, and its response to the environment. Microsc Res Tech 72:603–619. doi: 10.1002/jemt.20715. [DOI] [PubMed] [Google Scholar]
- 40.Nagarajan R, Ge Z, Sirbu B, Doughty C, Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR. 2013. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet 9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayashi S, Yoshida K, Matsui Y. 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 42.Dinant C, Houtsmuller AB, Vermeulen W. 2008. Chromatin structure and DNA damage repair. Epigenetics Chromatin 1:9. doi: 10.1186/1756-8935-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol 20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsukamoto Y, Kato J, Ikeda H. 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 45.Gottlieb S, Esposito RE. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 46.San-Segundo PA, Roeder GS. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97:313–324. doi: 10.1016/S0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 47.Getun IV, Wu ZK, Bois PRJ. 2012. Organization and roles of nucleosomes at mouse meiotic recombination hotspots. Nucleus 3:244–250. doi: 10.4161/nucl.20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bois PR. 2007. A highly polymorphic meiotic recombination mouse hotspot exhibits incomplete repair. Mol Cell Biol 27:7053–7062. doi: 10.1128/MCB.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu ZK, Getun IV, Bois PR. 2010. Anatomy of mouse recombination hot spots. Nucleic Acids Res 38:2346–2354. doi: 10.1093/nar/gkp1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. 2005. Crossover and noncrossover pathways in mouse meiosis. Mol Cell 20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 51.Steger DJ, Lefterova MI, Ying L, Stonestorm AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Biobel GA, Vakoc CR. 2008. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Karachentsev D, Sarma K, Reinberg D, Steward R. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev 19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kauppi L, Jasin M, Keeney S. 2013. How much is enough? Control of DNA double-strand break numbers in mouse meiosis. Cell Cycle 12:2719–2720. doi: 10.4161/cc.26079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smirnova NA, Romanienko PJ, Khil PP, Camerini-Otero RD. 2006. Gene expression profiles of Spo11−/− mouse testes with spermatocytes arrested in meiotic prophase I. Reproduction 132:67–77. doi: 10.1530/rep.1.00997. [DOI] [PubMed] [Google Scholar]
- 58.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998. doi: 10.1016/S1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 59.Kugel S, Mostoslavsky R. 2014. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci 39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen B, Petukhov PA, Jung M, Velena A, Eliseeva E, Dritschillo A, Kozikowski AP. 2005. Chemistry and biology of mercaptoacetamides as novel histone deacetylase inhibitors. Bioorg Med Chem Lett 15:1389–1392. doi: 10.1016/j.bmcl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Konsoula Z, Cao H, Velena A, Jung M. 2009. Pharmacokinetics-pharmacodynamics and antitumor activity of mercaptoacetamide-based histone deacetylase inhibitors. Mol Cancer Ther 8:2844–2851. doi: 10.1158/1535-7163.MCT-09-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konsoula R, Jung M. 2008. In vitro plasma stability, permeability and solubility of mercaptoacetamide histone deacetylase inhibitors. Int J Pharm 361:19–25. doi: 10.1016/j.ijpharm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Costi R, Di Santo R, Artico M, Miele G, Valentini P, Novellino E, Cereseto A. 2007. Cinnamoyl compounds as simple molecules that inhibit p300 histone acetyltransferase. J Med Chem 50:1973–1977. doi: 10.1021/jm060943s. [DOI] [PubMed] [Google Scholar]
- 64.Li L, Braiteh FS, Kurzrock R. 2005. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer 104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 65.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 66.Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E, Celeste A, Pagani M, Opravil S, Rosa-Velazquez IA, Espejo A, Bedford MT, Nussenzweig A, Busslinger M, Jenuwein T. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. 1995. Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 68.Ayton P, Sneddon SF, Palmer DB, Rosewell IR, Owen MJ, Young B, Presley R, Subramanian V. 2001. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis 30:201–212. doi: 10.1002/gene.1066. [DOI] [PubMed] [Google Scholar]
- 69.Bell EL, Nagamori I, Williams EO, Del Rosario AM, Bryson BD, Watson N, White FM, Sassoni-Corsi P, Guarente L. 2014. SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141:3495–3504. doi: 10.1242/dev.110627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toiber D, Erdel F, Bouzoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, D'Urso A, Näär AM, Kingston R, Rippe K, Mostoslavsky R. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51:454–468. doi: 10.1016/j.molcel.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci U S A 110:16061–16066. doi: 10.1073/pnas.1315204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lesch BJ, Page DC. 2014. Poised chromatin in the mammalian germ line. Development 141:3619–3626. doi: 10.1242/dev.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye J, Ai X, Eugeni EE, Zhang L, Carpenter LR, Jelinek MA, Freitas MA, Parthun MR. 2005. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell 18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malkova A, Klein F, Leung W-Y, Haber JE. 2000. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc Natl Acad Sci U S A 97:14500–14505. doi: 10.1073/pnas.97.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. 2004. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 76.Hajdu I, Ciccia A, Lewis SM, Elledge SJ. 2011. Wolf-Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc Natl Acad Sci U S A 108:13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greeson NT, Sengupta R, Arida AR, Jenuwein T, Sanders SL. 2008. Di-methyl H4 lysine 20 targets the checkpoint protein Crb2 to sites of DNA damage. J Biol Chem 283:33168–33174. doi: 10.1074/jbc.M806857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wakeman TP, Wang Q, Feng J, Wang XF. 2012. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J 31:2169–2181. doi: 10.1038/emboj.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. 2011. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeng J, Yi SV. 2014. Specific modifications of histone tails, but not DNA methylation, mirror the temporal variation of mammalian recombination hotspots. Genome Biol Evol 6:2918–2929. doi: 10.1093/gbe/evu230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 82.Sachs M, Onodera C, Blaschke K, Ebata KT, Song JS, Ramalho-Santos M. 2013. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3:1777–1784. doi: 10.1016/j.celrep.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shogren-Knaak M, Ishi H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 84.Tang YA, Wen WL, Chang JW, Wei TT, Tan YH, Salunke S, Chen CT, Chen CS, Wang YC. 2010. A novel histone deacetylase inhibitor exhibits antitumor activity via apoptosis induction, F-actin disruption and gene acetylation in lung cancer. PLoS One 5:e12417. doi: 10.1371/journal.pone.0012417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perrella G, Consiglio MF, Aiese-Cigliano R, Cremona G, Sanchez-Moran E, Barra L, Errico A, Bressan RA, Franklin FC, Conicella C. 2010. Histone hyperacetylation affects meiotic recombination and chromosome segregation in Arabidopsis. Plant J 62:796–806. doi: 10.1111/j.1365-313X.2010.04191.x. [DOI] [PubMed] [Google Scholar]
- 86.Merker JD, Dominska M, Greenwell PW, Rinella E, Bouck DC, Shibata Y, Strahl BD, Mieczkowski P, Petes TD. 2008. The histone methylase Set2p and the histone deacetylase Rpd3p repress meiotic recombination at the HIS4 meiotic recombination hotspot in Saccharomyces cerevisiae. DNA Repair (Amst) 7:1298–1308. doi: 10.1016/j.dnarep.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yelina NE, Choi K, Chelysheva L, Macaulay M, de Snoo B, Wijnker E, Miller N, Drouaud J, Grelon M, Copenhaver GP, Mezard C, Kelly KA, Henderson IR. 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet 8:e1002844. doi: 10.1371/journal.pgen.1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sasaki H, Matsui Y. 2008. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9:129–140. doi: 10.1038/ni1560. [DOI] [PubMed] [Google Scholar]
- 89.De Felici M. 2011. Nuclear reprogramming in mouse primordial germ cells: epigenetic contribution. Stem Cells Int 2011:425863. doi: 10.4061/2011/425863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. 2007. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 91.Hart A. 2001. Mann-Whitney test is not just a test of medians: differences in spread can be important. BMJ 323:391–393. doi: 10.1136/bmj.323.7309.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kauppi L, May CA, Jeffreys AJ. 2009. Analysis of meiotic recombination products from human sperm. Methods Mol Biol 557:323–355. doi: 10.1007/978-1-59745-527-5_20. [DOI] [PubMed] [Google Scholar]
- 93.Wang D, Veena MS, Stevenson K, Tang C, Ho B, Suh JD, Duarte VM, Faull KF, Mehta K, Srivatsan ES, Wang MB. 2008. Liposome-encapsulated curcumin suppresses growth of head and neck squamous cell carcinoma in vitro and in xenografts through the inhibition of nuclear factor kappaB by an AKT-independent pathway. Clin Cancer Res 14:6228–6236. doi: 10.1158/1078-0432.CCR-07-5177. [DOI] [PubMed] [Google Scholar]
- 94.Dubrova YE, Plumb M, Brown J, Fennelly J, Bois P, Goodhead D, Jeffreys AJ. 1998. Stage specificity, dose response, and doubling dose for mouse minisatellite germ-line mutation induced by acute radiation. Proc Natl Acad Sci U S A 95:6251–6255. doi: 10.1073/pnas.95.11.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Searle AG. 1974. Mutation induction in mice. Adv Radiat Biol 4:131–207. doi: 10.1016/B978-0-12-035404-7.50010-3. [DOI] [Google Scholar]
- 96.Hess RA, Renato de Franca L. 2008. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol 636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 97.Eddy EM. 2002. Male germ cell gene expression. Recent Prog Horm Res 57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 98.França LR, Avelar GF, Almeida FF. 2005. Spermatogenesis and sperm transit through the epididymis in mammals with emphasis on pigs. Theriogenology 63:300–318. doi: 10.1016/j.theriogenology.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 99.Nightingale KP, Baumann M, Eberharter A, Mamais A, Becker PB, Boyes J. 2007. Acetylation increases access of remodelling complexes to their nucleosome targets to enhance initiation of V(D)J recombination. Nucleic Acids Res 35:6311–6321. doi: 10.1093/nar/gkm650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis D. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.